Journal of Cancer Therapy
Vol. 4 No. 5 (2013) , Article ID: 33781 , 11 pages DOI:10.4236/jct.2013.45112
The Influence of Gene Polymorphisms on Tobacco and Alcohol-Induced Oral Cancer Risk
![]()
1Department of Head and Neck Surgery and Otorhinolaryngology of the Heliópolis Hospital, São Paulo, Brazil; 2Department of Legal Medicine, Bioethics and Occupational Health, School of Medicine, São Paulo, Brazil; 3Head and Neck Surgeon of São José Hospital, São Paulo, Brazil; 4Department of Head and Neck Surgery, Hospital das Clínicas, São Paulo School of Medicine, University of São Paulo, São Paulo, Brazil.
Email: *curioni@usp.br
Copyright © 2013 Otávio A. Curioni et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 17th, 2013; revised June 16th, 2013; accepted June 23rd, 2013
Keywords: Mouth Neoplasms; Head and Neck Neoplasms; Polymorphism; Genetic; Smoking; Alcohol Drinking
ABSTRACT
Aims: This study examined whether genetic polymorphisms of tobacco and alcohol-related metabolic genes such as GSTM1, GSTT1, GSTP1, CYP1A1, CYP2E1 and DNA repair genes (XRCC1 194Trp, XRCC1 399Gln, and XRCC3 Met) contribute to the risk of developing OSCC. Methods: Patients eligible for inclusion were over 18 years, had pathologically confirmed OSCC and were followed prospectively for at least two years or until death, from December 2000 to December 2004. Ninety-two OSCC patients were included along with 244 subjects from the same hospital, evaluated in the same period as patients without cancer, as the control group. Results: GSTM1 null and XRCC1-194Trp alone increased the risk of OSCC (OR, 2.15; 95% CI, 1.2 - 3.6 and OR, 2.02; 95% CI, 1.01 - 4.03, respectively). The joint effect of GSTM1 null with CYP1A1 or CYP2E1 polymorphism increased the risk two to threefold. Similar results were observed when XRCC1-194Trp was combined with GSTM1 null or the CYP2E1 polymorphism. By contrast, XRCC1- 399Gln was associated with protection against OSCC. Gene-gene and gene-environmental interactions were mainly detected for CYP1A1 and GSTP1 associated with more than 20 p/y of tobacco and XRCC1-194Trp when more than 30 g/L/d of alcohol was consumed (OR, 8.8; 95% CI; 1.3 - 45.7). Conclusions: The drug metabolizing and DNA repair enzyme polymorphisms may be informative for clinicians in the preventive management of patients at risk, particularly those with strong smoking and drinking habits.
1. Introduction
Oral squamous cell carcinoma (OSCC) is the eighth most frequent cancer in the world among males and the fourteenth among females, accounting for nearly 3% of all cancer cases worldwide. The main risk factors related to OSCC are tobacco and alcohol use, independent of the type of tobacco or alcoholic beverage, with the association being synergistic. Dietary factors, human papillomavirus (HPV) infection, genetic factors, hot mate, and poor oral hygiene are reported as risk factors, as well. Social inequalities are related to OSCC risk by being linked to factors directly affecting behaviour and lifestyle [1].
In Brazil, OSCC is the fourth most common cancer and 14,170 new cases of OSCC are estimated for the year 2012 (9990 and 4180 for men and women, respectively) [2]. Although the oral cavity is more accessible for complete examination, most cases are diagnosed in clinical stages III or IV due to negligence or inaccessibility to medical care.
After exposure to xenobiotics some inactive products are metabolically converted into active products (phase I enzymes) with high affinity for DNA, RNA and/or proteins resulting in adduct formation and DNA damage. In a dynamic cellular process most carcinogens are detoxified by drug-metabolizing enzymes (phase II enzymes).
The oxidative metabolism or phase I, mediated by cytochrome P450 enzymes and microsomal epoxide hydrolases that convert many highly reactive metabolites to carcinogenic compounds. The phase I cytochrome P450 superfamily of enzymes is encoded by CYP genes, which include CYP1A1 and CYP2E1 subfamilies. They play a critical role in the biotransformation of drugs, carcinogens, steroid hormones, and environmental toxicants leading to direct activation and production of more active metabolites [3].
The second, enzyme conjugate or phase II, consists of reactions that involve GSTs and other enzyme families that inactivate the products of phase I [4]. GST family genes are grouped in eight classes based in the structure, specific substrate and immunological properties: alpha, mu, kappa, pi, sigma, theta, zeta and omega [5]. Glutathione S-transferase pi-GSTP1 gene encodes an enzyme expressed in placental, spleen, heart and lung tissues. There are two genetic variations in GSTP1 gene: A313G (Ile105Val) and C341T (Ala114Val) that result in significant alterations in enzyme activity and it may be associated with the levels of DNA adducts [6]. These polymorphisms are associated with increased risk for head and neck squamous cell carcinoma-HNSCC [3]. Glutathione S-transferase mu-GSTM1 gene is polymorphic in human polulation (GSTM1*0) and Glutathione Stransferase theta-GSTT1 gene is also polymorphic in human population, presenting null phenotype by deletion (GSTT1*0) and consequently complete loss of enzyme activity [7].
As most of phase I and II enzymes have polymorphic sites, which may affect their activity, the strength and balance of these activities may differ according to the individual genotype of these metabolic genes [8]. In fact, inter-individual variation in the metabolism of xenobiotics may be related to differences in the risk for different types of cancer, and evidence exists suggesting their association with an increased risk of cancer [9].
As well as polymorphisms on metabolizing genes, the mechanisms of DNA damage repair are also important factors protecting cells against carcinogenesis due to environmental exposure. XRCC1 (X-ray repair cross-complementing) gene plays an important role in the base excision repair (BER) pathway and participates as scaffolding intermediate by interacting with ligase III, DNA polymerase b and poly(ADPribose) polymerase (PARP) in the C-terminal, N-terminal, and central regions of XRCC1, respectively [10-13]. XRCC3 participates in DNA double-strand break/recombination repair and is a member of an emerging family of RAD-51-related proteins [14,15]. The protein XRCC3 acts in the homologous pathway of double-stranded DNA repair. This pathway plays a substantial role in preventing chromosomal fragmentation, translocation, and deletion, which can lead to carcinogenesis [16,17]. Polymorphisms in DNA repair genes may alter their functions and their capacity to repair DNA damage, increasing genetic instability and the risk of developing cancer [18].
It has been suggested that a detailed knowledge of the risks to a particular gene-exposure status could be used to provide personalized prevention and follow-up of patients, mainly when there is high consumption of tobacco and alcoholic beverages. Further studies in these highrisk individuals could also provide an insight into applying these preventive approaches to the average risk population and lead to a better understanding of the multi-step carcinogenic process.
