Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2009, 2, 637-643 doi: 10.4236/jbise.2009.28093 Published Online December 2009 (http://www.SciRP.org/journal/jbise/ JBiSE ). Published Online December 2009 in SciRes.http://www.scirp.org/journal/jbise Application of SOM neural network in clustering Soroor Behbahani1, Ali Moti Nasrabadi2 1Biomedical Engineering Department, Science and Research Branch, Islamic Azad University, Tehran, Iran; 2Biomedical Engineering Department, Faculty of Engineering, Shahed University, Tehran, Iran. Email: soroor_behbahani@yahoo.com; a_m_nasrabadi@yahoo.com Received 11 June 2009; revised 29 June 2009; accepted 27 July 2009. ABSTRACT The Self-Organizing Map (SOM) is an unsupervised neural network algorithm that projects high-dimen- sional data onto a two-dimensional map. The projec- tion preserves the topology of the data so that similar data items will be mapped to nearby locations on the map. One of the SOM neural network’s applications is clustering of animals due their features. In this paper we produce an experiment to analyze the SOM in clustering different species of animals. Keywords: SOM Neural Network; Feature; Clustering; Animal 1. INTRODUCTION The Self-Organizing Map (SOM) is a fairly well-known neural network and indeed one of the most popular un- supervised learning algorithms. Since its invention by Finnish Professor Teuvo Kohonen in the early 1980s, more than 4000 research articles have been published on the algorithm, its visualization and applications. The maps comprehensively visualize natural groupings and relationships in the data and have been successfully ap- plied in a broad spectrum of research areas ranging from speech recognition to financial analysis. The Self-organ- izing Map performs a non-linear projection of multidi- mensional data onto a two-dimensional display. The mapping is topology-preserving, meaning that the more alike two data samples are in the input space, the closer they will appear together on the final map. The SOM belongs to the class of Neural Network algorithms. This is a group of algorithms based on analogies to the neural structures of the brain. The SOM in particular was in- spired by an interesting phenomenon: as physicians have discovered, some areas of brain tissue can be ordered according to an input signal. Basically, the SOM is a computer program simulating this biological ordering process. Applied to electronic datasets, the algorithm is capable of producing a map that shows similar input data items appearing close to each other. There are numerous applications involving the SOM algorithm but the most widespread use is the identification and visualization of natural groupings in the data. The process of finding similar items is generally referred to as clustering. Compared to the k-means clustering algorithm, the SOM exemplifies a robust and structured self-organizing neu- ral networks are based on the principle of transforming a set of p-variate observations into a spatial representation of smaller dimensionality, which may allow a more ef- fective visualization of correlations in the original data [4]. 