Advances in Bioscience and Biotechnology
Vol.2 No.5(2011), Article ID:7835,16 pages DOI:10.4236/abb.2011.25054
Doubling of crop yield through permutation of metabolic pathways
![]()
CARC, Prairie View A & M University, Prairie View, USA.
E-mail: *goosuji@pvamu.edu; cropyielddoublingbiotechnology@yahoo.com
Received 27 July 2011; revised 25 August 2011; accepted 13 September 2011.
Keywords: Peanut; Fatty Acid Bioreactor; Glutamate Dehydrogenase; Yield-Specific mRNAs
ABSTRACT
Hunger and food insecurity can be minimized by doubling crop yield without increasing cultivated land area and fertilizer applied. Since plant breeding has not genetically doubled photosynthesis per unit leaf area, an approach for doubling crop yield would be through a biotechnology that reprograms metabolic pathways in favor of photosynthesis. The anchor of this biotechnology is glutamate dehydrogenase (GDH) including the RNAs it synthesizes. Peanut was treated with stoichiometric combinations of mineral salt solutions to synchronize the GDH subunit polypeptides. Matured seeds were analyzed for fats by HPLC; the RNA biosynthetic activity of GDH, and mRNAs encoding yield-specific enzymes by Northern hybridization. In the PK-treated peanut, the GDH-synthesized RNAs silenced the mRNAs encoding granule-bound starch synthase, phosphoglucomutase (glycolysis), glucosyltransferase (cellulose biosynthesis), and nitrate reductase leaving unaffected the mRNAs encoding acetylcoenzyme A carboxylase (fatty acid biosynthesis), phosphate translocator, and NADH-glutamate synthase resulting to double seed (4342 kg/ha), cellulose (1829 kg/ha), and fat (1381 kg/ha) yields compared with the controls. Down-regulation of phosphate translocator and acetylcoenzyme A carboxylase caused decreased pod yields. GDH-synthesized RNAs that were homologous to yield-specific mRNAs shared extensive plus/plus and plus/minus sequence similarities, and they reprogrammed metabolism by permuting the partially down-regulated, not down-regulated, and down-regulated yield-specific pathways. Control peanut produced 70, NPKS-treated produced 420, NS-treated produced 1680, and PK-treated produced 280 probable rearrangements of the pathways. Therefore, down-regulation of metabolic reactions followed by permutation of yield-related pathways, and redistribution of metabolite load to molecularly connected pathways controls crop yield. Operating as efficient bioreactor, peanut can be maximized to 10,000 kg pod/ha, more than enough vegetable oil for nine billion people.
1. INTRODUCTION
Feeding a hungry humanity that will soon exceed nine billion people is a major challenge to governments and farmers, but an opportunity to the scientific community [1]. More than 850 million people worldwide lack the food they need [2] for normal growth, development and an active life. Prospects for a continuation of the food productivity growth seen between 1940 and 1980 [3,4] are hindered by climate change, land degradation, and strained water resources. Low crop yields lead to hunger and to high cost of food [5]. The short cut to minimize hunger, food insecurity, and cost of agricultural production is to double crop yield without increasing the cultivated land area, man hour input, fertilizer, pesticides, and water applied.
The metabolic pathways elegantly show that crop yield is controlled by many enzymes, but do not show pathway discrimination and integration for the doubling of crop yield. The biotechnological opportunity is to enhance those pathways that increase and to knock down those that hinder storage product accumulation. Much of such monogenic trait and polygenic control biotechnologies have been addressed with success through plant breeding, crop protection and plant genetic engineering approaches [6-11]. The biotechnological logic adopted hereunder was different being based on permutation of the metabolic pathways. Since plant breeding has not resulted in a genetic doubling of photosynthesis per unit leaf area [4], a direct approach for doubling crop yield would be through the manipulation of mineral ion-regulation of carbon and nitrogen intermediary metabolism [12], and photosynthesis. The advantage of this approach is that since there is complicated plethora of enzyme steps in carbon and nitrogen metabolism, the permutations of the variable enzyme activities leading to maximized storage product accumulation are virtually unlimited. The basis of the yield-maximizing biotechnology is that mineral ions [12-14] acting as electromagnets, will alter the oxidative state in the plant cells [15-17] thereby reprogramming the activities of those enzyme steps that regulate and/or coordinate metabolite load redistribution throughout intermediary metabolism with consequent doubling of respective metabolic end-products. The anchor of the biotechnology is glutamate dehydrogenase (GDH) including the RNAs it synthesizes which constitute the new gateway into biology. GDH charge isomers constitute a special electric field in the cell [18]. They are induced to isomerize and to synthesize some RNAs when they interface with electromagnets [19]. The RNAs so synthesized interfere with the translation of their homologous mRNAs [20]. GDH isomerization and the resultant RNAs have been applied to increase yields in soybeans, corn, alfalfa, peanut etc. [21,22]. But the ability of the GDH-synthesized RNAs to permute metabolic pathways in order to maximize and double crop yield was not described.
Peanut was chosen for this study because it is a universal food, fiber and oil crop grown on about 42 million acres worldwide, with India, China, USA, and Nigeria producing about 80% of the world crop [23]. The oil is being favorably evaluated for the manufacture of biodiesel. Peanut is ideal for this study because the responses of its yield to applied sulfate, potash, nitrogen, and phosphate fertilizers [23-27] are so complicated and conflicting that they have defied biochemical explanation. Metabolic pathway permutation may illuminate the bioscience of mineral ion reactivity.
In the yield-doubling biotechnology presented hereunder, peanut carbohydrate and fatty acid metabolic pathways acted in concert with the pathways for nitrogen assimilation through their permutation sequentially and spatially by the GDH-synthesized RNAs as influenced by mineral ions to produce record high quantities of fat and cellulose.
2. EXPERIMENTAL PROCEDURES
Treatment of peanuts with mineral ion solutions: Peanut (Arachis hypogaea L. Cv. Virginia) seeds were planted in 243.84 × 243.84 × 30.48 cm (width × length × depth) boxes, each filled with 18 bags of Metro mix 700 peat moss. Each box was set up on level ground in the field on Dewitt earth mat, a permeable weed-blocking fabric. About 100 - 110 seeds were planted per box. There was replanting to make up for ungerminated seeds. The applied mineral ion compositions were based on the model com-binations in Table 1 to mimick the binomial subunit poly
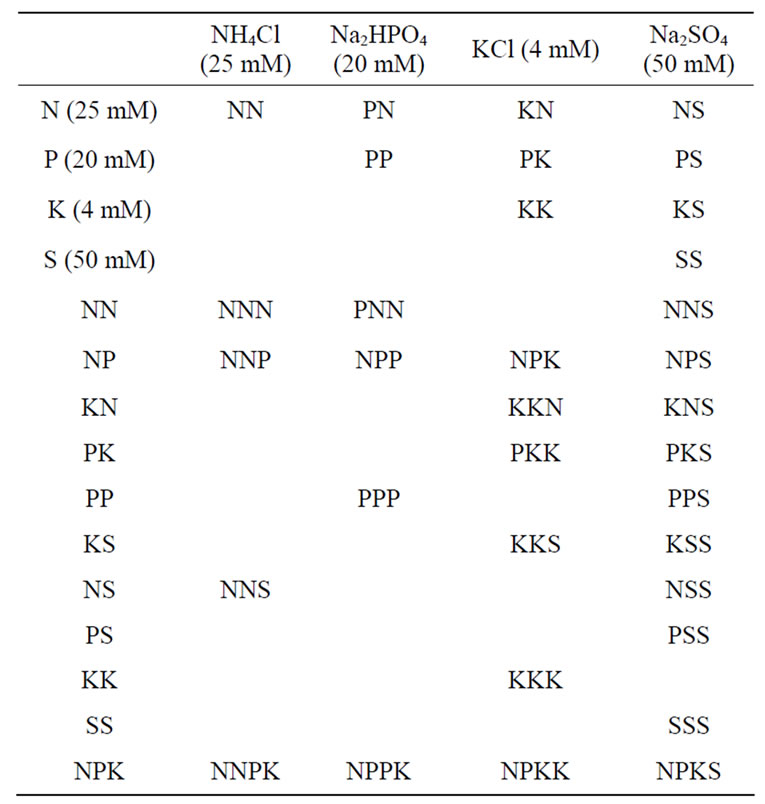
Table 1. Some mineral salt combinations mimicking peanut GDH subunit compositions.
peptide compositions of the GDH isoenzymes [22]. The first box was left as the untreated control; the second box (N) was treated with 1 L of NH4Cl solution (25 mM), the third box (Pi) was treated with 1 L of Na3PO4 solution (20 mM); the fourth box (S) was treated with 1 L of Na2SO4 solution (50 mM); the fifth box (K) was treated with 1 L KCl solution (4 mM); the sixth box (NPKS) was treated with 1 L of combined NH4Cl (25 mM), Na3PO4 (20 mM), Na2SO4 (50 mM), and KCl (4 mM) solution; the seventh box (PK) was treated with 1 L of combined Na3PO4 (20 mM) and KCl (4 mM) solution; the eighth box (NS) was treated with 1 L of combined NH4Cl (25 mM) and Na2SO4 (50 mM) solution; the ninth box (PN) was treated with 1 L of combined Na3PO4 (20 mM) and NH4Cl (25 mM) solution; the tenth box (PS) was treated with 1 L of combined Na3PO4 (20 mM) and Na2SO4 (50 mM) solution. The boxes were watered every other day. Mineral nutrient solutions were applied sequentially, first at pre-flowering stage (2 weeks after seed germination), second at flowering, and third at post-flowering. When the leaves turned yellow (peanut maturity), pods were harvested, allowed to dry on the greenhouse floor for about 2 weeks, weighed, shelled by hand, and the kernels (seeds) and shells weighed separately. Seeds were stored at –30˚C.