The objective of this study was to examine whether genetic polymorphisms of tobacco and alcohol-related metabolic genes and DNA repair genes contribute to susceptibility to carcinogenic exposure and the development of OSCC. The contribution of alcohol and tobacco consumption habits to the risk of developing OSCC was also evaluated.
2. Materials and Methods
A total of 336 participants in this study were recruited from Heliópolis Hospital, São Paulo, Brazil (Table 1). All 92 consecutive patients with histologically confirmed oral squamous cell carcinoma were preoperatively selected in the Department of Head and Neck Surgery and Otorhinolaryngology of the Hospital Heliópolis, and invited to participate after being informed about the research. Eligible patients comprised 82% men (81/92) and 12% women (11/92), with ages varying from 24 to 81 years old (53 median ±9.3 years old). Seventy-one percent of the patients (65/92) were diagnosed with floor of mouth or oral tongue cancer, 20% (19/92) with gingival/retromolar cancer, and 9% (8/92) with lip cancer. Approximately 74% of the study cohort consisted of patients diagnosed with TNM stage III-IV disease. The patients were followed prospectively from date of diagnosis until death or until July 2012. Follow-up data of the cohort members were collected by a prospective longitudinal tracking system. Information on vital status, primary site, TNM stage (consistent with the American Joint Committee on Cancer, AJCC) lymph node status, disease recurrence, and treatment were collected and updated at least every six months from clinician notes, pathology reports, laboratory reports, and death certificates.
The control group was composed of 244 patients without any malignancy, 92% men (225/244) and 8% women (19/244), varying from 20 to 82 years old (median 53.6 ± 10.5 years old), who were selected in the same hospital and in the same period as the patients with OSCC. The underlying causes of hospitalization of the control patients were grouped into large diagnostic categories (following the tenth version of the International Classification of Diseases): these patients mainly had non-cancerous digestive system diseases (44%) and cardiovascular diseases (33%).
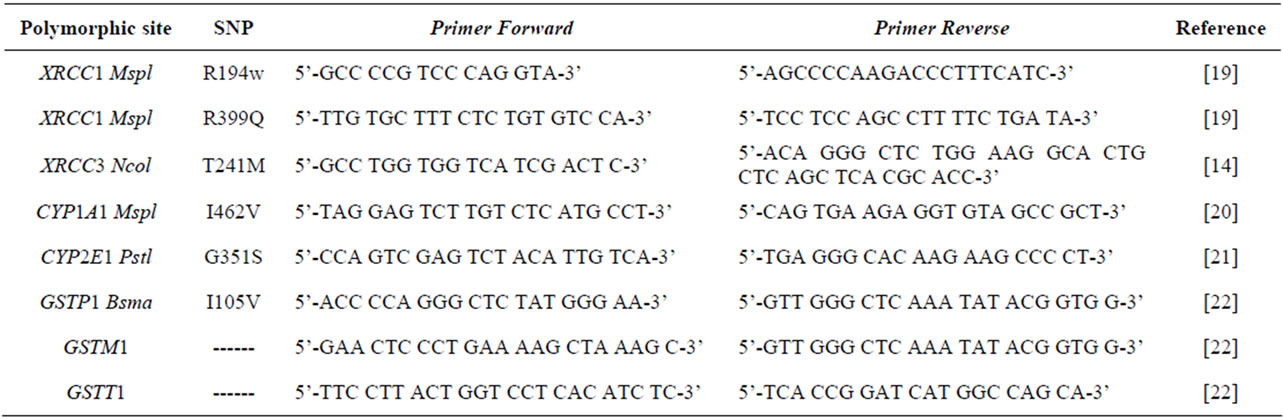
Table 1. Summary of the polymorphic sites of the genes, including used primers.
All patients with OSCC and the controls were interviewed using a standardized questionnaire regarding smoking habits, alcohol drinking history, dietary habits, and occupational activities. Data on the frequency and amount of alcohol consumption were estimated for all types of alcoholic beverages, including beer (5% ethanol), wine (12% ethanol), and hard liquor or cachaça (a spirit made from sugar cane and vastly consumed in Brazil), which is 41% ethanol.
Genomic DNA for genotype evaluation was isolated from lymphocytes in 5 ml of peripheral blood samples by means of a non-organic DNA extraction procedure. The CYP2E1, CYP1A1, GSTP1, XRCC1 and XRCC3 polymorphisms were detected by restriction fragment length polymorphism (RFLP) analysis as previously described [14,19-21]. A single assay using a multiplex PCR protocol was performed for simultaneous GSTM1/GSTT1 gene amplification, including a positive control to identify the GSTM1 and GSTT1 null genotype polymorphisms [22]. This technique does not distinguish between heterozygote and homozygote GSTM1 or GSTT1-positive genotypes, but it conclusively identifies null genotypes. Related polymorphisms were evaluated in polyacrylamide or agarose gel depending on fragment size. Negative controls were included in every run to test for contamination and all genotyping results were determined blind to casecontrol evaluation, compared to positive controls (Table 2).
The study protocol and the questionnaire were approved by the Institutional Ethics Committees, and all patients gave written informed consent to participate.
The statistical significance of associations between OSCC and SNP polymorphisms was analysed using Fisher’s exact test [23]. Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated as approximations of relative risk [24].
The risk of oral cancer was estimated by comparing subjects with GSTM1 and GSTT1 null genotypes with those positive for at least one allele, and for CYP2E1, CYP1A1, GSTP1, XRCC1-194, XRCC1-399, and XRCC3 the risk was considered by comparing individuals with mutations (homozygous and/or heterozygous) against those without mutations (wild type). Alcohol consumption was evaluated based on categories of cumulative consumption, expressed according to estimated dose as grams per litre per day (g/L/d) and tobacco consumption was evaluated according to packs per year (p/y); the risk point for alcohol consumption was found using a receiver operating characteristics (ROC) curve. The OR estimates were obtained by non-conditional logistic regression modelling adjusted for potential confounders. Statistical significance was assessed by the maximum likelihood ratio test. The disease effect of variables that showed p < 0.02 in the univariate analysis was compared using multiple logistic regression analysis. The statistical signifi cance of associations was determined by chi-square tests using the statistical computer software SPSS (version 15.0), and the critical level of rejection on the null hypothesis was considered to be 5%.
3. Results
Table 1 summarizes the demographic characteristics of the 92 patients with OSCC and the corresponding controls. The study groups were very similar regarding gender and age; both consisted mainly of men aged 60 years old or less. Regarding skin color, 67% of OSCC patients and 60% of the controls were classified as white skin color, as shown in Table 1.
Among the OSCC cases, only 2% reported that they did not smoke, compared to 23% in the control group. The difference was considered significant (p < 0.001), resulting in a more than fifteen times increased risk of cancer among smokers (OR, 16.72; 95% CI, 3.84 - 102.27), as shown in Table 1.
