Journal of Water Resource and Protection
Vol. 2 No. 8 (2010) , Article ID: 2459 , 6 pages DOI:10.4236/jwarp.2010.28088
16-23S rRNA Spacer Region Polymorphism in Gangetic River Water Isolates of Salmonella
Animal Biotechnology Centre, Govind Ballabh Pant University of Agriculture & Technology, Pantnagar, India
E-mail: mumtesh@rediffmail.com
Received March 4, 2010; revised June 7, 2010; accepted June 15, 2010
Keywords: Ganges River, Salmonella, Spacer Region Polymorphism, 16-23s rRNA
ABSTRACT
Salmonella is one of the major pathogenic bacteria present in contaminated water. 16-23S rRNA spacer region has been reported to be polymorphic at serovar level in Salmonella. Salmonella isolates obtained from Ganges river water were studied for 16-23S rRNA spacer region polymorphism. Thirty three isolates belonging to eight serovars (S. Typhimurium, S. Abuja, S. Pantypridd, S. Lagos, S. Chinkual, S. Zwickau, S. Goldenberg and S. Oritamerin) were studied for the polymorphism. Out of 33 isolates, 15 different profiles were observed no serovar specific profile. Our findings indicate that 16-23S rRNA spacer region is not specific at serovar level, but can be used for differentiation of different Salmonella isolates.
1. Introduction
The Ganges is a major river in Indian sub-continent and is considered to be Sacred and has been declared as National River by Indian Government in Year 2008. The fertile soil of the river basin measuring around one million square kilometers is a key to agricultural economy of the country and supports one of the highest human density populations. The large quantity of pollutant such as untreated sewage material in large volume (approximately one billion liters per day) [1] are disposed off in river Ganges that leads to accumulation of hazardous pathogens like Shistosoma, Escherichia coli O157:H7, Shigella and Salmonella [1-3] and expose the surrounding population to these diseases.
Salmonella is considered a dreaded pathogen causing several diseases such as endocarditis [4], Typhoid and Paratyphoid fever [5], Pneumonia [6] and meningitis [7]. Identification and serovar differentiation are prime requisite for controlling Salmonellosis. Conventional serovar typing is cumbersome and time consuming process [8], but with the development of molecular biology techniques like PCR genome has been targeted for this purpose. In the genome of the bacteria rRNA region has been extensively used for taxonomic purpose [9,10]. In Salmonella, 16-23S rRNA region has been studied and reported to be serovar specific [11], as he differentiated many serovars of Salmonella using 16-23S rRNA polymorphism. The basis of differentiation of serovars was based on variation in number and type of t-RNA sequences found within the spacer region of 16-23S rRNA [12]. The present study was undertaken to differentiate eight serovars of Salmonella isolated from Ganges river water by targeting 16-23S rRNA region.
2. Material and Method
Thirty three isolates of Salmonella were obtained from six different locations (stations) viz, Haridwar, Hastinapur, Kanpur, Garhmukteshwar, Varanasi and Narora. These isolates were serotyped at National Salmonella Centre I.V.R.I. (Izatnagar) India, and were characterized as S.Typhimurium, S. Abuja, S. Pantypridd, S. Lagos, S. Chinkual, S. Zwickau, S. Goldenberg and S. Oritamerin (Table 1). Genomic DNA was isolated from all isolates by C-TAB method [13].
3. 16-23 S rRNA Spacer Region PCR
Five μl (40ng) of genomic DNA was used for 50μl of PCR reaction mixture, containing 20 pmol of each primer (Primer1 (5’ GAA GTC GTA ACA ACG 3’) and Primer2 (5’CAA GGC ATC CAC CGT 3’), 200 μM of dNTPs, 3.0U of Taq polymerase and 1.5 mM final concentration of MgCl2. All chemicals used in PCR were procured from MBI-fermentas. PCR program was as follows-initial denaturation (94℃ for 5 min.) followed


Table 1. Description of Salmonella isolates (serovars, place of isolation, band profile and Molecular type) isolated from various locations from river Ganges.