Analyses for peanut yield: Dry and milled (composited) seeds (100 g) per experimental treatment, sent to SGS North America Inc., St. Rose, Louisiana, USA were custom analyzed by HPLC for percent oil weight. Milled shells (250 g) per experimental treatment, sent to Universal Testing, Quincy, IL, USA were custom analyzed for dry matter, and cellulose (neutral detergent fiber, and acid detergent fiber) using standard gravimetric method.
Purification and assay of GDH: GDH charge isomers were purified by electrophoresis as described before [17] from peanut seeds harvested from the control or mineral ion-treated boxes. RNA synthetic activity of GDH isoenzymes [22] was assayed in combined deamination and amination substrate solutions of 0.1 M Tris-HCl buffer (pH 8.0) containing the four NTPs (0.6 mM each), CaCl2 (3.5 mM), L-glu (3.23 µM), NAD+ (0.375 µM), NH4Cl (0.875 mM), α-ketoglutarate (10.0 mM), NADH (0.225 mM), 5 Units RNase inhibitor, 1 Unit DNase 1, and 5 µg of actinomycin D. Reaction was started by adding 0.2 mL of whole gel-eluted GDH charge isomers containing 3 - 9 µg protein per mL. Final volume of the reaction was brought to 0.4 mL with 0.1M Tris-HCl buffer pH 8.0. Reactions were incubated at 16˚C overnight and stopped by phenol-chloroform (pH 5.5) extraction of the enzyme. RNA was precipitated with ethanol, and dissolved in minimum volume of molecular biology quality water. RNA yield and quality were determined by photometry and by agarose gel electrophoresis. Assays were carried out in duplicate to verify the reproducibility of the results.
Total RNA: Total RNA was extracted from peanut seeds harvested from the control or mineral-treated boxes using the acidic phenol/chloroform (pH 4.5) method [28].
cDNA synthesis, cloning, characterization, and probe selection: cDNAs were synthesized with 2 µg of each product RNA synthesized by the whole gel-eluted GDH charge isomers using random hexamer primer. Restriction fragment PCR amplification; adapter ligation; sequencing gel fractionation; and purification of cDNA fragments [22] were conducted according to the methods of Display Systems Biotech, Vista, CA, USA. Selected cDNA fragments were subcloned into pCR4-TOPO vector and transformed into TOP10 One Shot Chemically Competent Escherichia coli (Invitrogen, Carlsbad, CA), followed by overnight growth on selective plates. Up to ten positive transformant colonies were picked per plate and cultured overnight in LB medium containing 50 µg/mL of kanamycin. Plasmid DNA was purified with a plasmid kit (Novagen, Madison, WI). The insert cDNA was sequenced with T3 and T7 primers by Genemed Synthesis, Inc. (South San Francisco, CA, USA), and Functional Biosciences, Inc. (Madison, WI, USA). To identify the GDH-synthesized RNAs that were homologous to mRNAs encoding the crop yield-related enzymes, the cDNA sequences were used as queries to search the NCBI nucleotide-nucleotide (excluding ESTs) BLAST (blastn), and non-redundant protein translation (blastx) databases. cDNAs that displayed the highest alignment scores with mRNAs encoding the enzymes of photosynthesis, nitrogen assimilation, glycolysis, cellulose, fatty acid, and nucleotide biosyntheses [20,29,30] were selected as the probes.
Northern blot analysis: Equal amounts (10 µg) of total RNA, and RNAs synthesized by GDH charge isomers from the control and mineral nutrients-treated peanuts were loaded, briefly electrophoresed on 2% agarose gels, stained with ethidium bromide, and photographed to verify RNA quality. RNA was electro-transferred from the electrophoresed gel onto Brightstar-Plus nylon membrane (Applied Biosystems, Foster City, CA, USA) as described before [30].
The cDNAs that were used as Northern probes were those homologous to mRNAs encoding phosphate translocator, granule-bound starch synthase (GBSS), phosphoglucomutase (PGM), glucosyltransferase, acetyl CoA carboxylase (ACCase), NADH-glutamate synthase (GOGAT), nitrate reductase (NR), glycinamide ribonucleotide (GAR) synthetase/GAR transformylase. For the labeling of the cDNA probes, cDNA inserts were amplified by PCR from the corresponding plasmids (15 ng) using M13 forward and M13 reverse primers (2 µM each), [32P]-dATP (6000 Ci/mmol, 20 mCi/mL), dCTP/ dGTP/TTP mix 50 mM, (2 µL), and Taq polymerase (1U), in a final volume of 50 µL. Amplification was according to Display Systems Biotech (Vista, CA, USA) “touch-down” PCR procedure (denature: 94˚C, 1 min for the first 10 cycles: 94˚C, 30 sec; anneal: 60˚C, 30 sec for the first cycle, then reduced the temperature 0.5˚C each cycle until an annealing temperature of 55˚C was reached after 10 cycles; extension: 72˚C, 1 min. Continued another 25 cycles with 94˚C, 30 sec; 55˚C, 30 sec; 72˚C, 1 min; final extension 72˚C, 5 min). Nylon membranes with immobilized RNA were prehybridized with ULTRAhyb buffer and hybridized with 32P-labeled cDNA inserts as probes overnight at 68˚C as described before [30]. Solutions of labeled cDNA were first heated in boiling water bath for 10 min before adding to the prehybridized membrane. After hybridization, the membranes were washed (30 min, 68˚C) with NorthernMax (Applied Biosystems, Foster City, CA, USA) low stringency wash solution followed by NorthernMax high stringency wash solution (30 min, 68˚C). The membrane was autoradiographed by exposure to X-ray film within intensifying screens at –80˚C. Northern band intensities were digitalized using UN-SCAN-IT gel digitalizing software (Silk Scientific, Inc., Orem, Utah, USA).
3. RESULTS
1) Mineral ion treatments: as was done previously [15, 18,22], mineral ion compositions were formulated to mimic the stoichiometric ratios of, and to synchronize the subunit polypeptide compositions of GDH (Table 1). This comprehensive rainbow of minerals made for internal repeats in the mineral ion compositions thus limiting stochastic variability in peanut plot treatments, imposed a firm control on the number of experimental repeats, and consolidated the biochemical comprehensiveness of the project design [30]. These conditions assured the most efficient production metabolism of the peanuts as natural bioreactor through synchronization in the isomerization of GDH, synthesis of RNA by GDH, silencing of mRNAs by homologous RNAs synthesized by GDH, structural and spatial integration of the yield-related metabolic pathways leading at crop maturity to the doubling of pod yield.
2) Mineral ions, GDH isomerization and RNA synthesis: isoelectric focusing of GDH followed by native polyacrylamide gel electrophoresis fractionated the charge isomers to their isoenzymes. The isoenzymes focused in almost the same 8 - 9 Rotofor chambers (Figures 1(a)-(k)) irrespective of mineral treatments thus confirming process reproducibility and the firm control imposed on random variability in treatments and experiments. The GDH isoenzyme population patterns were also generally in agreement with the expected binomial distribution of the three subunits in the hexameric isoenzymes, on the basis of the twin nonallelic GDH1 and GDH2 gene structure, with the gene (GDH1) encoding the more acidic subunits (a and α) being heterozygous and codominant, whereas the other gene (GDH2) encoding the less acidic subunit (β) is homozygous [31]. Although the isoenzyme distribution patterns appeared to be identical, the RNAs that the isoenzyme population synthesized were dramatically different. The abundant RNA bands in the arrays of GDH-synthesized RNAs were the 4.5 kb, 2 kb, and 0.1 kb bands [22]. There was a fourth one of very high molecular weight (>10 kb). The proportions of the four bands differed for each mineral-treated peanut (Figures 1(i)-(x)). The differences in the GDH isoenzyme populations and in the RNAs that the isoenzymes synthesized constituted the molecular basis for the yield doubling biotechnology. The GDH isoenzyme population taken together with the array of RNA synthesized per mineral regiment (Figure 1) described the biochemical comprehensiveness of the research approach.
3) Northern hybridization: the Northern hybridization results described the molecular mechanism of metabolic pathway permutation. The Northern blots determined the population distribution of probe per array of GDH-synthesized RNA, the threshold quantities of the probes per GDH-synthesized RNA, the mRNAs that were homologous to the probes, GDH-synthesized RNA present in total RNA; and confirmed absence of total RNA from GDH-synthesized RNA preparations. The control peanut without mineral ion treatment was the negative control; the NPKS-treated peanut with the full mineral ion composition and concentration was the positive control for verification of the bioscience of silencing by GDH-synthesized RNA [29]. The probes (Table 2) were cDNAs of the GDH-synthesized RNAs that were homologous to mRNAs. Some of the cDNA probes (for the mRNAs encoding Pi translocator, glucosyltransferase, and nitrate reductase) have been reported before [15,22]. Two probes (3E, and 48Q) were used for detection/quantitation of the GDH-synthesized RNA and mRNAs encoding glycinamide ribonucleotide (GAR) synthetase/(GAR) transformylase (Table 2). The two probes targeted different sequences in the mRNAs encoding GARS/GART.
In many respects, each Northern blot population of bands was a reflection of the distribution of the GDHsynthesized RNA bands on the agarose gel landscape (Figures 1(i)-(x)). The tenacious binding of the complimentary strands of the probes to their targets in mRNAs, and GDH-synthesized RNAs showed that the GDHsynthesized RNAs were very specific in their silencing of mRNAs. The Northern band population (Figure 2) for the GDH-synthesized RNAs of the control peanut, showed that the probes for the mRNAs encoding Pi translocator, NR, PGM, and GARS/GART hybridized to the 4.5 kb, and 2 kb RNA bands, without hybridizing to the >10 kb, and the 0.1 kb RNA bands (Figure 1(i)). Residual quantities of GDH-synthesized RNAs, as well as of the mRNAs encoding Pi translocator, GARS/ GART, NR, or PGM were not detected in the respective total RNA lanes. This suggested that the GDH-synthesized RNAs knocked out their homologous mRNAs. Northern assays for the mRNAs encoding GBSS, glucosyltransferase, ACCase, and NADH-GOGAT gave no bands meaning that the GDH of the control peanut discriminated by not synthesizing RNAs that were homologous to the four mRNAs.