The known risk factors for head and neck cancer such as tobacco and chronic alcohol intake were confirmed. As shown in Table 1, OSCC patients reported a median
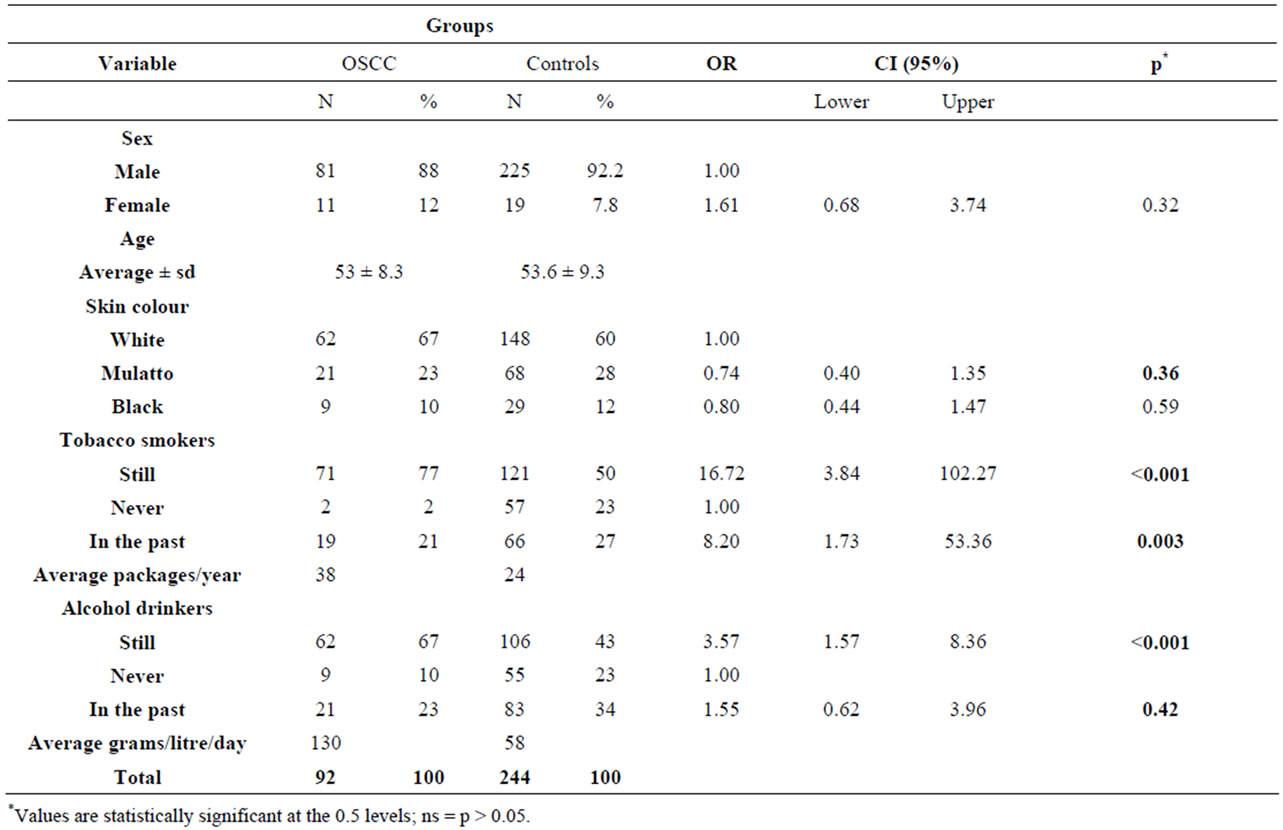
Table 2. Distribution of demographic factors in the OSCC patients and controls.
alcohol consumption of 130 g/L/day, which was higher than that declared by the control group (58 g/L/day) and resulted in a more than three times increased risk of cancer among alcoholics (OR, 3.57; 95% CI, 1.57 - 8.360).
As identified in others studies, alcoholics are also often smokers, which makes it difficult to analyse the effects of each one of these habits individually. To assess the combined effect of tobacco and alcohol on cancer, all possible comparisons between alcohol and tobacco use were evaluated. The consumption of cigarettes combined with the use of alcohol increased the risk of OSCC by more than eleven times (OR, 11.49; CI 95%, 1.50 - 241.44), as shown in Table 3.
Using a ROC curve, it was possible to estimate the amount of alcohol and tobacco that maximized the risk of developing OSCC. It was found that individuals who reported consuming over 30.655 g/L/day of alcohol and 27.5 p/y of tobacco had an approximately four times greater risk of developing OSCC compared with controls (Table 4).
Table 5 shows the genotype frequencies for the different GSTs, CYPs and DNA repair genes in the patients and corresponding controls. As evident from the table, the GSTM1 null genotype was found to be present in a higher frequency (56.5%) of the patients when compared to the controls (37.7%), which resulted in significant association between OSCC and GSTM1 null genotype (OR, 2.15; CI, 1.28 - 3.6). Likewise, the XRCC1-194Trp genotype was also found to be more prevalent in patients than controls, revealing a significant association between OSCC and a polymorphism of the DNA repair gene XRCC1-194Trp (C > T) at exon 6 (OR, 2.02; 95% CI, 1.01 - 4.03). As shown in Table 5, the XRCC1-399Gln heterozygous genotype (52%) and homozygous genotype (12.3%) were found to be inversely associated (39.1% in controls and 6.5% in OSCC), which resulted in a decreased risk of OSCC (OR, 0.46; 95% CI, 0.28 - 0.75).
The risk of OSCC associated with multiple at-risk genotypes was also studied in the controls as showed on Table 6. All possible polymorphism combinations of the genes studied were assessed but only the selected results are presented. First, the frequencies of present or null variants of GSTM1 combined with the other tested gene variants were analyzed. The frequency of GSTM1 null combined with the CYP1A1 mutated genotype was higher in OSCC cases (72.8%) than in the controls (58.2%), and this result was significant (OR, 1.93; 95% CI; 1.10 - 3.37). A similar significance was observed with the joint effect of GSTM1 and CYP2E1Pst I polymorphisms in OSCC patients when compared to the controls, which increased the risk of OSCC (OR, 2.29; 95% CI; 1.36 - 3.87), and the GSTM1 null and XRCC1-194

Table 3. Alcohol and tobacco considered together in the risk of OSCC.

Table 4. The diary alcohol intake and tobacco consumption and the risk of OSCC considering 92 patients and 244 controls.
polymorphism was significantly more frequent in OSCC cases than in controls (OR, 2.44; 95% CI 1.44 - 4.14). Similar results were observed when the XRCC1-194Trp genotype was combined with the CYP2E1 PstIgenotype (OR, 2.0; 95% CI; 1.10 - 3.62).