 (a)
(a)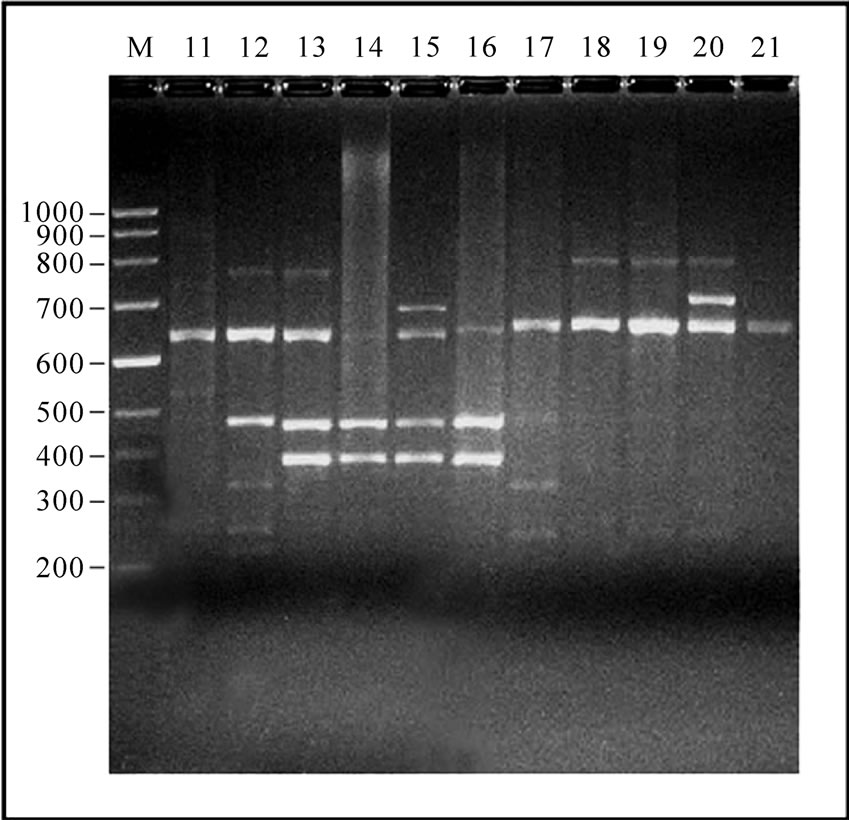 (b)
(b)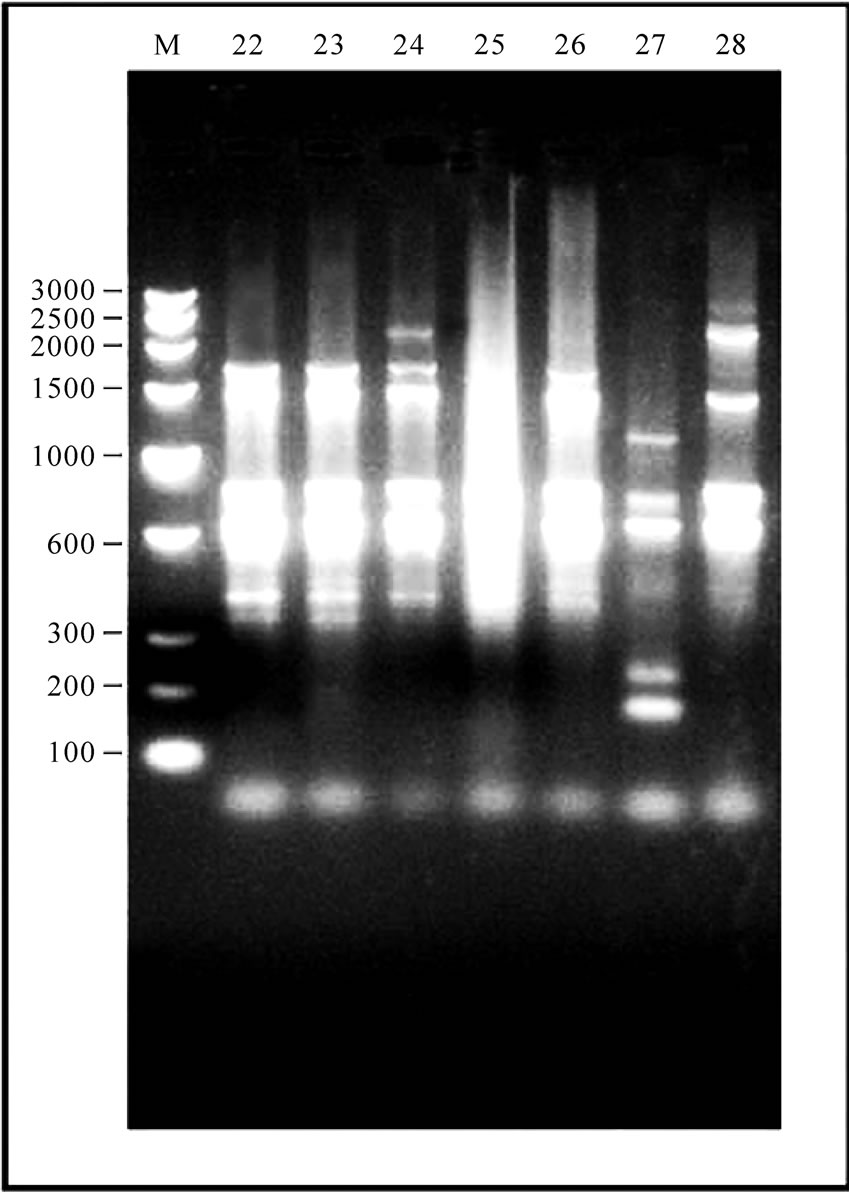 (c)
(c)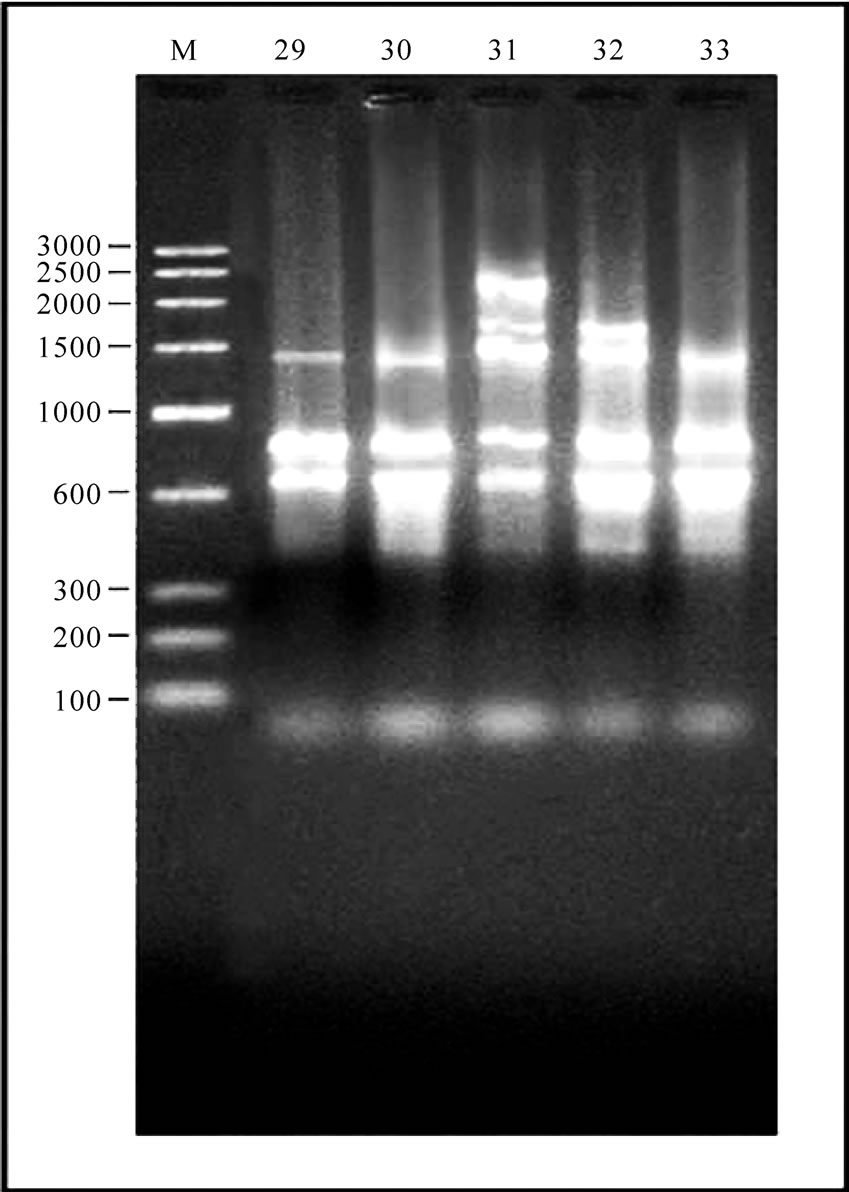 (d)
(d)
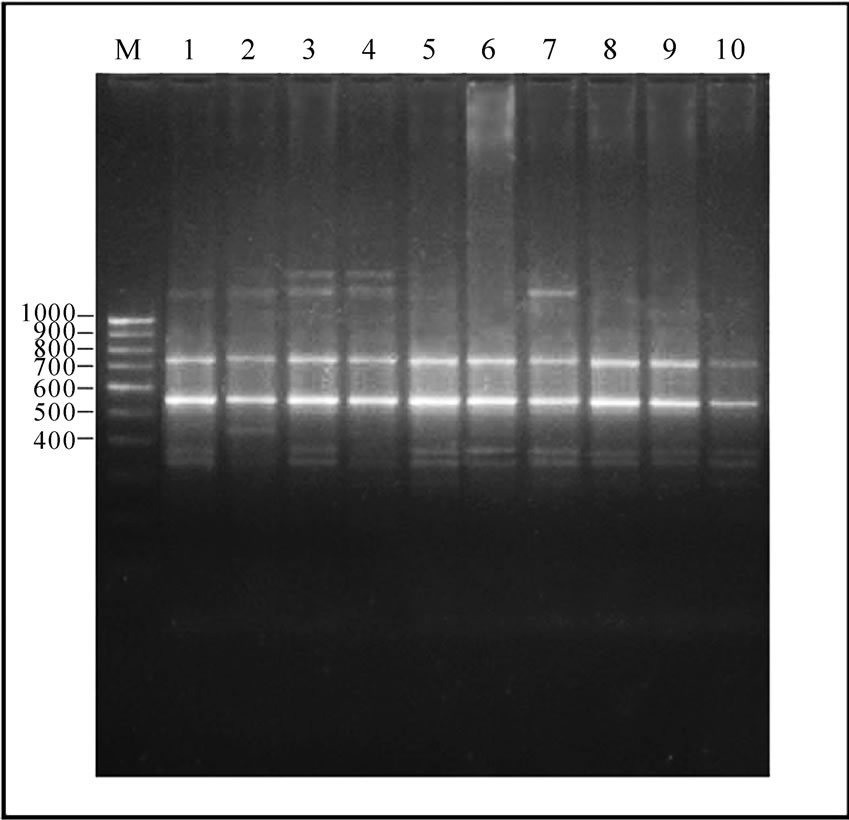
Figure 1. 12-23 SrRNA Spacer region amplicon of Gangatic isolates of Salmonella. (a) M: Marker 100 bp ladder (Baglore genei, India) Lane 1-10 Salmonella isolates (G29, G33, G36, G40, G23, G28, G35, G37, G38, G47); (b) M: Marker 100 bp (Baglore genei, India) Lane 11-21 Salmonella isolates (G4, G6, G7, G12, G13, G15, G17, G19, G22, G25, G39); (c) M: DNA Marker (100-3000) bp low range DNA rular (Baglore genei, India) Lane 22-28 Salmonella isolates (G1, G10, G14, G11, G16, G20, G21); (d) M: DNA Marker (100-3000) bp low range DNA rular (Baglore genei, India) Lane 29-33 Salmonella isolates (G45, G46, G50, G52, G55).
by 30 cycles composed of denaturation (94℃ for 1 min.), annealing (54℃ - 2min) and polymerization (72℃ - 2 min.). Final extension was carried out at 72℃ for 10 minutes. Five μl of amplified product was loaded on 1.5% agarose gel and electrophoresis was conducted in 1XTAE at 5V/cm. for two hours. Each sample was amplified thrice with negative control to avoid possibility of artifacts. Molecular weight of amplicon was measured by comparing 100bp ladder (Banglore Genei) using Bio1-D Software.