2. SELF-ORGANIZING MAP The Self-Organizing Map belongs to the class of unsu- pervised and competitive learning algorithms. It is a sheet-like neural network, with nodes arranged as a regular, usually two-dimensional grid. As explained in the previous section on Neural Networks, we usually think of the node connections as being associated with a vector of weights. In the case of Self-Organizing Maps, it is easier to think of each node as being directly associ- ated with a weight vector. The items in the input data set are assumed to be in a vector format. If n is the dimension of the input space, then every node on the map grid holds an n-dimensional vector of weights: mi = [mi1, mi2, mi3, . , min] (1) The basic principle of the Self-Organizing Map is to adjust these weight vectors until the map represents a picture of the input data set. Since the number of map nodes is significantly smaller than the number of items in the dataset, it is needless to say that it is impossible to represent every input item from the data space on the map. Rather, the objective is to achieve a configuration in which the distribution of the data is reflected and the most important metric relationships are preserved. In particular, we are interested in obtaining a correlation between the similarity of items in the dataset and the distance of their most alike representatives on the map. In other words, items that are similar in the input space should map to nearby nodes on the grid [4].  638 S. Behbahani et al. / J. Biomedical Science and Engineering 2 (2009) 637-643 SciRes Copyright © 2009 JBiSE 2.1. Image’s Characteristics To represent the 3D image of 12 lead ECG, three axes for time, temporal and spatial are needed witch temporal axis represented the time domain of the cardiac signal and the spatial axis represented the locations of the limb and thoracic leads. The data axis is represented two ex- tracted features of cardiac signal contains amplitude and wavelet coefficients. 6 leads are used to represent the image obtained by thoracic leads and 6 leads of 12 are used to represent the image obtained by limb leads. In order to determine the information between con- secutive leads in the spatial axis, an interpolation tech- nique was used to witch could cause to homogeneous- ness of the image. 