After stratification for the potential confounding variables mentioned in Table 1, some interesting differences in genotype frequencies were noted between the patients and controls (Table 7). Patients that consumed less tobacco had a higher frequency of the GSTM1 null genotype when compared to patients with a higher tobacco intake, although the sample size was a limiting factor for statistical significance. For the GSTP1 BsmA polymorphism, there was an increasing trend in this mutant genotype in patients with a high tobacco intake which became statistically significant when more than 39 p/y of tobacco was consumed (OR, 5.0; 95% CI; 1.9 - 12.4 (Table 6)). Similar results were observed for the CYP1A1 MspI polymorphism, which increased the risk of OSCC by almost four times with increased tobacco use (Table 5). For drinkers, the mutated genotype for XRCC1-399 and XRCC3 seemed to protect against OSCC when the amount of alcohol consumed was between 5 - 30 g/L/d (OR, 0.1; 95% CI; 0.03 - 0.7 (Table 5)), and XRCC1-194 increased the risk of OSCC by almost nine times when more than 30 g/L/d of alcohol was consumed (OR, 8.8; 95% CI; 1.3 - 45.7 (Table 7))
4. Discussion
Tobacco and alcohol consumption have been described as the most important risk factors associated with head and neck squamous cell carcinoma HNSCC [25]. More
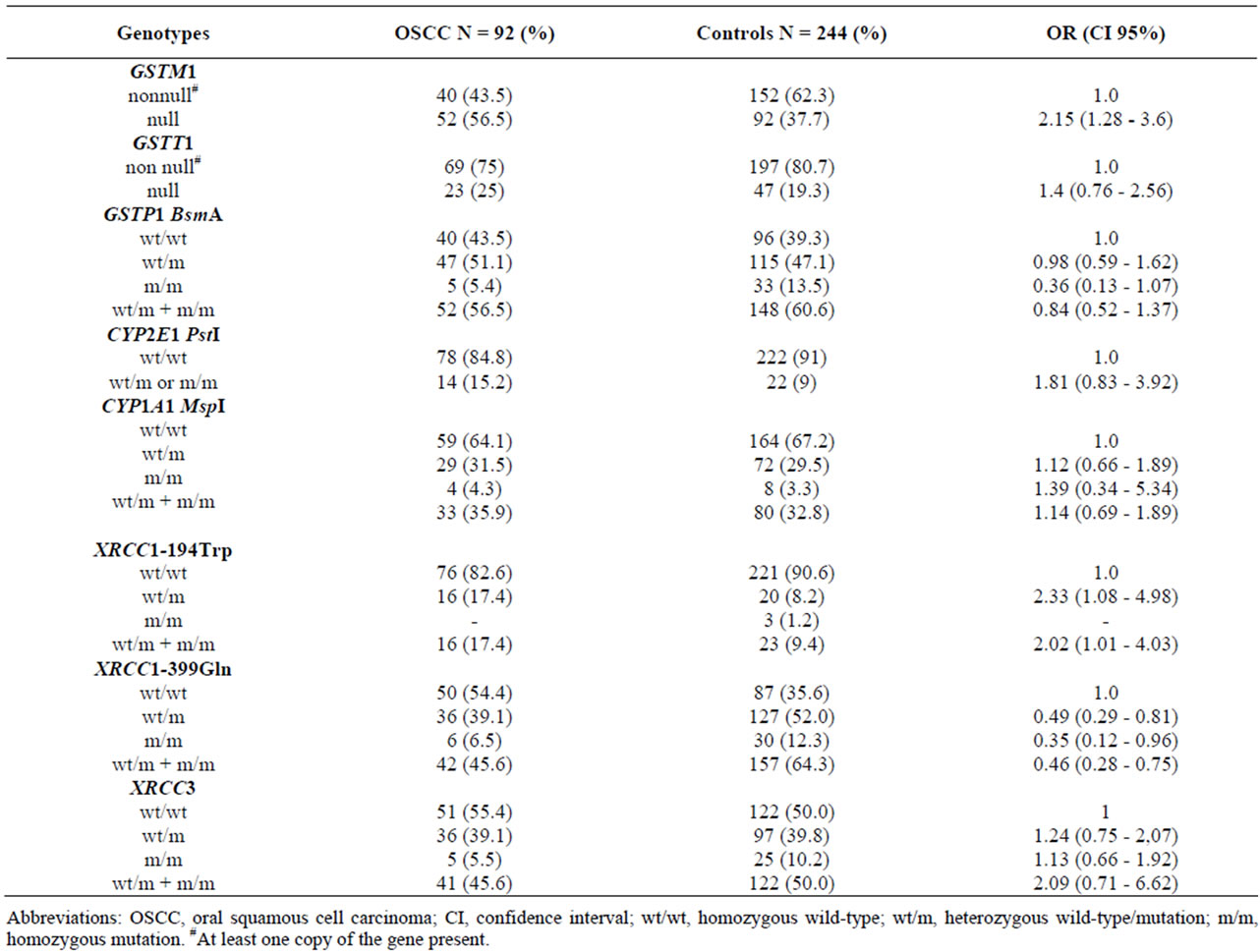
Table 5. The genotype frequencies and risk (estimated by odds ratio (OR) and 95% CI) of genotypes GSTM1, GSTT1, GSTP1, CYP2E1, CYP1A1, XRCC1-194, XRCC1-399, XRCC3 in the OSCC patients and controls.
recently, the results from case-control studies of several phenotypic and genotypic assays supported the hypothesis that genetic susceptibility or predisposition plays an important role in head and neck tumour aetiology [26]. It has been hypothesized that susceptibility to disease development is based on inherited differences in the efficiencies of carcinogen metabolism, DNA repair and cell cycle control or a combination of these.
In our series, only a few individuals reported that they had never smoked or consumed alcoholic beverages. Alcohol and tobacco consumption were more frequently observed in patients with OSCC than in the control subjects (p < 0.001). The risk of development of different types of cancer is dependent on inter-individual variability in sensitivity towards carcinogens. In this context, in the present study we evaluated the association between the drug-metabolizing gene polymorphisms GSTM1, GSTT1, GSTP1 BsmA, CYP1A1 and CYP2E1 and the DNA repair gene polymorphisms XRCC1-194, XRCC1- 399 and XRCC3. The results indicated that the GSTM1 null genotype confers a 2.15-fold increase in the risk of oral cancer. A similar result was observed for the XRCC1-194 genotype, which also confers a 2.02-fold increase in the risk of OSCC. The other DNA repair gene polymorphism, XRCC1-399 and XRCC3, however, appeared to protect against oral cancer.