The Northern band population for the GDH-synthesized RNAs of the NPKS-treated peanut, showed that the probes for the mRNAs encoding Pi translocator, PGM, glucosyltransferase, GARS/GART, and NR hybridized to the high molecular weight (>10 kb) RNA bands (Figure 1(ii)), whereas the probe for the mRNA encoding GBSS hybridized to the 4.5 kb and 2 kb GDH-synthesized RNA bands. Residual quantities of GDH-synthesized high molecular weight (>10 kb) RNAs, as well as the mRNAs encoding Pi translocator, and GARS/GART were detected in the Northern blots in the corresponding total RNA lanes. This suggested that in those cases, the GDHsynthesized RNAs only knocked down the homologous mRNAs. But the probes for the mRNAs encoding GBSS, PGM, and glucosyltransferase did not detect residual mRNA in the total RNA lanes. Northern assays for the

Figure 1. Responses of GDH isoenzyme (a)-(k), and RNA arrays (i)-(x) synthesized by the isoenzymes to mineral treatments of peanut. M is RNA molecular weight marker; t is total RNA.

Table 2. cDNA sequences of peanut GDH-synthesized RNAs used as probes. Bold and underlined sequences indicate internally repeated sequences.
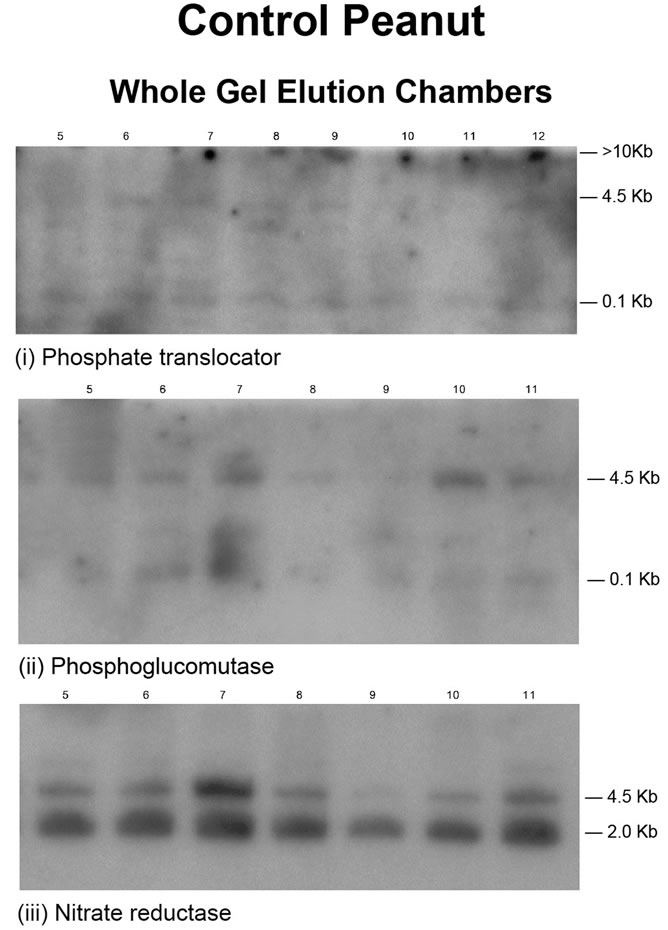
Figure 2. Population distribution of Northern bands for the RNAs synthesized by the GDH charge isomers of control peanut. Total RNA from control peanut was the marker. Probes were cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated yield-specific enzymes (Figure 2(i)-(iii)).
mRNAs encoding ACCase, and NADH-GOGAT gave no bands meaning that the GDH of NPKS-treated peanut discriminated by not synthesizing RNAs that were homologous to the two mRNAs.
The Northern band populations for the GDH-synthesized RNAs of the NS-treated peanut showed that the probes for the mRNAs encoding PGM, ACCase, and glucosyltransferase hybridized to the >10 kb RNA bands, whereas the probe for the mRNA encoding NR hybridized to the 4.5 kb and 2 kb GDH-synthesized RNA bands in agreement with the population pattern of the GDH-synthesized RNA (Figure 1(iii)). Residual quantities of GDH-synthesized 4.5 kb, and 2 kb RNAs, as well as 0.1 kb mRNA fragments were detected in the Northern blots in the respective total RNA lanes. Northern assays for the mRNAs encoding Pi translocator, GBSS, GARS/GART, and NADH-GOGAT gave no bands meaning that the GDH of the NS-treated peanut exercised discrimination by not synthesizing RNAs that were homologous to the four mRNAs.
The Northern band population patterns (Figure 3) for the GDH-synthesized RNAs of the Pi-treated peanut, showed that the probes for the mRNAs encoding PGM, glucosyltransferase, ACCase, GARS/GART, and NADHGOGAT hybridized to the >10 kb RNA bands. There were no 4.5 kb and 2 kb Northern bands in agreement with the RNA population of the GDH-synthesized RNA (Figure 1(iv)). Residual quantities of GDH-synthesized high molecular weight (>10 kb) RNAs, as well as the mRNAs encoding PGM, glucosyltransferase, and ACCase were detected in the Northern blots in the respective total RNA lanes. This suggested that in those cases, the GDH-synthesized RNAs only knocked down the homologous mRNAs. The probes for the mRNAs encoding GARS/GART, and NADH-GOGAT did not detect residual mRNA in the total RNA lanes (Figure 3). Northern assays for the mRNAs encoding Pi translocator, GBSS,

Figure 3. Population distribution of Northern bands for the RNAs synthesized by the GDH charge isomers of phosphate-treated peanut. Total RNA from phosphate-treated peanut was the marker. Probes were cDNAs of the GDH-synthesized RNAs that were homologous to the mRNAs encoding the indicated yield-specific enzymes (Figures 3(i)-(v)).
and NR gave no bands meaning that the GDH of the Pitreated peanut discriminated by not synthesizing RNAs that were homologous to the three mRNAs.
Visually, the ten treated peanuts grew excellently and the leaves were green to the same extent. There were no symptoms of mineral nutrient deficiency. However, comparison of the Northern blots for the control and Pitreated peanuts (Figures 2 and 3) give a clearer molecular differentiation. Whereas Figure 2 depicts the control peanut as a miserable low quality crop whose metabolism was uncoordinated, Figure 3 depicts the Pi-treated peanut as a robust crop whose primary metabolic pathways were under efficient coordination. These differences at the molecular level were confirmed by the different vegetable oil and cellulose yields. Therefore, Northern assays using GDH-synthesized RNAs as probes are good pictorial demonstrations of the efficiency of metabolic processes of crops.
The Northern band populations for the GDH-synthesized RNAs of the KCl-treated peanut showed that the probes for the mRNAs encoding glucosyltransferase, NR, and Pi translocator hybridized to the >10 kb, 4.5 kb, and 2 kb RNA bands (Figure 1(v)). Residual quantities of GDH-synthesized high molecular weight RNA bands were detected in the Northern blots in the corresponding total RNA lanes. Northern assays for the mRNAs encoding GBSS, PGM, ACCase, GARS/GART, and NADHGOGAT gave no bands meaning that the GDH of the KCl-treated peanut did not synthesize RNAs that were homologous to the five mRNAs.
The Northern band populations for the GDH-synthesized RNAs of the sulfate-treated peanut showed that the probe for the mRNA encoding Pi translocator hybridized to the >10 kb RNA bands (Figure 1(vi)), whereas the probe for the mRNA encoding NR hybridized to the 4.5 kb and >10 kb GDH-synthesized RNA bands. Residual quantities of GDH-synthesized high molecular weight RNA bands, as well as the mRNAs encoding Pi translocator, and NR were detected in the Northern blots in the respective total RNA lanes. The probe for the mRNA encoding GARS/GART hybridized to the >10 kb, and low molecular weight (0.1 kb) RNA bands to give a population of Northern bands that was identical to the pattern of GDH-synthesized RNA bands (Figure 1(vi)). Northern assays for the mRNAs encoding GBSS, PGM, glucosyltransferase, ACCase, and NADH-GOGAT gave no bands meaning that the GDH of the sulfate-treated peanut did not synthesize RNAs that were homologous to the five mRNAs.
The Northern bands for the GDH-synthesized RNAs of the PS-treated peanut showed that the probes for the mRNAs encoding Pi translocator, PGM, GBSS, glucosyltransferase, and GARS/GART hybridized to the >10 kb RNA bands (Figure 1(vii)); and in addition the probe for the mRNA encoding GARS/GART hybridized to the 0.1 kb GDH-synthesized RNA bands. Northern assays for the mRNA encoding ACCase gave no bands meaning that the GDH of the PS-treated peanut exercised discrimination by not synthesizing RNA that was homologous to the mRNA. Residual quantities of GDH-synthesized >10 kb RNA bands, as well as of mRNAs were detected in the Northern blots in the respective total RNA lanes. There were no 4.5 kb Northern bands just as there were no 4.5 kb RNA bands in the array of the RNAs that were synthesized by the GDH of PS-treated peanut (Figure 1(vii)).