4. Results and Discussion
In all serovars, 16-23S r-RNA Spacer region was amplified. After separation of PCR product on 1.5% agarose gel only intense bands were scored. The relative molecular weights of amplicons were compared to the band with Standard molecular weight marker (100 bp ladder B.Genei India). Size of bands ranged from 144 to 2435 bp. All the isolates could be grouped into 15 profiles (M1-M15). M1 was most common and observed in 11 isolates. Eight profiles (M1, M8, M9, M10, M11, M12, M14 and M15Profile) were observed in 24 isolates of Salmonella enteric serovar Typhimurium. Two profiles M2 and M3 were observed in four isolates of S.Abuja. Three isolates of S.Lagos exhibited three different profiles M4, M5 and M6. Two isolates of S.Chinkaul and one isolate of S. Oritamerin shared common profile M2.One isolate of S. Zwickau obtained from Narora had altogether different profile M7 (Figure 1, Table 1). Comparison of similar and dissimilar bands of amplicon with Jaccord’s coefficient value revealed genetic variability in different isolates. On cluster analysis using Unweighted Pair Group Methods with Arithmetic Mean (UPGMA) the genetic similarity between 33 different isolates of Salmonella ranged from 0 to 100% and five major clusters were produced five major clusters at the level of zero similarity index.
In this study eight serovars of Salmonella were isolated by molecular methods from Ganges river water at six different locations (stations). Salmonella enterica serovar Typhimurium is considered a life threatening pathogen with zoonotic importance [14,15] and is found to be commonly associated with multiple drug resistance in human clinical cases [16]. Salmonella Oritamerin which is common in other countries [17] but not so frequently reported in India were present in Ganges water possibly due to the congregation of vast human population from across the diverse geographical boundries during
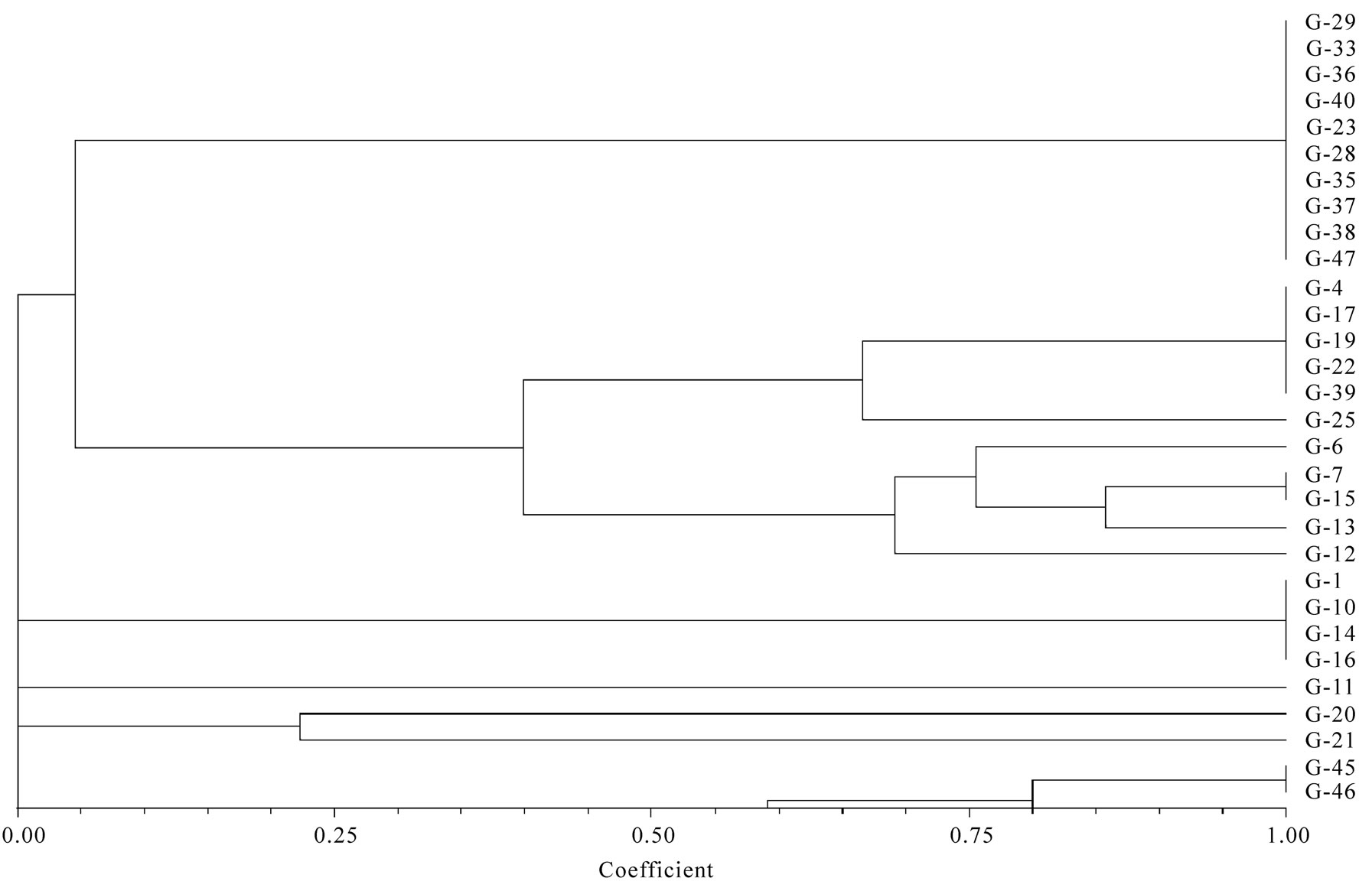
Figure 2. Dendrogram visualizing genetic variation in 16-23S rRNA spacer regions of isolates of Salmonella isolated from Ganges water.
festivals such as Kumbh.