The association between specific genetic polymorphisms and the risk of head and neck cancer has been evaluated in different populations all over the world [27]. Despite the fact that an increased frequency of patients with HNSCC were found to carry the GSTM1 null genotype when compared to patients without cancer, independent studies and subsequent metanalysis reports remain inconclusive about the association between HNSCC and GST variants [28]. The positive association between the GSTM1 null genotype and OSCC observed in this study is in agreement with other recent publications, but not with OSCC patients from Rio de Janeiro, Brazil [6]. A lack of the GSTM1 enzyme is thought to increase cancer susceptibility as a result of the decreased ability to detoxify reactive intermediates of tobacco carcinogens, including polycyclic aromatic hydrocarbons
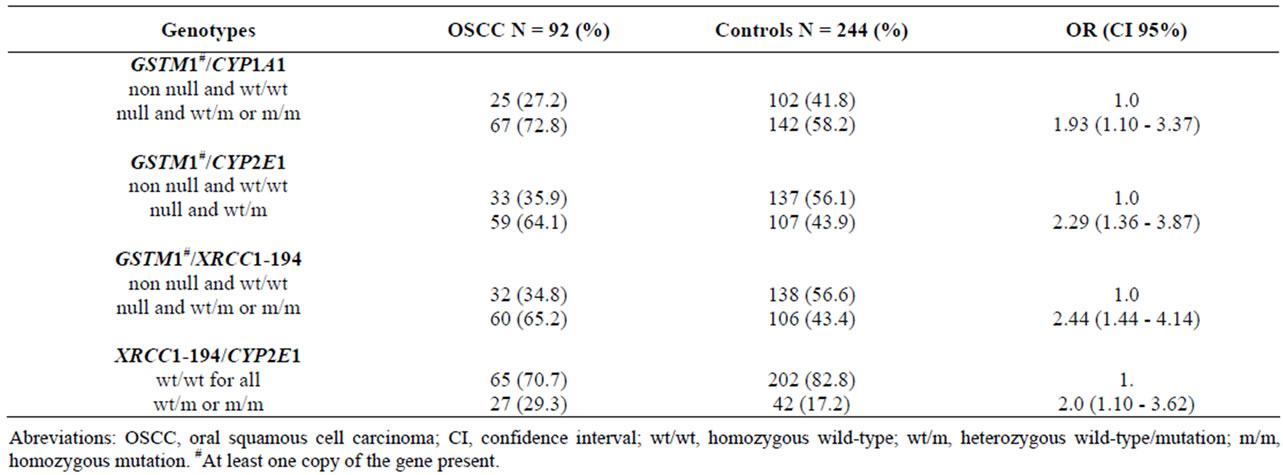
Table 6. Frequencies of the selected gene-gene combinations, the odds ratios, and 95% CI intervals in the OSCC patients and controls.
and aromatic amines and for regulating the DNA repair enzymes. In fact, GSTM1 levels in the peripheral whole blood of individuals with the GSTM1 null genotype are nine times lower, indicating that homozygous deletion of the GSTM1 gene associated with tobacco and alcohol habits can even increase the risk of OSCC [29]. In fact, one carcinogenic role attributed to alcohol is believed to be its ability to act as a solvent, facilitating the entry of tobacco carcinogens into oral tissues and explaining the synergy between tobacco and alcohol. In this investigation the two other detoxification enzyme polymorphisms, GSTT1 and GSTP1, did not show an association with OSCC risk; this result is in accordance with the results found by some other authors [30].
Unlike other authors that have studied tumours of the oral cavity, pharynx and larynx [31], our results show that the frequency of the CYP2E1 and CYP1A1 mutated alleles was higher in patients with OSCC compared to the controls, but the difference between the groups was not statistically significant. It is important to point out that the CYP1A1 gene codes for the enzyme aryl hydrocarbon hydroxylase, which is responsible for the first step in the metabolism of polycyclic aromatic hydrocarbons, and the CYP2E1 gene product is involved in the metabolism of ethanol. The CYP2E1 gene also catalyses oxidation and the DNA adduct formation of several compounds found in cigarette smoke, such as N-nitrosamines and benzene.
Moreover, there are not only individual differences in metabolic activation and detoxification processes, but DNA repair mechanisms may also affect the acquired status of the host and influence the risk of developing cancer. The DNA repair enzyme, XRCC1, has an effect on the base excision repair of genomic damage caused by exposure to carcinogens such as tobacco and alcohol [32]. Amino acid substitutions in the active protein binding domains may impair the efficiency of repairing DNA damage and affect its function. Associations between XRCC1 polymorphisms and different types of cancer such as lung, breast, colon and head and neck cancers have been investigated. However, the results are not consistent. In our study, we observed a significant difference between the frequencies of the XRCC1-194Trp polymorphism in patients compared to controls, which increased the risk of OSCC by more than twice. These data are in agreement with a report on patients with breast cancer [32], whereas they are in contrast to studies in which the XRCC1-194Trp polymorphism was not found to confer a significant risk of lung cancer in the Caucasian population [33]. These conflicting results may stem from the complexity of cancer aetiology with regard to exposure to carcinogens, DNA repair genotypes or other genetic factors, and to the different sample sizes evaluated.
In our study we also observed a significant difference between frequencies of the XRCC1-399Gln allele in patients. Although the reason for the reduction in OSCC risk for patients with the XRCC1-399Gln genotype is uncertain, a potential explanation is that this genotype leads to less efficient repair of the damage incurred by cigarette smoking which may prevent cells from progressing through the apoptotic pathways that increase the likelihood of progressive genomic instability and cancer recurrence. Since polymorphisms in the XRCC1 gene have been linked to a reduced capacity for the removal of DNA damage, our results indicating an association between the polymorphisms studied and oral cancer may imply that the base excision pathway might be involved in the repair of DNA damage that plays a role in the initiation of head and neck carcinogenesis.