3. AN EXAMPLE OF SOM NEURAL NETWORK APPLICATION More researches are performing in the field of SOM neural network applications in last two decades. One of the most important and famous examples of this applica- tion is clustering of animals due their features. General features are using in this example based on the Kohonen animal data base (Table 1). But the fact is that, these features are not sufficient for different species of animals. In previous experiments, it had been assumed that there were only one species for each animal, whereas there may be exist more than 10 species for each special animal. So, for analyzing the ability of SOM neural net- work we perform a new experiment and assumed more than one species for them and increase the number of features to invent better separability. These features con- sist of geographical dispersion, nourishing and habitat, etc (Table 2). The SOM size in this research is 7*7 and the initial weights are selected randomly. Although there are 3 animals that are not settle in right location in SOM map, and selected wrong neu- rons, the results shows that extracted features could well separate the different species of animals. This result shows that the selected features for these 3 ani- mals have not sufficient ability to separate them. This problem could be solved by adding extra features or choosing the features with more precise. One of the most important points in neural networks is the method of features extraction, but increasing the number of features could not always be the best solution for ap- proving the results, because sometimes increasing the features lead to derangement in network. Another rea- son of bad result in neural networks relates to number of inputs. Increasing the number of inputs (animal species) leads to spreading the SOM size and could decrease the ability of it, because there would be more correlation between inputs, so the statistic of error will be increased. 4. RESULTS Choosing suitable features for separating animal’s spe- cies lead to good results of SOM neural network .There were some similarity between some of the animal’s fea- ture in Kohonen data base. For example the features of Goose and Owl, as well as, horse and zebra are exactly the same. And this similarity leads to wrong results in clustering of these animals. Although there are some errors, in this new experiment, these errors occurred between different species of one animal not between different animals. So the more similarity between ani- mal’s species, the more errors will occur. Table 1. The animal data set. Animal Dove Hen Duck Goose Owl HawkEagleFox DogWolfCatTiger Lion Horse ZebraCow is Small 1 1 1 1 1 1 0 0 0 0 1 0 0 0 0 0 Medium 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 Big 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 has Two kgs 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 Four legs 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 Hair 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 Hooves 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 Mane 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 Feathers 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 likes to Hunt 0 0 0 0 1 1 1 1 0 1 1 1 1 0 0 0 Run 0 0 0 0 0 0 0 0 1 1 0 1 1 1 1 0 Fly 1 0 0 1 1 1 