The Northern band populations for the GDH-synthesized RNAs of the NH4Cl-treated peanut showed that the probes for the mRNAs encoding Pi translocator, GBSS, PGM, and glucosyltransferase hybridized to the >10 kb RNA bands (Figure 1(viii)), and there were Northern smears similar to the RNA smears observed in the array of RNAs synthesized by the GDH of NH4Cl-treated peanut (Figure 1(viii)). Residual quantities of GDHsynthesized high molecular weight RNA bands, as well as the mRNAs encoding Pi translocator, and GBSS were detected in the Northern blots in the corresponding total RNA lanes. Northern assays for the mRNAs encoding ACCase, GARS/GART, NR, and NADH-GOGAT gave no bands meaning that the GDH of the NH4Cl-treated peanut did not synthesize RNAs that were homologous to the four mRNAs.
The Northern bands for the GDH-synthesized RNAs of the PN-treated peanut showed that the probes for the mRNAs encoding Pi translocator, GBSS, PGM, and glucosyltransferase hybridized to the >10 kb RNA bands (Figure 1(ix)). In contrast to the RNA band pattern (Figure 1(ix)), there were no 4.5 kb, 2 kb, and 0.1 kb Northern bands. Residual quantities of GDH-synthesized high molecular weight RNA bands, as well as the mRNAs encoding Pi translocator, GBSS, and PGM were detected in the Northern blots in the corresponding total RNA lanes. Northern assays for the mRNAs encoding GARS/GART, NR, and NADH-GOGAT gave no bands meaning that the GDH of the PN-treated peanut exercised discrimination by not synthesizing RNAs that were homologous to the three mRNAs.
The Northern band populations for the GDH-synthesized RNAs of the PK-treated peanut showed that the probes for the mRNAs encoding GBSS, PGM, glucosyltransferase, and GARS/GART hybridized to the >10 kb RNA bands (Figure 1(x)), whereas the probe for the mRNA encoding NR hybridized to all the GDH-synthesized RNA bands. The Northern result for the nitrate reductase-encoded probe was remarkable in that all the GDH-synthesized RNA had sequences that were homologous to the probe. Residual quantities of GDH-synthesized high molecular weight RNA bands were present only in the total RNA lane of the GARS/GART Northern result. Northern assays for the mRNAs encoding Pi translocator, and NADH-GOGAT gave no bands meaning that the GDH of the PK-treated peanut did not synthesize RNA that was homologous to the four mRNAs.
Digital (UN-SCAN-IT) quantitation of the Northern bands showed that GDH synthesized the required RNA sequences in sufficient quantities to meet the thresholds for silencing the target homologous mRNAs. A minimum 2:1 ratio between the GDH-synthesized RNA to the target mRNA was the requisite threshold for the silencing reaction. The ratio was the baseline control platform applied for unbiased judgment of the Northern bands across experimental repeats and mineral ion treatments. The Northern analyses in which total RNAs were loaded side-by-side to the GDH synthesized RNAs (Figures 1(i)-(x)) made for easier interpretation of the efficiency of transcript silencing by the GDH-synthesized RNAs. The definitive detection of the high molecular weight (>10 kb) RNAs synthesized by GDH in the total RNA lanes in the Northern hybridizations was an evidence that GDH synthesizes RNAs in vivo. The Northern results further highlighted the importance of the preponderant >10 kb GDH-synthesized RNA in the efficiency of mRNA silencing. The >10 kb GDH-synthesized RNA assured a maximum structural interaction with the homologous mRNAs, a crucial tertiary stabilization for the initiation of silencing [32].
4) Sequential permutation by GDH-synthesized RNA: the Northern blot results demonstrated a spider web-like molecular connectivity between photosynthesis, glycolysis, cellulose biosynthesis, nitrogen assimilation, and fatty acid biosynthesis in the peanut. This connectivity described metabolic pathway integration at the molecular level. The GDH-synthesized RNA that is homologous to the mRNA encoding GBSS shared two-fold plus/plus sequence similarity with that homologous to the mRNA encoding ACCase. The plus/plus sequence similarities between GDH-synthesized RNAs ensured an ordered variation of the abundances of homologous mRNAs. The GDH-synthesized RNA that is homologous to the mRNA encoding GBSS also shared six-fold plus/minus sequence similarities with the GDH-synthesized RNA (plasmid 3E) that is homologous to the mRNAs encoding GARS/GART; two-fold plus/plus sequence similarities with that homologous to the mRNA encoding NR; and two-fold plus/minus sequence similarities with that homologous to the mRNA encoding NADH-GOGAT. This is a reciprocal regulation of the mRNAs which are homologous to the same GDH-synthesized RNA so that at least one in the group of mRNAs could not be knocked out with the other three. Reciprocal regulation of glycolysis and nitrogen assimilation was further enforced by the GDH-synthesized RNA that is homologous to the mRNA encoding PGM. The probe shared ten-fold plus/ minus sequence similarities with the GDH-synthesized RNA (plasmid 3E) that is homologous to the mRNAs encoding GARS/GART; and two-fold plus/minus sequence similarities with that homologous to the mRNA encoding NADH-GOGAT. This sequential permutation meant that at least one of the mRNAs could not suffer demise with the other two. The GDH-synthesized RNA that is homologous to the mRNA encoding ACCase shared 22-fold plus/plus sequence similarity with the probe (plasmid 3E) that is homologous to the mRNAs encoding GARS/GART, but 7-fold plus/minus sequence similarities with those homologous to the mRNAs encoding NR and NADH-GOGAT. This spider web-like coordination by the GDH-synthesized RNAs that are homologous to the mRNAs encoding NADH-GOGAT, GARS/GART, and NR provided all-round metabolic pathway integration so that all three of them could not suffer demise with the mRNA encoding ACCase under any applied mineral ion regimen. This explains the robust capability of peanut to produce oil even on acid sandy, infertile, droughty soil [33]. The GDH-synthesized RNA that is homologous to the mRNAs encoding the GARS/GART shared 6-fold plus/plus sequence similarities with that homologous to the mRNAs encoding NR; and 16-fold plus/plus similarity with that homologous to the mRNA encoding NADH-GOGAT. This structural integration meant that all the three mRNAs could not be knocked out simultaneously. These were important structural provisions that prevented the total shut-down of the pathways for nitrogen assimilation while metabolites were preferentially channeled towards saccharide and fatty acid metabolism. The cob web-like cross connections between metabolic pathways at the molecular level introduce the concept of probability or likelihood for metabolites to flow in one or alternative directions in peanuts treated with mineral ions.
5) Spatial permutation by GDH-synthesized RNAs: metabolic pathway integration at the molecular level was consolidated by statistical redistribution of metabolite load to molecularly connected pathways. GDH-synthesized RNAs are statistical in nature, their primary structure being dependent on the binomial subunit compositions of the GDH isoenzymes [30]. Therefore several probabilities arise under a prevailing mineral nutrient combination/concentration for the GDH-synthesized RNA to reprogram the mRNA abundances and consequently the rates of yield production. The reprogramming of the sequence of metabolism in response to the prevailing mineral ions in order to maximize crop yield can best be approached by permuting the yield-specific pathways guided by the enzyme systems that were not down-regulated, partially down-regulated, and fully down-regulated by GDH-synthesized RNAs per mineral environment, and limited to the eight metabolic pathways affected. The number of permutations varied widely: control peanut produced 70; NPKS-treated peanut produced 420; NStreated peanut produced 1680; Pi-treated peanut produced 560; PN, PK, and NH4Cl-treated peanuts, each produced 280; Na2SO4, PS, and KCl-treated peanuts, each produced 168 permutations. Therefore, there were many probable rearrangements of peanut metabolic pathways in response to any mineral ion regimen. This is the explanation for the contradictions and conflicts about the peanut yield responses to applied mineral nutrients [34- 37]. Frequency distribution plot of the metabolic pathways versus the number of permutations gave a positively skewed graph reminiscent of the GDH binomial isoenzyme population distribution pattern [22]. The mode (224) of the frequency distribution plot had a frequency of 4 suggesting that the yield of peanut could be increased further about 50% of the yield of the PKtreated peanut to about 10,000 kg·pod·ha–1 by other mineral nutrient treatments. With a doubled peanut yield of 10,000 kg·ha–1 and assuming about 50% oil content, 70% weight of peanut pod in the shell [33], peanut will have the potential to yield 880 - 1100 gallons of biodiesel·ha–1, which will be a remarkable contribution to bioenergy supply without cultivation of more land, and without application of more fertilizer and pesticides. There are many stoichiometric combinations of mineral nutrients for this purpose (Table 1).
6) Yield biochemistry of peanut: phosphate translocator: peanut photosynthetic reduction of CO2 produces triose phosphates, the key compounds used in starch synthesis in the chloroplast; or after leaving the chloroplasts to cytosol, in the sucrose synthesis in cytosol [12]. Pi translocator exports triose phosphates from the chloroplast to the cytosol in exchange for Pi import from the cytosol into the chloroplast [38]. Therefore, in PKtreated peanut where the mRNA encoding Pi translocator was not down-regulated by GDH-synthesized RNA, triose phosphates were maximally metabolized to acetyl CoA and channeled towards fatty acid biosynthesis thus resulting to maximum yields of seeds and pods (Figure 4). Conversely, in control peanut (Figure 2) where the mRNA encoding Pi translocator was down-regulated by GDH-synthesized RNA, Pi and triose phosphates were minimally mobilized with consequent decreased biosynthesis of fatty acids, and minimum yields of seeds, and pods (Figure 4).