Different GST isoforms exhibit overlapping substrate specificity and combinations of various unfavourable
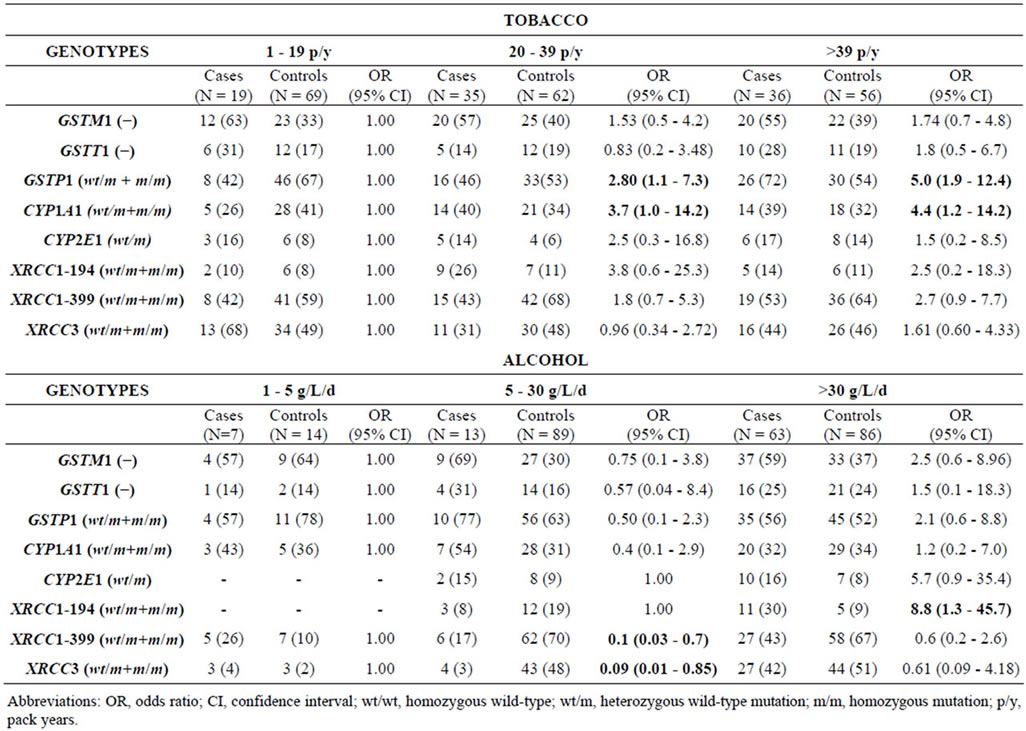
Table 7. GSTM1, GSTT1, GSTP1 BsmA, CYP1A1 MspI, CYP2E1 PstI, XRCC1-194Trp, XRCC1-399Gln mutated genotype frequencies in OSCC cases and controls stratified by tobacco (90 patients and 187 controls) and alcohol consumption (83 patients and 189 controls).
deletion genotypes may theoretically confer an even higher risk of cancer. Consistent with the previous study [34], our data also demonstrated a significantly elevated risk of OSCC in patients with combined high-risk genotypes. The simultaneous presence of the CYP1A1 and CYP2E1 polymorphic genes, which increase gene expression, and the lack of the GSTM1 enzyme, which makes elimination of the metabolic products of these genes more difficult, could result in the accumulation of electrophilic intermediates resulting in adduct formation [35]. In a recently published metanalysis study, a joint effect of the CYP1A1 and GSTM1 polymorphisms on cancer risk was observed, suggesting that tobacco use and genetic factors play significant roles in oral and pharyngeal cancer [36].
In this study, besides genotypic markers of susceptibility such as GST, CYP and DNA repair gene polymorphism, we also evaluated phenotypic markers of susceptibility to alcohol and tobacco intake. When we evaluated the different amounts of tobacco consumption and gene interactions it was observed that there was a four times greater risk of OSCC for patients who consumed more than 20 packets of tobacco per year and had the CYP1A1 variant genotype. The role of CYP1A1 in the metabolism of tobacco is well known and the tissue specific expression of this protein has been also demonstrated in oral buccal cells [37].
In the present study the GSTP1 polymorphism was associated with an increased risk of cancer when the tobacco consumption was higher than 39 p/y, but not when the patients and controls were first compared. Several studies examined the relationship between GSTP1 polymorphisms and smoking status in HNSCC and the results were not consistent. Ophuis et al. [38] found that HNSCC patients with the GSTP1 polymorphic genotype showed increased odds ratios when compared to the wild-type GSTP1 genotype. However, McWilliams et al. [39] found no association between GSTP1 genotypes and smoking status in HNSCC risk. The metabolic action of GST enzymes may differ with different cancer sites. The highest concentrations of GSTP1 were observed in oral and pharyngeal tissues, and the highest concentrations of GSTM1 were observed in laryngeal tissue, relative to the other GSTs [40].
When the associations between DNA repair genes and the established risk factors for OSCC were investigated, our results showed that high alcohol intake and the XRCC1-194Trp genotype increased cancer risk but the association with XRCC1-399Gln and XRCC3Met genotypes reduced OSCC risk. For each tumour type, the biological pathway responsible for the induction of apoptosis and the inactivation of this mechanism may impact on both the ability to detect and the direction of the XRCC1-disease association. This is likely to vary significantly, not only by carcinogen exposure but also by disease, ethnicity, and geography. There are not many gene-interaction studies in the medical literature and these preliminary data should be regarded with caution. The assessment of smoking and alcohol consumption is difficult, especially for the total accumulative exposure. Exposure usually takes place over many years and is not always consistent. In addition, different cigarette brands may yield completely different nicotine and tar exposures.
Finally, oral carcinogenesis is a complex multifocal process of multiclonal field carcinogenesis and intraepithelial clonal spread. Substantial evidence indicates genomic instability as a cause rather than a consequence of malignant transformation that points to a key role of aberrant DNA content in carcinogenesis. Considering that oral cancer is a disfiguring, potentially fatal disease which even nowadays continues to rise in incidence among young people and the elderly alike, controlling its devastating consequences will require intervention in at-risk people ideally before the disease becomes invasive, locally advanced or metastatic. So far it has been found that drug metabolizing enzymes and DNA repair enzyme polymorphisms are constitutive genotypes that may be informative for clinicians in the preventive management of patients at risk, particularly those with strong smoking and drinking habits.
5. Acknowledgements
The authors thank the biologist Salete Siraque Garcez for assistance with data collection and Prof. Dr Eduardo Massad for statistical assistance. The authors also thank Dr. Adhemar Longatto-Filho for suggestions and a critical review of the manuscript. This work was partially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and LIM-HC-FMUSP.
REFERENCES
- M. Camargo Cancela, L. Voti, M. Guerra-Yi, F. Chapuis, M. Mazuir and M. P. Curado, “Oral Cavity Cancer in Developed and in Developing Countries: PopulationBased Incidence,” Head & Neck, Vol. 32, No. 3, 2010, pp. 357-367.
- INCA—Instituto Nacional de Câncer, “Incidência de Câncer no Brasil/Estimativa,” 2012. http://www.inca.gov.br/estimativa/2012
- M. Lacko, M. B. Oude Ophuis, W. H. Peters and J. J. Manni, “Genetic Polymorphisms of Smoking-Related Carcinogen Detoxifying Enzymes and Head and Neck Cancer Susceptibility,” Anticancer Research, Vol. 29, No. 2, 2009, pp. 753-761.
- P. Jancova, P. Anzenbacherb and E. Anzenbacherovaa, “Phase II Drug Metabolizing Enzymes,” Biomedical Papers of the Medical Faculty of the University Palacky, Olomouc, Czech Republic, Vol 154, No. 2, 2010, pp. 103-116. doi:10.5507/bp.2010.017
- R. Whalen and T. Boyer, “Human Glutathione S-Transferase,” Seminars in Liver Disease, Vol. 18, No. 4, 1998, pp. 345-358. doi:10.1055/s-2007-1007169
- A. Hatagima, E. C. G. Costa, C. F. S. Marques, R. J. Koifman, P. Boffetta and S. Koifman, “Glutathione STransferase Polymorphisms and Oral Cancer: A CaseControl Study in Rio de Janeiro, Brazil,” Oral Oncology, Vol. 44, No. 2, 2008, pp. 200-207. doi:10.1016/j.oraloncology.2007.02.001
- J. Seigard and G. Ekstrom, “The Role of Human Gluthatione Transferases and Epoxide Hydrolases in the Metabolism of Xenobiotics,” Environmental Health Perspectives, Vol. 105, Supplement 4, 1999, pp. 791-799.