1 0 0 0 0 0 0 0 0 0 Swim 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 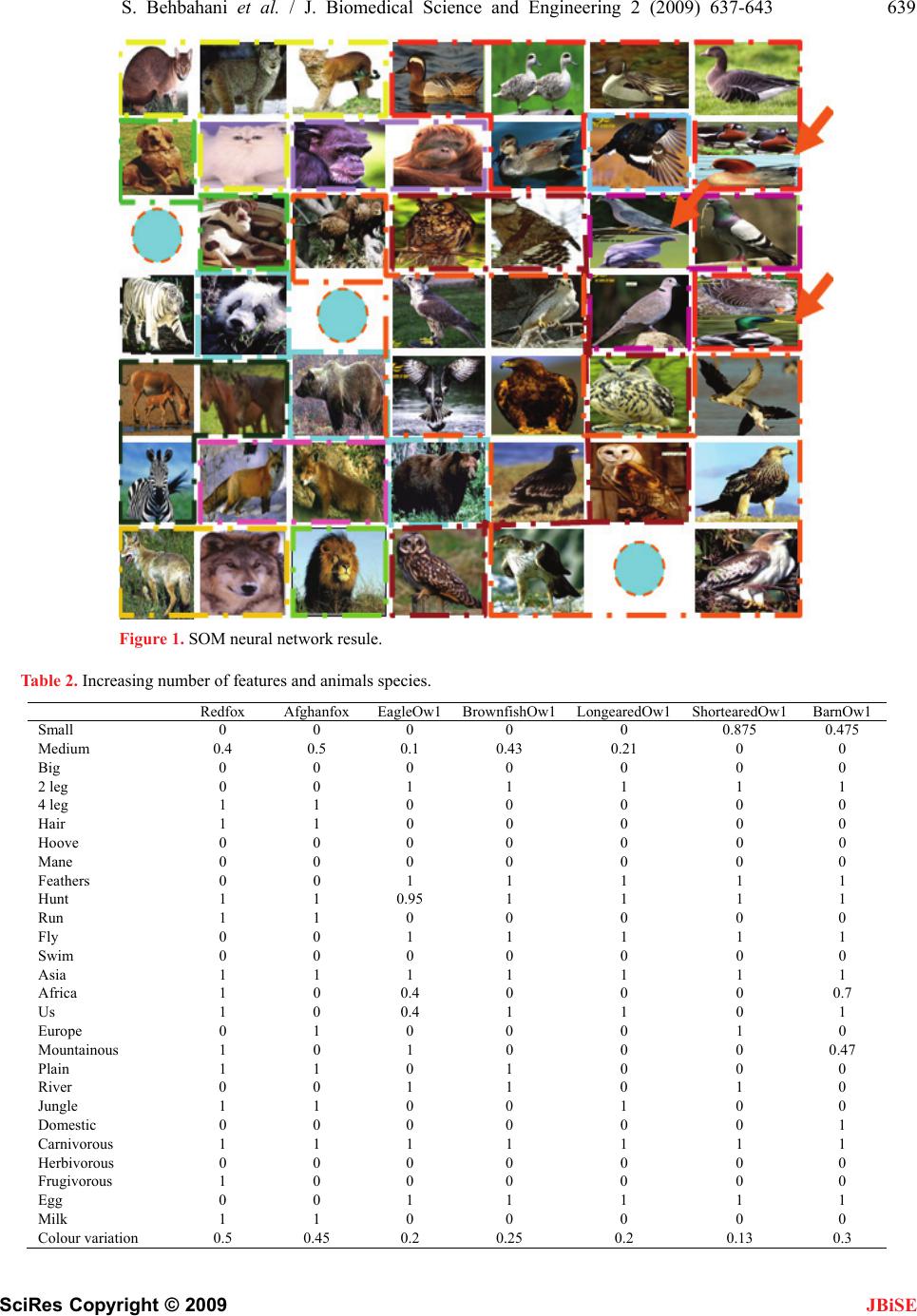 S. Behbahani et al. / J. Biomedical Science and Engineering 2 (2009) 637-643 639 SciRes Copyright © 2009 Figure 1. SOM neural network resule. Table 2. Increasing number of features and animals species. Redfox Afghanfox EagleOw1BrownfishOw1LongearedOw1ShortearedOw1 BarnOw1 Small 0 0 0 0 0 0.875 0.475 Medium 0.4 0.5 0.1 0.43 0.21 0 0 Big 0 0 0 0 0 0 0 2 leg 0 0 1 1 1 1 1 4 leg 1 1 0 0 0 0 0 Hair 1 1 0 0 0 0 0 Hoove 0 0 0 0 0 0 0 Mane 0 0 0 0 0 0 0 Feathers 0 0 1 1 1 1 1 Hunt 1 1 0.95 1 1 1 1 Run 1 1 0 0 0 0 0 Fly 0 0 1 1 1 1 1 Swim 0 0 0 0 0 0 0 Asia 1 1 1 1 1 1 1 Africa 1 0 0.4 0 0 0 0.7 Us 1 0 0.4 1 1 0 1 Europe 0 1 0 0 0 1 0 Mountainous 1 0 1 0 0 0 0.47 Plain 1 1 0 1 0 0 0 River 0 0 1 1 0 1 0 Jungle 1 1 0 0 1 0 0 Domestic 0 0 0 0 0 0 1 Carnivorous 1 1 1 1 1 1 1 Herbivorous 0 0 0 0 0 0 0 Frugivorous 1 0 0 0 0 0 0 Egg 0 0 1 1 1 1 1 Milk 1 1 0 0 0 0 0 Colour variation 0.5 0.45 0.2 0.25 0.2 0.13 0.3 JBiSE  640 S. Behbahani et al. / J. Biomedical Science and Engineering 2 (2009) 637-643 SciRes Copyright © 2009 JBiSE SakerFalcon LannerFalcon PeregrineFalconOspreyEagleBootedEagleBonelliEagle SpottedEagle Small 0 0 0.95000 0 Medium 0.083 0.33 0.13 0.08 0.08 0.41 0.52 Big 0 0 0 0 0 0 0 2 leg 1 1 1 1 1 1 1 4 leg 0 0 0 0 0 0 0 Hair 0 0 0 0 0 0 0 Hoove 0 0 0 0 0 0 0 Mane 0 0 0 0 0.066 0 0 Feathers 1 1 1 1 1 1 1 Hunt 1 0.9 0.91 1 1 1 1 Run 0 0 0 0 0 0 0 Fly 0.8 1 1 1 1 1 1 Swim 0 0 0 0 0 0 0 Asia 1 1 0.98 1 1 1 0.91 Africa 0.86 0 1 0 0.243 0 1 Us 0 0.5 1 0.5 0.7 0.61 0 Europe 1 0.95 0 0 1 1 1 Mountainous 0.5 0.27 1 1 0 1 0.5 Plain 1 1 0 0 0 0.5 0 River 0 0 0.96 0 0 0 1 Jungle 0 0.032 0 0 1 0 1 Domestic 0 0 0 0 0 0 0 Carnivorous 1 1 1 1 1 0.95 0.89 Herbivorous 0 0 0 0 0 0 0 Frugivorous 0 0 0 0 0 0 0 Egg 1 1 1 1 1 0.92 1 Milk 0 0 0 0 0 0 0 Colour variation 0.24 0.54 0.35 0.24 0.4 0.33 0.21 LesserspottedEagle ImperialEagle GoldenEagleRedbreastedGooseGreylagGooseWithefrontedGoose SantherbertDog Small 0 0 0 0 0 0 0 Medium 0.58 0.61 0.71 0.23 0.68 0.31 0.76 Big 0 0 0 0 0 0 0 2 leg 1 1 1 1 1 1 0 4 leg 0 0 0 0 0 0 1 Hair 0 0 0 0 0 0 1 Hoove 0 0 0 0 0 0 0 Mane 0 0 0 0 0 0 0 Feathers 1 1 1 1 1 0.898 0 Hunt 1 0.99 1 0 0 0 0 Run 0 0 0 0 0.0363 0 1 Fly 0.95 1 1 1 1 1 0 Swim 0 0 0 1 0.99 0.85 0 Asia 1 0 0.87 1 0 0 0 Africa 0 1 1 0 0.9 0 1 Us 0.43 1 0 0 1 0 0 Europe 0.4 0 1 0.441 0 0 0.515 Mountainous 0 0.5 1 0 0.85 1 0 Plain 0.35 1 0 1 0 0.2 0 River 0 1 0 1 1 1 0.9 Jungle 0.9 0 0.03 0 0 0 0 Domestic 0 0 0 0 1 0.5 0 Carnivorous 1 1 1 0 0 0 0.858 Herbivorous 0 0 0 0 0 0 0.02 Frugivorous 0 0 0 0.98 1 1 0.04 Egg 1 1 1 1 1 1 0 Milk 0 0 0 0 0 0.021 0.88 Colour variation 0.4 0.16 0.13 0.5 0.1 0.15 0.12  S. Behbahani et al. / J. Biomedical Science and Engineering 2 (2009) 637-643 641 SciRes Copyright © 2009 JBiSE PointerDog WoodPigeon StockDove RockDove CollaredDave Wolf Kaiot Small 0 