On amplification of spacer region the product size varied from 529 to 2435 bp, which is very similar to the product size range reported by Jensen and Hubner [18]. The Variation of rRNA loci in Salmonella genome [19] and high genetic diversity of intergenic spacer region [20] could be attributed as possible reasons for high genetic variation shown by the isolates. We did not observed any serovar specific profile in the present study. S. Abuja and S. Chinkaul serovars shared the common profile M2 and S, Typhimurium the most common serovars observed in this study exhibited eight profiles. With these findings we suggest that 16-23S rRNA spacer region is not serovar specific but has a high genetic variation can be used for molecular typing of field isolates of Salmonella.
5. Acknowledgements
The Department of Science & Technology Government of India is highly acknowledged for providing financial assistance for this work. We are highly thankful to Prof. B. D. Lackchura (Ex-Head Department of Veterinary Biochemistry) for his guidance during this work.
REFERENCES
- S. Hamner, S. C. Broadway, V. B. Mishra, A. Tripathi, R. K. Mishra, E. Pulcini., B. H. Pyle and E. F. Timothy, “Isolation of Potentially Pathogenic E. Coli O157:H7 from Ganges River,” Applied and Environmental Microbiology, Vol. 73, No. 7, April 2007, pp. 2369-2372.
- S. M. Faruque, R. Khan, M. Kamruzzaman, Y. Shinji, Q. S. Ahmad, T. Azim, G. B. Nair, Y. Tekeda and A. S. David, “Isolation of Shigella dysenteriae Type 1 and S. flexneri Strains from Surface Waters in Banglades: Comparative Molecular Analysis of Environmental Shigella Isolates Versus Clinical Strains,” Applied and Environmental Microbiology, Vol. 68, No. 8, August 2002, pp. 3908-3913.
- A. Tikko, A. K Tripathi, S. C. Verma, N. Agrawal and G. Nath, “Application of PCR Fingerprinting Techniques for Identification and Discrimination of Salmonella Isolates,” Current Science, Vol. 80, No. 8, 2001, pp. 1049-1052.
- J. G. Moreno, C. Moar, F. Roman, R. Perez-Maestu and J. M. Lopez de Letona “Salmonella Endocarditis Presenting a Cerebral Hemorrhage,” European Journal of Internal Medicine, Vol. 11, No. 2, April 2000, pp. 96-97.
- R. Hasan, F. J. Cooke, S. Nair, B. N. Harish and J. Wain, “Typhoid and Paratyphoid Fever,” The Lancet, Vol. 366, No. 9497, November 2005, pp. 1603-1604.