- W. W. Au, “Usefulness of Biomarkers in Population Studies: From Exposure to Susceptibility and to Prediction of Cancer,” International Journal of Hygiene and Environmental Health, Vol. 210, No. 3-4, 2007, pp. 239-246. doi:10.1016/j.ijheh.2006.11.001
- E. Reszka, P. Czekaj, J. Adamska and W. Wasowicz, “Relevance of Glutathione S-Transferase M1 and Cytochrome P450 1A1 Genetic Polymorphisms to the Development of Head and Neck Cancers,” Clinical Chemistry and Laboratory Medicine, Vol. 46, No. 8, 2008, pp. 1090-1096.
- J. J. Hu, T. R. Smith, M. S. Miller, H. W. Mohrenweiser, A. Golden and L. D. Case, “Amino Acid Substitution Variants of APE1 and XRCC1 Genes Associated with Ionizing Radiation Sensitivity,” Carcinogenesis, Vol. 22, No. 6, 2001, pp. 917-922. doi:10.1093/carcin/22.6.917
- R. A. Nash, K. W. Caldecott, D. E. Barnes and T. Lindahl, “XRCC1 Protein Interacts with One of Two Distinct Forms of DNA Ligase III,” Biochemistry, Vol. 36, No. 17, 1997, pp. 5207-5211. doi:10.1021/bi962281m
- Y. Kubota, R. A. Nash, A. Klungland, P. Schar, D. E. Barnes and T. Lindahl, “Reconstitution of DNA Base Excision-Repair with Purified Human Proteins: Interaction between DNA Polymerase Beta and the XRCC1 Protein,” The EMBO Journal, Vol. 15, No. 23, 1996, pp. 6662-6670.
- M. Masson, C. Niedergang, V. Schreiber, S. Muller, J. Menissier de Murcia and G. de Murcia, “XRCC1 is Specifically Associated with Poly(ADP-Ribose) Polymerase and Negatively Regulates Its Activity Following DNA Damage,” Molecular and Cellular Biology, Vol. 18, No. 6, 1998, pp. 3563-3571.
- G. Matullo, D. Palli, M. Peluso, S. Guarrera, S. Carturan, E. Celentano, V. Krogh, A. Munnia, R. Tumino, S. Polidoro, A. Piazza and P. Vineis, “XRCC1, XRCC3, XPD Gene Polymorphisms, Smoking and (32)P-DNA Adducts in a Sample of Healthy Subjects,” Carcinogenesis, Vol. 22, No. 9, 2001, pp. 1437-1445. doi:10.1093/carcin/22.9.1437
- N. Liu, J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones and L. H. Thompson, “XRCC2 and XRCC3, New Human Rad51- Family Members, Promote Chromosome Stability and Protect against DNA Cross-Links and Other Damages,” Molecular Cell, Vol. 1, No. 6, 1998, pp. 783-793. doi:10.1016/S1097-2765(00)80078-7
- S. L. Winsey, N. A. Haldar, H. P. Marsh, M. Bunce, S. E. Marshall, A. L. Harris, F. Wojnarowska and K. I. Welsh, “A Variant within the DNA Repair Gene XRCC3 is Associated with the Development of Melanoma Skin Cancer,” Cancer Research, Vol. 60, No. 20, 1998, pp. 5612-5616.
- R. Kanaar, J. H. Hoeijmakers and D. C. van Gent, “Molecular Mechanisms of DNA Double Strand Break Repair,” Trends in Cell Biology, Vol. 8, No. 12, 1998, pp. 483-489. doi:10.1016/S0962-8924(98)01383-X
- T. Sugimura, H. Kumimoto, I. Tohnai, T. Fukui, K. Matsuo, S. Tsurusako S, K. Mitsudo, M. Ueda, K. Tajima and K. Ishizaki, “Gene-Environment Interaction Involved in Oral Carcinogenesis: Molecular Epidemiological Study for Metabolic and DNA Repair Gene Polymorphisms,” Journal of Oral Pathology & Medicine, Vol. 35, No. 1, 2006, pp. 11-18. doi:10.1111/j.1600-0714.2005.00364.x
- S. Z. Abdel-Rahman, A. S. Soliman, M. L. Bondy, S. Omar, S. A. El-Badawy, H. M. Khaled, I. A. Seifeldin and B. Levin, “Inheritance of the 194Trp and the 399Gln Variant Alleles of the DNA Repair Gene XRCC1 Are Associated with Increased Risk of Early-Onset Colorectal Carcinoma in Egypt,” Cancer Letters, Vol. 159, No. 1, 2000, pp. 79-86. doi:10.1016/S0304-3835(00)00537-1
- U. Carstensen, A. K. Alexandrie, D. T. B. Hogste, A. Rannug, I. Bratt and L. Hagmar, “Band T-Lymphocyte Micronuclei in Chimney with Respect to Genetic Polymorphism for CYP1A1 and GSTT1 (Class Mu),” Mutation Research, Vol. 289, No. 2, 1993, pp. 178-195. doi:10.1016/0027-5107(93)90069-R
- S. Kato, P. G. Shields, N. E. Caporaso, R. N. Hoover, B. F. Trump, H. Sugimura, A. Weston and C. C. Harris, “Cytochrome P450IIE1 Genetic Polymorphisms, Racial Variation, and Lung Cancer Risk,” Cancer Research, Vol. 52, No. 23, 1992, pp. 6712-6715.
- S. Z. Abdel-Rahman, R. A. El-Zein, W. A. Anwar and W. W. Au, “A Multiplex PCR Procedure for Polymorphic Analysis of GSTM1 and GSTT1 Genes in Population Studies,” Cancer Letters, Vol. 107, No. 2, 1996, pp. 229-233. doi:10.1016/0304-3835(96)04832-X
- A. Agresti, “A Survey of Exact Inference for Contingency Tables,” Statistical Science, Vol. 7, No. 1, 1992, pp. 131-153. doi:10.1016/0304-3835(96)04832-X
- N. E. Breslow and N. E. Day, “Statistical Methods in Cancer Research,” In: N. E. Breslow and N. E. Day, Eds., The Analysis of Case-Control Studies, International Agency for Research on Cancer, Lyon, 1980, IARC Scientific Publications No. 32.