1 0.8 0.8 0.675 0 0 Mediu m 0.31 0.211 0.020 00.48 0.825 Bi g 0 0 0000 0 2 le g 0 1 1110 0 4 le g 1 0 0001 1 Hai r 1 0 0001 1 Hoove 0 0 0000 0 Mane 0 0 0000 0 Feathers 0 1 1110 0 Hun t 0 0 0001 1 Run 0.89 0 0000.88 0.88 Fl y 0 1 10.96510 0 Swi m 0.88 1 0000 0 Asia 0 1 0.910.870.921 1 Africa 0.66 1 0.50.110.50 0 Us 1 0 110.190.17 1 Euro p e 0.79 1 1011 0 Mountainous 0 0.2 0 0.95 0.210 0.4 Plain 0 1 1001 1 Rive r 0.78 0 110.20.11 0 Jun g le 0.023 1 0001 0 Domestic 0 1 110.950.032 0 Carnivorous 1 0 0001 1 Herbivorous 0 0 000.10 0 Fru g ivorous 0.1 1 1110 0 E gg 0 0 1111 0 Mil k 0 1 0000 1 Colour variation 0.54 0.67 0.20.30.470.31 0.21 Tiger Lion Horse IranianZebraZebra MarbledCat ChinchilaCat Small 0 0 0 0 0 0 0 Medium 0 0 0.85 0.62 0.967 0.45 0.16 Big 0.88 1 1 0 0 0 0 2 leg 0 0 0 0 0 0 0 4 leg 1 1 1 1 1 1 1 Hair 1 1 1 1 1 0 0 Hoove 0 0 1 1 1 0 0 Mane 0 1 1 1 1 0 0 Feathers 0 0 0 0 0 0 0 Hunt 1 0.89 0 0 0 1 0 Run 1 1 0.84 0.87 1 0.99 0.88 Fly 0 0 0 0 0 0 0.96 Swim 0 0 0 0 0 1 1 Asia 1 1 1 1 0.78 0 0 Africa 1 0 0.416 0 0 1 1 Us 0.9 0.3 0 0.91 1 0 0 Europe 1 0.78 0.445 0 0.93 0.35 0.032 Mountainous 1 0.95 0.985 0 0 1 1 Plain 0 1 0.95 0.35 0.64 0.033 0 River 0 0 0 0.019 0 0 1 Jungle 1 1 0.5 1 1 0 0 Domestic 0 0 1 0 0 0.91 1 Carnivorous 1 1 0 0 0 1 1 Herbivorous 0 0 1 1 0.8 0 0 Frugivorous 0.21 0.0033 0 0.1 0 0.2 0 Egg 0 0 0 0 0 0 0 Milk 1 1 1 1 1 1 1 Colour variation 0.3 0 0.92 0.54 0.75 0.21 0.3  642 S. Behbahani et al. / J. Biomedical Science and Engineering 2 (2009) 637-643 SciRes Copyright © 2009 JBiSE AsiangolodenCat BlackearCatCaucasianblackGrouseMallardDuck GadwellDuck WigeonDuck PanitalDuck Small 0 0.818 1 0 0 0 0 Medium 0.55 0.8 0.03 0.28 0.16 0.083 0.25 Big 0.01 0 0 0 0 0 0 2leg 0 0 1 1 1 1 1 4leg 1 1 0 0 0 0 0 Hair 1 1 0 0 0 0 0 Hoove 0 0 0 0 0 0 0 Mane 0 0 0 0 0 0 0 Feathers 0 0 1 1 1 1 1 Hunt 1 1 0 0 0 0 0 Run 1 1 0 0 0 0 0 Fly 0 0 1 1 1 0.95 1 Swim 0 0 0 0 0 0 0 Asia 1 1 1 1 0.91 1 1 Africa 0 1 0 0.8 0.4 0 1 Us 0.33 0 0.5 1 0 1 0 Europe 0 1 0 0 1 0 0 Mountainous 0 1 0.99 1 0.45 0 0 Plain 1 0 0.5 0 0 1 0 River 1 0 0 1 0.3 1 1 Jungle 1 1 0.5 0 0 0 0 Domestic 1 0 0.95 0 0.5 0 0.75 Carnivorous 1 1 0 0 0 0 0 Herbivorous 0 0 0 0 0 0 0 Frugivorous 0 0 1 1 1 1 1 Egg 0 0 1 1 1 1 1 Milk 1 1 0 0 0 0 0 Colour variation 0.4 0.7 0.1 0.21 0.3 0.24 0.5 GarganeyDuck MarbledtealDuckBlackBearGrizliBearPandaBearOrangotanMonky ShampainMonky Small 0 0.95 0 0 0 0 0 Medium 0.191 0 0 0 0 0 0.65 Big 0 0 0.01 0.28 0.17 0.45 0.23 2leg 0 1 0 0 0 1 1 4leg 1 0 1 1 1 0 0 Hair 0 0 1 1 1 1 1 Hoove 0 0 0 0 0 0 0 Mane 0 0 0 0 0 0 0 Feathers 0 1 0 0 0 0 0 Hunt 1 0 1 1 1 1 1 Run 0 0 1 1 1 0 0 Fly 0 0.95 0 0 0 0 0 Swim 0 0 0 0 0 0 0 Asia 1 1 0 1 0.85 0.95 0 Africa 1 0.033 0 0 0 1 0.89 Us 0 0 1 1 0 0 0.045 Europe 1 1 0 1 1 1 0 Mountainous 0 0 0 0 1 0.21 0 Plain 0 0.5 0 1 0 0 0.9 River 1 1 1 1 0 0.89 0 Jungle 1 1 1 1 1 1 0.8 Domestic 0 0 0 0 0 0 0 Carnivorous 0 0 1 1 1 0 0.14 Herbivorous 0 0 0 1 1 1 