Starch synthase: granule-bound starch synthase 1 (GBSS1) is responsible for the synthesis of linear glucan (amylase) in the starch granule [39], whereas another isoform, GBSSII is important in the synthesis of the branched (amylopectin) component [40]. In the peanut chloroplast, the export of triose phosphates to the cytosol by Pi translocator competed with their utilization in the chloroplast for starch biosynthesis. Therefore, in PKtreated peanut where the mRNAs encoding the GBSS isoforms were down-regulated and in addition the mRNA encoding Pi translocator was not down-regulated by GDH-synthesized RNAs, triose phosphates and Pi were maximally mobilized and metabolized to acetyl CoA and channeled towards fatty acid biosynthesis thus resulting to maximum yields of seeds and pods (Figure 4). Here, we observed at the molecular level the sequential combination of two metabolic pathways by GDH-synthesized RNAs to double crop yield (Figure 4). Conversely, in the Pi-treated (Figure 3), and NS-treated peanuts where the mRNAs encoding the GBSS isoforms and phosphate translocator were not down-regulated by GDH-synthesized RNAs, starch biosynthesis and phosphate translocation competed for the triose phosphates with consequent decreased biosynthesis of fatty acids resulting to minimum yields of fats, seeds, and pods (Figure 4).
Phosphoglucomutase (PGM, EC 2.7.5.1) generates glucose-1-phosphate (Glc-1-P) from glucose-6-phosphate (Glc-6-P). It is localized both in the plastids and cytosol [41]. The plastidic PGM is essential for starch synthesis to store photosynthates in leaves during the day, and in the degradation of starch [42]. The cytosolic PGM is involved in sucrose catabolism to provide intermediates for glycolysis etc [43]. Deficiency in PGM activity resulted in “starchless” and wrinkled seed phenotypes accompanied by significantly increased seed lipid contents [44]. Therefore, PGM plays an important role in the distribution of Glc-6-P between glycolytic, starch, and fatty acid pathways. In KCl-treated peanut where the mRNA encoding PGM was not down-regulated by GDH-synthesized RNA, the pod yield was double that of NPKS-treated peanut where the mRNA encoding PGM was down-regulated (Figure 4). In the control (Figure 2) and NS-treated peanuts where the mRNA encoding GBSS was not down-regulated but that encoding PGM was down-regulated, pod yields (Figure 4) were the lowest meaning that PGM activity exercised a stronger influence than GBSS on starch synthesis and crop yield in agreement with the role of PGM in the supply of Glc-6-P utilized in the starch pathway [45]. Therefore, combination of down-regulated PGM and GBSS doubled the yields in the NH4Cl, and PK-treated peanuts (Figure 4). In PN-treated peanut where the mRNAs encoding PGM, GBSS, and Pi translocator were only partially down-regulated, pod, seed and fatty acid yields were 14% - 24% higher than in sulfate-treated peanut where
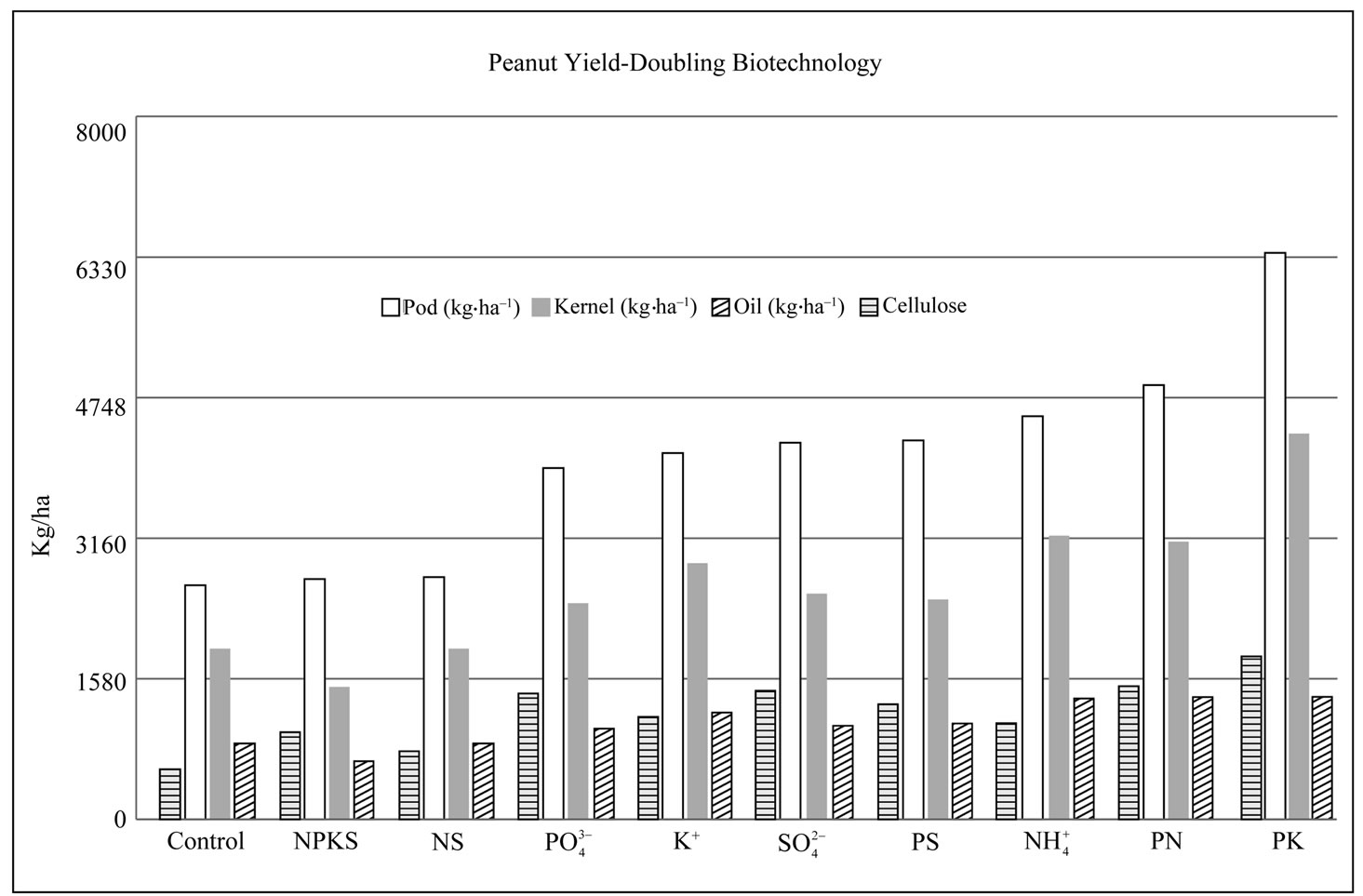
Figure 4. Yield (pod, kernel, oil, and cellulose) responses of peanut to stoichiometric combinations of mineral salt solutions that synchronize GDH synthesis of RNA and GDH isomerization.
the mRNAs encoding PGM and GBSS were not downregulated and in addition that encoding Pi translocator was partially down-regulated. Variable reprogramming of mRNA enables fine-tuning of encoded enzyme for efficient modulation of metabolite flow through the metabolic pathway. Variable partial down-regulation of mRNA by metabolic non-genomic RNA (GDH-synthesized RNA) is a unique mechanism of metabolic coordination in biology.
Glucosyltransferases catalyze the formation of glucosidic linkages between monosaccharide residues from UDP-glucose and specific acceptors [46] in the initiation of cellulose biosynthesis [47]. Peanut shell biomass is dense in cellulose, is an important solid biofuel with a relatively high energy content of 16 Mj/kg [48], and readily convertible to liquid fuel (cellulosic ethanol). Mineral-treated peanuts yielded high percentages (71% - 90%) of shell neutral detergent fiber (NDF). Shell NDF (cellulose) yield of the control peanut was at least doubled in the PS, Na2SO4, Pi, PN, and PK-treated peanuts (Figure 4). At the molecular level, these doubling of shell NDF yields were accounted for by the sequential permutation of the abundances of the mRNA encoding glucosyltransferase, GBSS, PGM, and Pi translocator. In the control (Figure 2) and NS-treated peanuts where the mRNA encoding GBSS was not down-regulated, the mRNA encoding PGM was down-regulated, and the mRNA encoding glucosyltransferase was partially or not down-regulated, the shell cellulose yields were the lowest compared with Pi (Figure 3), Na2SO4, and PK-treated peanuts where the mRNAs encoding GBSS, and glucosyltransferase were not down-regulated, partially or fully down-regulated, the shell cellulose yields were at least doubled (Figure 4). The metabolic control of cellulose by the pathways of starch synthesis, and glycolysis is understandable based on the reaction steps catalyzed by GBSS and PGM in the respective pathways. The influence of Pi translocator on peanut cellulose relies on the plus/plus sequence similarity between the GDH-synthesized RNAs that are homologous to the mRNAs encoding glucosyltransferase and phosphate translocator. The mRNAs encoding Pi translocator and glucosyltransferase cannot be completely knocked out simultaneously under any applied mineral nutrient environment [20]. The GDH-synthesized RNAs make for permutation of the mRNAs homologous to them and in that way the encoded enzymes are fine-tuned to maximize metabolite production. This is the structural permutation of metabolic pathways for the doubling of crop yield.
Acetyl-coenzyme A carboxylase (ACCase EC 6.4.1.2) catalyzes the first committed reaction in lipid biosynthesis with production of malonyl-CoA. The regulation of the enzyme is an important control point in lipid biosynthesis [49]. ACCase controls the production of fats in peanut [22] similar to soybean [50]. In PK-treated peanut where the mRNA encoding ACCase was not downregulated by GDH-synthesized RNAs, the kernel yield was double that (Figure 4) where the mRNA was downregulated as in NS-treated peanut. Because triose phosphates, starch, Glc-6-P, and acetyl coenzyme A are involved in fatty acid metabolism, it is necessary to understand the effects of the permutation of the activities of phosphate translocator, GBSS, PGM, and ACCase on peanut yield. In the NPKS-treated peanut where the mRNA encoding Pi translocator was partially downregulated, the mRNAs encoding PGM, and GBSS were down-regulated but the mRNA encoding ACCase was not down-regulated, the kernel and fatty acid yields were the lowest (Figure 4) compared with the PK-treated peanut with doubled kernel, pod, and fatty acid yields where the mRNAs encoding Pi translocator and ACCase were not down-regulated, and the mRNAs encoding PGM and GBSS were down-regulated, by the GDH-synthesized RNAs. In the PK-treated peanut, because the mRNA encoding Pi translocator was not down-regulated, there was maximum phosphate translocator activity to import Pi into the chloroplast for maximum synthesis of triose phosphates; the mRNA encoding GBSS was down-regulated, the triose phosphates were not expended in starch synthesis; the mRNA encoding PGM was down-regulated, Glc-6-P was sufficiently channeled to glycolysis; and finally the mRNA encoding the ACCase was not down-regulated thereby ensuring maximum utilization of acetyl-Co A from citric acid cycle to synthesize maximum quantity of fatty acids. This is an aspect of the process by which GDH-synthesized RNA sequentially coordinates and permutes metabolic pathways at the molecular level to maximize end product formation and doubling of crop yield.