- S. M. N. Garcia, O. A. Curioni, M. B. Carvalho and G. J. F. Gattás, “Polymorphisms in Alcohol Metabolizing Genes and the Risk of Head and Neck Cancer in a Brazilian Population,” Alcohol & Alcoholism, Vol. 45, No. 1, 2010, pp. 6-12. doi:10.1093/alcalc/agp078
- E. Negri, P. Boffetta, J. Berthiller, X. Castellsague, M. P. Curado, L. Dal Maso, et al., “Family History of Cancer: Pooled Analysis in the International Head and Neck Cancer Epidemiology Consortium,” International Journal of Cancer, Vol. 124, No. 2, 2009, pp. 394-401. doi:10.1002/ijc.23848
- E. S. Peters, M. D. McClean, C. J. Marsit, B. Luckett and K. T. Kelsey, “Glutathione S-Transferase Polymorphisms and the Synergy of Alcohol and Tobacco in Oral, Pharyngeal, and Laryngeal Carcinoma,” Cancer Epidemiology, Biomarkers & Prevention, Vol. 15, No. 11, 2006, pp. 2196-2202. doi:10.1158/1055-9965.EPI-06-0503
- M. Hashibe, P. Brennan, R. C. Strange, R. Bhisey, I. Cascorbi, P. Lazarus, M. B. O. Ophuis, S. Benhamou, W. D. Foulkes, T. Katoh, C. Coutelle, M. Romkes, L. Gaspari, E. Taioli and P. Boffeta, “Metaand Pooled Analyses of GSTM1, GSTT1, GSTP1, and CYP1A1 Genotypes and Risk of Head and Neck Cancer,” Cancer Epidemiology, Biomarkers & Prevention, Vol. 12, No. 12, 2003, pp. 1509-1517.
- D. Konig-Greger, H. Riechelmann, U. Wittich and S. Gronau, “Genotype and Phenotype of Glutathione-STransferase in Patients with Head and Neck Carcinoma,” Otolaryngology—Head and Neck Surgery, Vol. 130, No. 6, 2004, pp. 718-725.
- A. J. Evans, W. D. Henner, K. M. Eilers, M. A. Montalto, E. M. Wersinger, P. E. Andersen, J. I. Cohen, E. C. Everts, J. E. McWilliams and T. M. Beer, “Polymorphisms of GSTT1 and Related Genes in Head and Neck Cancer Risk,” Head & Neck, Vol. 26, No. 1, 2004, pp. 63-70. doi:10.1002/hed.10342
- N. M. Cury, A. Russo, A. L. S. Galbiatti, M. A. T. Ruiz, L. S. Raposo, J. V. Maniglia, E. C. Pavarino and E. N. Goloni-Bertollo, “Polymorphisms of the CYP1A1 and CYP2E1 Genes in Head and Neck Squamous Cell Carcinoma Risk,” Molecular Biology Reports, Vol. 39, No. 2, 2012, pp. 1055-1063. doi:10.1007/s11033-011-0831-1
- S. Ramachandran, K. Ramadas, R. Hariharan, R. Rejnish Kumar and M. Radhakrishna Pillai, “Single Nucleotide Polymorphisms of DNA Repair Genes XRCC1 and XPD and Its Molecular Mapping in Indian Oral Cancer,” Oral Oncology, Vol. 42, No. 4, 2006, pp. 350-362. doi:10.1007/s11033-011-0831-1
- D. Butkiewicz, M. Rusin, L. Enewold, P. G. Shields, M. Chorazy and C. C. Harris, “Genetic Polymorphisms in DNA Repair Genes and Risk of Lung Cancer,” Carcinogenesis, Vol. 22, No. 4, 2001, pp. 593-597. doi:10.1093/carcin/22.4.593
- P. Chacko, B. Rajan, T. Joseph, B. S. Mathew and M. R. Pillai, “Polymorphisms in DNA Repair Gene XRCC1 and Increased Genetic Susceptibility to Breast Cancer,” Breast Cancer Research and Treatment, Vol. 89, No. 1, 2005, pp. 15-21. doi:10.1007/s10549-004-1004-x
- G. J. F. Gattás, M. B. Carvalho, M. S. Siraque, O. A. Curioni, P. Kohler, J. Eluf-Neto and V. Wunsch-Filho, “Genetic Polymorphisms of CYP1A1, CYP2E1, GSTM1, and GSTT1 Associated with Head and Neck Cancer,” Head & Neck, Vol. 28, No. 9, 2006, pp. 819-826. doi:10.1007/s10549-004-1004-x
- L. Varela-Lema, E. Taioli, A. Ruano-Ravina, J. M. Barros-Dios, D. Anantharaman, S. Benhamou, S. Boccia, R. A. Bhisey, G. Cadonu, E. Capoluogos, C.-J. Chen, W. D. Foulkes, E. N. Goldoni-Bertollo, A. Hatagima, R. B. Hayes, T. Katoh, S. Koifman, P. Lazarus, J. J. Manni, M. Mahimkar, S. Morita, J. Park, K.-K. Park, E. C. P. Bertelli, E. M. S. Ribeiro, B. Roy, M. R. Spitz, R. C. Strange, O. Wei and C. C. Ragin, “Meta-Analysis and Pooled Analysis of GSTM1 and CYP1A1 Polymorphisms and Oral and Pharyngeal Cancers: A HuGE-GSEC Review,” Genetics in Medicine, Vol. 10, No. 6, 2008, pp. 369-384. doi:10.1097/GIM.0b013e3181770196
- D. Anantharaman, P. M. Chaubal, S. Kannan, R. A. Bhisey and M. B. Mahimkar, “Susceptibility to Oral Cancer by Genetic Polymorphisms at CYP1A1, GSTM1 and GSTT1 Loci among Indians: Tobacco Exposure as a Risk Modulator,” Carcinogenesis, Vol. 28, No. 7, 2007, pp. 1455-1462. doi:10.1093/carcin/bgm038
- M. B. Oude Ophuis, H. M. Roelofs, P. A. Van Den Brandt, W. H. Peters and J. J. Manni, “Polymorphisms of the Glutathione S-Transferase P1 Gene and Head and Neck Cancer Susceptibility,” Head & Neck, Vol. 25, No. 1, 2003, pp. 37-43. doi:10.1002/hed.10182
- J. E. McWilliams, A. J. Evans, T. M. Beer, P. E. Andersen, J. I. Cohen and E. C. Everts, “Genetic Polymorphisms in Head and Neck Cancer Risk,” Head & Neck, Vol. 22, No. 6, 2000, pp. 609-617. doi:10.1002/1097-0347(200009)22:6<609::AID-HED10>3.0.CO;2-L
- S. Landi, “Mammalian Class Theta GST and Differential Susceptibility to Carcinogens: A Review,” Mutation Research, Vol. 463, No. 3, 2000, pp. 247-283. doi:10.1002/1097-0347(200009)22:6<609::AID-HED10>3.0.CO;2-L
NOTES
*Corresponding author.