0 Frugivorous 1 1 0 1 0 1 1 Egg 1 1 1 0 0 0 0 Milk 0 0 0 1 1 0.87 1 Colour variation 0.4 0.3 0.4 0.13 0.36 0.04 0.351  S. Behbahani et al. / J. Biomedical Science and Engineering 2 (2009) 637-643 643 SciRes Copyright © 2009 5. CONCLUSIONS SOM is a highly useful multivariate visualization me- thod that allows the multidimensional data to be dis- played as a 2-dimensional map. This is the main advan- tage of SOM. The map units clustering makes it easy to observe similarities in the data. Through our experiment, we demonstrated that the possibility of quick observa- tion of relationship between component (feature) and the class as well as the relationship among different compo- nent (feature) of the dataset from the visualization of a dataset. SOM is also capable of handling several types of classification problems while providing a useful, interac- tive, and intelligible summary of the data. JBiSE However, SOM also has some disadvantages. For example, adjacent map units point to adjacent input data vector, so sometimes distortions are possible because high dimensional topography can not always be repre- sented in 2D. To avoid such phenomenon, training rate and the neighborhood radius should not be reduced too quickly. Hence, SOM usually need many iterations of training. And SOM also does not provide an estimation of such map distortion. Alternatives to the SOM have been de- veloped in order to overcome the theoretical problems and to enable probabilistic analysis. Current research showed a simple application of SOM neural network in clustering. This method can be used in many applications that need classification and one of them could be disease clustering. As seen in current re- search, we used fuzzy method to determine the features of each animal. Similar to this research, we can determine the features of diseases. This method could help the physician in their diagnosis. We can use the sign of diseases as the input of SOM neural network. As some classes of dis- ease have similar symptoms the SOM neural network can show a limitation of neighbor diseases that have such symptoms, so the physician can focus on them to diagnose the patient’s disease with more accuracy. Fuzzy features can increase the ability of SOM neural network if they choose carefully with more accuracy and of course it need some trail and error methods to find a rule to relate a membership function to each disease and its symptoms. REFERENCES [1] A. Forti, (2006) Growing hierarchical tree SOM: An unsupervised neural network with dynamic topology, , Gian Luca Foresti, Neural Networks, 19, 1568–1580. [2] S. Haykin, (1999) Neural networks a comprehensive foundation (2nd ed.), Prentice Hall. [3] R. G. Adams, K. Butchart and N. Davey, (1999) Hierar- chical classification with a competitive evolutionary neural tree, Neural Networks, 12, 541–551. [4] J. Li, Information visualization of self organizing maps. |