Nitrogen assimilation: three metabolic pathways cooperated to assimilate nitrogen in peanut. They are purine biosynthesis, nitrate reduction, and glutamine synthetase-glutamate synthase (GS-GOGAT) cycle. The purine metabolic pathway is employed in tropical legumes to assimilate and detoxify ammonia [39]. GAR synthase catalyzes the second step, and GAR transformylase catalyzes the third step in purine biosynthetic pathway. GDH-synthesized RNA, probes 3E and 48Q (Table 2) were found to be homologous to the mRNAs encoding GARS and GART [22]. NR catalyzes the reduction of nitrate to nitrite which is then reduced by nitrite reductase to  ion [51], followed by incorporation into amino acids by GS-GOGAT cycle [52]. NADH-GOGAT is an important enzyme in the salvage of
ion [51], followed by incorporation into amino acids by GS-GOGAT cycle [52]. NADH-GOGAT is an important enzyme in the salvage of  ion [53]. The role was confirmed in the peanut because most of the mineral treatments (except Pi, and PS treatments) failed to induce the GDH synthesis of the RNA homologous to the mRNA encoding NADH-GOGAT, the induction of which in the Pi (Figure 3), and PS-treated peanuts caused up to 38% decrease in crop yield compared with the PK-treated peanut where NADH-GOGAT was not down-regulated. In control peanut (Figure 2) where the GDH-synthesized RNA down-regulated the mRNAs encoding GARS, GART, and NR but the mRNA encoding NADH-GOGAT was not, pod yield was the lowest as compared with the PK-treated peanut where the mRNAs encoding GARS/GART was only partially downregulated, the mRNA encoding NR was down-regulated, and the mRNA encoding NADH-GOGAT was not downregulated, the pod yield was increased by 2.4 folds (Figure 4). Therefore purine biosynthesis is important in salvaging nitrogen and doubling peanut yield.
ion [53]. The role was confirmed in the peanut because most of the mineral treatments (except Pi, and PS treatments) failed to induce the GDH synthesis of the RNA homologous to the mRNA encoding NADH-GOGAT, the induction of which in the Pi (Figure 3), and PS-treated peanuts caused up to 38% decrease in crop yield compared with the PK-treated peanut where NADH-GOGAT was not down-regulated. In control peanut (Figure 2) where the GDH-synthesized RNA down-regulated the mRNAs encoding GARS, GART, and NR but the mRNA encoding NADH-GOGAT was not, pod yield was the lowest as compared with the PK-treated peanut where the mRNAs encoding GARS/GART was only partially downregulated, the mRNA encoding NR was down-regulated, and the mRNA encoding NADH-GOGAT was not downregulated, the pod yield was increased by 2.4 folds (Figure 4). Therefore purine biosynthesis is important in salvaging nitrogen and doubling peanut yield.
7) Bioscience of GDH-synthesized RNA: as a new gateway into biology, procedures for analyzing transcript silencing by GDH-synthesized RNAs are slightly different from classical siRNA approaches in that there are no transfection and reverse transcription steps, and the GDHsynthesized RNAs are endogenously elicited. Transcript silencing by GDH-synthesized RNA takes place as a biochemical reaction at the molecular level; and it is inducible at-will in crops. Experimentation on GHD-synthesized RNA embodies many layers of technical repeats, normalization, and standardization that assure result reproducibility across experiments. GDH purification by whole gel electro-elution into the 14 chambers of BioRad’s mini whole gel eluter give 14 instead of one preparation of the enzyme, an infinite opportunity for verifying the RNA synthetic activity 14 times (Figure 1) instead of once per treated-peanut. The binomial assembly of the subunit polypeptides in the hexameric isoenzymes did not permit the combination of the 14 fractions to make a single preparation. Equal weights (10 µg) of RNA synthesized per the 14 GDH samples were agarose gel-electrophoresed, trans-blotted after electrophoresis, followed by Northern hybridization (Figures 2 and 3). Therefore, in these protocols, there were 14 repeats of the purified GDH, 14 repeats of RNA synthesis, and 14 repeats of RNA loading into gel. More than 90% of the GDH and RNA lanes (Figure 1) showed that protein and RNA repetitive loadings onto gels were consistent, efficient and reproducible across mineral-treated peanuts. Northern blots were performed in duplicate using each cDNA of GDH-synthesized probe, thereby making a total of 28-repeat RNA tracks per treated peanut. There were several evidence for internal control reactions in the transcript silencing including the plus/plus and/or plus/ minus sequence similarities among the GDH-synthesized RNAs, partial silencing of mRNAs encoding distinct enzymes, GDH-synthesized RNA present in total RNA, absence of total RNA from GDH-synthesized RNA etc. Residual quantities of mRNAs encoding PGM and ACCase in the Northern blots (Figure 3) of Pi-treated peanut confirmed the sequence similarities between the GDH-synthesized RNAs homologous to the mRNAs encoding PGM, NADH-GOGAT, ACCase, and GARS/ GART. Absence of total RNA from GDH-synthesized RNA (Figure 1(i)-(x)) assured that Northern bands were not false positives. Messenger RNAs encoding several housekeeping enzymes were not useful references in the Northern assays because Northern results [29] in which GDH-synthesized RNAs were used as probes suggested the housekeeping mRNAs were also reprogrammed under the experimental regimen. Therefore, a ratio of 2:1 between the GDH-synthesized RNA and the target mRNA was adopted as the minimum normalization and standardization factor for silencing. Although the ideal reference internal mRNA is one that does not vary as a function of experimental treatment [54], it has been difficult to identify a single mRNA that meets the criterion. Where the mRNAs were fully silenced and there were no residual mRNAs as in the Northern blots for GARS/ GART (Figure 3(iv)), and NADH-GOGAT (Figure 3(v)) it was assumed that the 2:1 ratio was upheld. Where the mRNAs were partially silenced as in the Northern blots for PGM (Figure 3(i)), glucosyltransferase (Figure 3(ii)), and ACCase (Figure 3(iii)) it was assumed that the residual mRNA constituted 50% of the total mRNA. The full amount of mRNA was determined when the GDH of the peanut did not synthesize the homologous RNA and the mRNA was the lone band in the Northern assay. The limits of the bioscience of the GDH-synthesized RNA experimentation were defined by the peanut without any mineral ion treatment representing the negative control; and the peanut with the full mineral ion concentration and composition (NPKS) representing the positive control (Figure 1). The positive control is important to provide the line of verification for the molecular mechanism of GDH action, and for result verification, confirmation and validation.
4. DISCUSSION
Metabolic variants of peanut: ability of GDH to spontaneously isomerize and synthesize RNAs that silence homologous mRNAs constitutes one aspect of the functions of the enzyme. The other aspect is the signal integration and discrimination function [19] and is implemented through the permutation of the metabolic pathways in sequence and in space, by the GDH-synthesized RNAs. The two functions of GDH constitute the new gateway into biology. GDH-synthesized RNAs are metabolic and non-genomic [29], and so they do not suffer from mutational changes. The two functions introduce the concept of metabolic variants as different from genetic variants. The yield-doubling variants with respect to the control, and NPKS-treated peanut included the Pi,  , and PK-treated peanuts.
, and PK-treated peanuts.
Pathway discrimination and integration in Control metabolic variant: the control peanut yield metabolism was unique because GBSS, ACCase, NADH-GOGAT, and glucosyltransferase were not down-regulated; but Pi translocator, NR, GARS/GART, and PGM were downregulated leading to low production of pods (2637 kg·ha–1), kernel (1922 kg·ha–1), and fatty acids (855 kg·ha–1). There were no partially down-regulated yieldspecific enzyme steps. This is the kind of low yield (Figure 4) that peasant farmers achieve in the developing countries when they cultivate peanut according to the recommended tradition of crop rotation technology [25]. The low efficiency of the metabolism of the control peanut confirmed that the nitrogen fixing ability of the rhizobia was not sufficient for the growth and yield doubling of the crop (Figure 4) compared with mineraltreated variants.
Pathway discrimination and integration in NPKS metabolic variant: GARS/GART, and Pi translocation were partially down-regulated. Nitrate reductase, glucosyltransferase, and GBSS were down-regulated. But ACCase and NADH-GOGAT were not down-regulated. The consequences were decreases in fatty acids (656 kg·ha–1), and kernel (1491 kg·ha–1) yields (Figure 4). This is the kind of low yield that farmers obtained when peanuts followed a heavily fertilized small grain crop in the rotation, or when peanut is planted on soils with high residual NPKS fertility [55]. The low efficiency of the metabolism of the NPKS-treated peanut confirmed that the NPKS combination of mineral ions was not an effective mix for the induction of the maximum production metabolism of the crop.