- J. R. Gezen, D. M. Towle and J. D. Kravetz, “Salmonella Typhimurium Pulmonary Infection in an Immunocompitent Patient,” Connecticut Medicine, Vol. 72, July 2008, pp. 139-142.
- K. Swe, G. Nage, M. Van and A. Hoosen, “Salmonella Typhimurium Meningitis in an Adult Patient with AIDS,” Journal of Clinical Pathology, Vol. 61, No. 1, 2008, pp. 138-139.
- J. Baudart, K. Lamarchand, A. Brisabois and P. Lebaron, “Diversity of Salmonella Strains Isolated from Aquatic Environment as Determined by Serotyping and Amplification of the Ribosomal DNA Spacer Regions,” Applied Environmental Microbiology, Vol. 66, No. 4, April 2000, pp. 1544-1552.
- N. González, J. Romero and R. T. “Espejo Comprehensive Detection of Bacterial Populations by PCR Amplification of the 16S–23S rRNA Spacer Region,” Journal of Microbiological Methods, Vol. 55, No. 1, October 2003, pp. 91-97.
- A. A. Fadl, A. V. Nguyen and M. I. Khan, “Analysis of Salmonella enteritidis Isolates by Arbitrarily Primed PCR,” Journal of Clinical Microbiology, Vol. 33, No. 4, 1995, pp. 987-989.
- M. A. Jensen, J. A. Webster and N. Straus, “Rapid Identification of Bacteria on the Basis of Polymerase Chain Reaction Amplified Ribosomal DNA Spacer Polymorphism,” Applied Environmental Microbiology, Vol. 59, No. 4, April 1993, pp. 945-957.
- Brosius, et al., “Gene Organization of Primary Structure of Ribosomal RNA Operon from E. Coli,” Journal of Molecular Biology, Vol. 148, No. 2, 1981, pp. 107-127.
- K. Wilson, “Preparation of Genomic DNA from Bacteria” Current protocols in Molecular Biology, Unit 2.4.1, Wiley, New York, 1987.
- W. Rabsch, H. Tschäpea and A. J. Baumlerb, “Non-Typhoidal Salmonellosis: Emerging Problems,” Microbes and Infection, Vol. 3, No. 3, 2001, pp. 237-247.
- S. L. Foley, D. G. White, P. F. McDermott, R. D. Walker, B. Rhodes, P. J. Fedorka-Cray, S. Simjee and S. Zhao, “Comparison of Subtyping Methods for Differentiating Salmonella Enterica Serovar Typhimurium Isolates Obtained from Food Animal Sources,” Journal of Clinical Microbiology, Vol. 44, No. 10, October 2006, pp. 3569- 3577.
- E. J. Threlfall, “Antimicrobial Drug Resistance in Salmonella: Problems and Perspectives in Food and Water Borne Infections,” FEMS Microbiology Reviews, Vol. 26, No. 2, January 2006, pp. 141-148.
- L. Ronold, Y. Anderson, J. Biritta and P. Norberg, “A Summary of Reported Food Borne Diseases Incidents in Sweden, 1992 to 1997,” Journal of Food Protection, Vol. 63, No. 10, October 2000, pp. 1315-1320.
- M. A. Jensen and R. J. Hubner, “Use of Homoduplex Ribosomal DNA Spacer Amplification Products and Heteroduplex Cross Hybridization Products in Identification of Salmonella Serovars,” Applied and Environmental Microbiology, Vol. 62, No. 8, August 1996, pp. 2741- 2746.
- V. Gürtler, “Typing of Clostridium Difficile Strains by PCR-Amplification of Variable Length 16S-23S rDNA Spacer Regions,” Journal of General Microbiology, Vol. 139, No. 12, 1993, pp. 3089-3097.
- C. Lagatolla, L. Dolzani, E. Tonin, A. Lavenia, M. D. Michele, T. Tommasini and C. Monti-Bragadin, “PCR Ribotyping for Characterizing Salmonella Isolates of Different Serotypes,” Journal of Clinical Microbiology, Vol. 34, No. 10, June 1996, pp. 2440-2443.