Pathway discrimination and integration in Phosphate metabolic variant: Na3PO4-treated peanut had the downregulation of GARS/GART, and NADH-GOGAT; partial down-regulation of PGM, ACCase, and glucosyltransferase; leaving Pi translocator, GBSS, and NR not down-regulated with the advantage that pod (3955 kg·ha–1), kernel (2433 kg·ha–1), and fatty acid (1022 kg·ha–1) yields were increased (Figure 4) compared with control peanut. Phosphorus fertilizer is cautiously recommended for peanuts because it did not increase fatty acid yields on sandy soils [25,56].
Pathway discrimination and integration in NH4Cl metabolic variant: fertilizer nitrogen is generally not recommended for peanut because rhizobia assist to fix atmospheric nitrogen for the crop [36]. Comparison of the control peanut yield with that of NH4Cl-treated peanut shows the overwhelming superiority and doubling of yield by the NH4Cl variant (Figure 4).The NH4Cl treatment instantaneously relieved the demise of the purine nucleotide biosynthesis, and NR in the untreated peanut with dramatic increases in pod (4541 kg·ha–1), kernel (3245 kg·ha–1), and fatty acid (1361 kg·ha–1) yields (Figure 4). PGM, and glucosyltransferase were down-regulated; Pi translocator, and GBSS were partially down-regulated; leaving ACCase, GS-GOGAT cycle, NR, and GARS/ GART not down-regulated. Crop yield doubling biotechnology calls for fresh recommendations for nitrogen fertilization of leguminous crop species.
Pathway discrimination and integration in PK metabolic variant: irrespective of soil type, location, and peanut variety, the most popular mineral ion recommendation for peanut production worldwide is the PK combination [34,35], although detrimental quantities of potash were generally applied [25]. The production metabolism of the PK variant attests to this popularity because GBSS suffered demise, all the triose phosphates were used for glycolysis; PGM suffered demise, all the Glc-6-P was used for glycolysis; glucosyltransferase suffered partial demise, the amount of ATP utilized in UDP-Glc synthesis was minimized; GARS/GART suffered partial demise, there was sufficient ATP to power glucose metabolism; and NR suffered demise, the amount of NADH utilized in intermediary metabolism was very low. But NADHGOGAT did not suffer demise thereby allowing nitrogen assimilation to proceed in support of amino acid and purine nucleotide biosyntheses; and Pi translocator did not suffer demise, so all the triose phosphates were channeled to acetyl coenzyme A production. Finally, ACCase was not down-regulated giving the peanut variant the advantage that the maximum photosynthates were priority-channeled to fatty acid, cellulose, and amino acid production. The PK-treated peanut was therefore the most efficient metabolic variant, the pod (6377 kg·ha–1), kernel (4342 kg·ha–1), oil (1381 kg·ha–1), and cellulose (1829 kg·ha–1) yields being doubled compared with the controls (NPKS, and untreated control) that were the most inefficient for yield production (Figure 4).
From the foregoing, demise (full down-regulation) of metabolic reaction steps followed by permutation of yield-related metabolic pathways and redistribution of metabolite load to molecularly connected metabolic pathways controls crop yield. Because the metabolic variants are distinct bioreactors, the yields of pods, fatty acids etc are also distinct, there being no biochemical justification for calculating average yields (Figure 4) between two or more variants.
5. CONCLUSIONS
Many research projects had investigated a genetic base for crop yield [10,11]. Crop yield-doubling biotechnology is different, being based on metabolic pathway demise, statistical redistribution of metabolite load to molecularly connected pathways, coordination of metabolic pathways at the molecular level and concomitant alteration of yield by the GDH-synthesized RNAs, which constitute a new gateway into biology. Different metabolic variants of peanut exist based on the discrimination and integration (permutation) of metabolic pathways and altered crop yield. Metabolic variants prove that peanut is a natural bioreactor for the production of large quantities of fatty acids through the discrimination against saccharide metabolism. Some of the peanut bioreactors (PK, PN, PS, Pi, NH4Cl, and  -treated) are more energy efficient than others (control, NS, NPKS, and KCl-treated). The demise of pathways, statistical redistribution of metabolite load, and alteration of crop yield are programmable by mineral ions. The yield doubling biotechnology is a manifestation of the property of crops as programmable bioreactors. A maximum yield of 10,000 kg·pods·ha–1 is achievable based on the bioreactor property of peanut. This is a biotechnological approach for doubling crop yield [57] to feed a human population that will soon be nine billion people. Farmers’ optimal yield of peanut was about 3400 kg·ha–1 [58], suggesting that yields can be tripled by cultivating the crop as an energy efficient bioreactor rather than by crop rotation, dependence on biological nitrogen fixation, or by application of kg amounts of fertilizer [24]. The bioreactor peanut yield-doubling biotechnology requires mM amounts of mineral salts per hectare, a thousand-fold saving in financial input by farmers. Crop yield doubling biotechnology has the promise to reformulate fertilizer compositions, and minimize not only the cost of agricultural production but also food insecurity.
-treated) are more energy efficient than others (control, NS, NPKS, and KCl-treated). The demise of pathways, statistical redistribution of metabolite load, and alteration of crop yield are programmable by mineral ions. The yield doubling biotechnology is a manifestation of the property of crops as programmable bioreactors. A maximum yield of 10,000 kg·pods·ha–1 is achievable based on the bioreactor property of peanut. This is a biotechnological approach for doubling crop yield [57] to feed a human population that will soon be nine billion people. Farmers’ optimal yield of peanut was about 3400 kg·ha–1 [58], suggesting that yields can be tripled by cultivating the crop as an energy efficient bioreactor rather than by crop rotation, dependence on biological nitrogen fixation, or by application of kg amounts of fertilizer [24]. The bioreactor peanut yield-doubling biotechnology requires mM amounts of mineral salts per hectare, a thousand-fold saving in financial input by farmers. Crop yield doubling biotechnology has the promise to reformulate fertilizer compositions, and minimize not only the cost of agricultural production but also food insecurity.
REFERENCES
- Fraley, R. (2010) Sustainable solutions for doubling crop productivity by 2030. Plenary Lecture XII, 12th IAPB Congress, St. Louis, 8 June 2010.
- FAO (2006) The state of food insecurity in the world. Rome.
- Brown, L.R. (1997) Facing the challenge of food scarcity: Can we raise grain yields fast enough? In: Ando, T., Fujita, K., Mae, T., Matsumoto, H., Mori, S. and Sekiya, J., Eds., Plant Nutrition for Sustainable Food Production and Environment, Kluwer Academic Publishers, Dordrecht/ Boston/London, 15-24.
- Richards, R.A. (2000) Selectable traits to increase crop photosynthesis and yield of grain crops. Journal of Experimental Botany, 51, 447-458. doi:10.1093/jexbot/51.suppl_1.447
- Carpenter, C. (2011) World food prices climb to record as UN sounds alarm on further shortages. http://www.bloomberg.com/news/2011-03-03/
- Hinchee, M.A.W., Padgette, S.R., Kishore, G.M., Delannay, X. and Fraley, R.T. (1993) Herbicide-tolerant crops. In: Kung, S. and Wu, R., Eds., Transgenic Plants, Academic Press, San Diego, 243-263.
- Gould, F. (1998) Sustainability of transgenic insecticidal cultivars: Integrating pest genetics and ecology. Annual Review of Entomology, 43, 701-726. doi:10.1146/annurev.ento.43.1.701
- Horton, P. (2000) Prospects for crop improvement through the genetic manipulation of photosynthesis: Morphological and biochemical aspects of light capture. Journal of Experimental Botany, 51, 475-585. doi:10.1093/jexbot/51.suppl_1.475
- Century, K., Reuber, T.L. and Ratcliffe, O.J. (2008) Regulating the regulators: The future prospects of transcription factor-based agricultural biotechnology products. Plant Physiology, 147, 20-29. doi:10.1104/pp.108.117887
- Reynolds, M., Foulkes, M.J., Slafer, G.A., Berry, P., Parry, M.A.J., Snape, J.W. and Angus, W.J. (2009) Raising yield potential in wheat. Journal of Experimental Botany, 60, 1899-1918. doi:10.1093/jxb/erp016
- Xing, Y. and Zhang, Q. (2010) Genetic and molecular bases of rice yield. Annual Review of Plant Biology, 61, 421-442. doi:10.1146/annurev-arplant-042809-112209
- Cakmak, I. and Engels, C. (1999) Role of mineral nutrients in photosynthesis and yield formation. In: Rengal, Z., Ed., Mineral Nutrition of Crops, Food Products Press, New York, London, Oxford, 144-168.
- McDonald, M.B. and Copeland, L. (1997) Seed production principles and practices. Chapman and Hall, New York.
- Cianzio, S.R. (1999) Breeding crops for nutrient efficiency: Soybean and wheat as case studies. In: Rengal, Z., Ed., Mineral Nutrition of Crops, Food Products Press, New York, London, Oxford, 267-287.
- Osuji, G.O., Reyes, J.C. and Mangaroo, A.S. (1998) Glutamate dehydrogenase isomerization: A simple method for diagnosing nitrogen, phosphorus, and potassium sufficiency in maize (Zea mays L.). Journal of Agricultural and Food Chemistry, 46, 2395-2401. doi:10.1021/jf971065x
- Osuji, G.O., Braithwaite, C., Fordjour, K., Madu, W.C., Beyene, A., Roberts, P.S. and Wright, V. (2003) Purification of glutamate dehydrogenase isoenzymes and characterization of their substrate specificities. Preparative Biochemistry & Biotechnology, 33, 13-28. doi:10.1081/PB-120018366
- Osuji, G.O., Konan, J. and M’Mbijjewe, G. (2004) RNA synthetic activity of glutamate dehydrogenase. Applied Biochemistry and Biotechnology, 119, 209-228. doi:10.1007/s12010-004-0003-z
- Osuji, G.O. and Madu, W.C. (1995) Ammonium iondependent isomerization of glutamate dehydrogenase in relation to glutamate synthesis in maize. Phytochemistry, 39, 495-503. doi:10.1016/0031-9422(94)00976-Z
- Osuji, G.O., Braithwaite, C., Pointer, R. and Reyes, J.C. (1999) Pesticide inactivation of peanut glutamate dehydrogenase: Biochemical basis of the enzyme’s isomerization. Journal of Agricultural and Food Chemistry, 47, 3345-3351. doi:10.1021/jf980531v
- Osuji, G.O. and Brown, T. (2007) Role of the RNAs synthesized by glutamate dehydrogenase in the coordinate regulation of metabolic processes. The Icfai Journal of Biotechnology, 1, 37-48.
- Osuji, G.O., Mangaroo, A.S., Reyes, J., Bulgin, A. and Wright, V. (2003/4) Biomass enhancement in maize and soybean in response to glutamate dehydrogenase isomerization. Biologia Plantarum, 47,45-52.
- Osuji, G.O., Brown, T.K. and South, S.M. (2010) Optimized fat and cellulosic biomass accumulation in peanut through biotechnology. International Journal of Biotechnology and Biochemistry, 3, 455-476.
- Putnam, D.H., Oplinger, E.S., Teynor, T.M., Oelke, E.A., Kelling, K.A. and Doll, J.D. (1991) Peanut. Alternative Field Crop Manual. mhtml:file://E:\Peanut yield.mht
- Walker, M.E. and Ethredge J. (1974) Effects of N rate and application on Spanish peanut (Arachis hypogaea L.) yield and seed grade, N and oil. Peanut Science, 1, 45-47. doi:10.3146/i0095-3679-1-2-2
- Mitchell, C.C. and Adams, J.F. (1994) Phosphorus and Potassium Chapter 5, in research-based soil testing interpretation and fertilizer recommendations for peanuts on coastal plain soils. Southern Cooperative Series Bulletin. http://www.ag.auburn.edu/aaes/communications/380site/contents.htm accessed 10/26/2009
- Kaur, P. and Hundal, S.S. (1999) Forecasting growth and yield of groundnut with a dynamic simulation model “PNUTGRO” under Punjab conditions. Journal of Agricultural Science, Cambridge, 133, 167-173.
- Jamal, A., Fazli, I.S., Ahmad, S., Kim, K.-T., Oh, D.-G. and Abdin, M.Z. (2006) Effect of sulfur on nitrate reductase and ATP sulfurylase activities in groundnut (Arachis hypogeal L.). Journal of Plant Biology, 49, 513- 517. doi:10.1007/BF03031134
- Grierson, D., Slater, A., Speirs, J. and Tucker, G.A. (1985) The appearance of polygalacturonasem RNA in tomatoes: One of a series of changes in gene expression during development and ripening. Planta, 163, 263-271. doi:10.1007/BF00393517
- Osuji, G.O., Brown, T.K. and South, S.M. (2008) Discovery of the RNA synthetic activity of glutamate dehydrogenase and its application in drug metabolism. The Open Drug Metabolism Journal, 2, 1-13. doi:10.2174/1874073100802010001
- Osuji, G.O., Brown, T.K. and South, S.M. (2009) Nucleotide-dependent reprogramming of mRNAs encoding acetylcoenzyme A carboxylase and lipoxygenase in relation to fat contents of peanut. Journal of Botany, 2009, 278324. doi: 10.1155/2009/278324
- Cammaerts, D. and Jacobs, M. (1983) A study of the polymorphism and the genetic control of the glutamate dehydrogenase isoenzymes in Arabidopsis thaliana. Plant Science Letters, 31, 67-73. doi:10.1016/0304-4211(83)90130-X
- Klahre, U., Leuenberger, S.A., Iglesias, V.A. and Meins, F. (2002) High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proceedings of the National Academy of Sciences of the United States of America, 10, 1073- 1078.
- Wright, D.L. (2008) Production of biofuels crops in Florida: Peanut. mhtml://E:\peanut yield 2.mht.
- Henning, R.J., Allison, A.H. and Tripp, L.D. (1982) Cultural practices. In: Pattee, H.E. and Young, C.T., Eds., Peanut Science and Technology, American Peanut Research and Education Society Inc. Publisher, Yoakum, 126-137.
- Nasr-Alla, A.E., Fatma, A.A.O. and Soliman, K.G. (1998) Effects of increased phosphorus and potassium or sulfur application in their different combinations on yield, yield components, and chemical composition of peanut in a newly reclaimed sand soil. Zagazig Journal of Agricultural Research, 25, 557-579.
- Kidder, G. (1994) Nitrogen and sulfur. In researchbased soil testing information and fertilized recommendation for peanuts on coastal plain soils. Southern Cooperative Series Bulletin. http://www.auburn.edu/aaes/communications/380site/chapterthree.htm. accessed 10/26/2009.
- Jamal, A. (2010) Enzyme activity assessment of peanut under slow-release sulfur fertilization. mhtml: file:// E:\peanut yield 1.mht
- Stitt, M. and Quick, W.P. (1989) Photosynthetic carbon partitioning: Its regulation and possibilities for manipulation. Physiologia Plantarum, 77, 633-641. doi:10.1111/j.1399-3054.1989.tb05402.x
- Smith, P.M.C. and Atkins, C.A. (2002) Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiology, 128, 793-802. doi:10.1104/pp.010912
- Edwards, A., Marshall, J., Denyer, K., Sidebottom, C., Visser, R.G.F., Martin, C. and Smith, A.M. (1996). Evidence that a 77-kilodalton protein from the starch of pea embryos is an isoform of starch synthase that is both soluble and granule-bound. Plant Physiology, 112, 89-97. doi:10.1104/pp.112.1.89
- Streb, S., Elgi, B., Eicke, S. and Zeeman, S. (2009) The debate on the pathway of starch synthesis: A closer look on low-starch mutants lacking plastidial phosphoglucomutase supports the chloroplast-localized pathway. Plant Physiology, 151, 1769-1777. doi:10.1104/pp.109.144931
- Hattenbach, A. and Heineke, D. (1999) On the role of chloroplastic phosphoglucomutase in the regulation of starch turn over. Planta, 207, 527-532. doi:10.1007/s004250050513
- Manjunath, S., Lee, C.H.K., VanWinkle, P. and BaileySerres, J. (1998) Molecular and biochemical characterization of cytosolic phosphoglucomutase in maize expression during development and in response to oxygen deprivation. Plant Physiology, 117, 997-1006. doi:10.1104/pp.117.3.997
- Periappuram, C., Steinhauer, L., Barton, D.L., Taylor, D.C., Chatson, B. and Zou, J. (2000) The plastidic phosphoglucomutase from Arabidipsis. A reversible enzyme reaction with an important role in metabolic control. Plant Physiology, 122, 1193-1199. doi:10.1104/pp.122.4.1193
- Harrison, C.J., Mould, R.M., Leech, M.K., Johnson, S.A., Turner, L., Schreck, S.L., Baird, K.M., Jack, P.L., Rawsthorne, S., Hedley, C.L. and Wang, T.L. (2000) The rug3 of pea encodes plastidial phosphoglucomutase. Plant Physiology, 122, 1187-1192. doi:10.1104/pp.122.4.1187
- Keegstra, K. and Raikhel, N. (2001) Plant glycosyltransferases. Current Opinion in Plant Biology, 4, 219-224. doi:10.1016/S1369-5266(00)00164-3
- Saxena, I.M. and Brown, R.M. (1997) Identification of cellulose synthases in higher plants: Sequence analysis of processive β-glycosyltransferases with the common motif ’D, D, D35Q(R,Q)XRW. Cellulose, 4, 33-49. doi:10.1023/A:1018411101036
- Roberson, R. (2006) Peanut biodiesel promising but costly alternative fuel. http://www.renewableenergyworld.com
- Page, R.A., Okada, S. and Harwood, J.L. (1994) AcetylCoA carboxylase exerts a strong flux control over lipid synthesis in plants. Biochimica et Biophysica Acta, 1210, 369-372.
- Reverdatto, S., Beilinson, V. and Nielsen, N.C. (1999) A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiology, 119, 961-978. doi:10.1104/pp.119.3.961
- Kleinhofs, A., Warner, R.L. and Melzer, J.M. (1989) Genetics and molecular biology of higher plant nitrate reductase. In: Poulton, J.E., Romeo, J.T. and Conn, E.E. Eds., Plant Nitrogen Metabolism, Plenum Publishing Corporation, New York, 117-155. doi:10.1007/978-1-4613-0835-5_4
- Vance, C.P., Miller, S.S., Gregerson, R.G., Samac, D.A., Robinson, D.L. and Gantt, J.S. (1995) Alfalfa NADHdependent glutamate synthase: Structure of the gene and importance in symbiotic N2 fixation. Plant Journal, 8, 345-358. doi:10.1046/j.1365-313X.1995.08030345.x
- Chen, F. and Cullimore, J.V. (1988) Two isoenzymes of NADH-dependent glutamate synthase in root nodules of Phaseolus vulgaris L. Plant Physiology, 88, 1411-1417. doi:10.1104/pp.88.4.1411
- Schmittgen, T.D. and Zakrajsek, B.A. (2000) Effect of experimental treatment on housekeeping gene expression: Validation by real-time quantitative RT-PCR. Journal of Biochemical and Biophysical Methods, 46, 69-81. doi:10.1016/S0165-022X(00)00129-9
- Sturkie, D.G. and Buchanan, G.A. (1973) Cultural practices. In peanuts: Culture and uses. APREA, Stillwater, 299-326.
- Gobarah, M.E., Mohamed, M.H. and Tawfik, M.M. (2006) Effect of phosphorus fertilizer and foliar spraying with Zinc on growth, yield, and quality of groundnut under reclaimed sandy soil. Journal of Applied Sciences Research, 2, 491-496.
- Fageria, N.K., Baligar, V.C. and Jones, C. (1997) Growth and mineral nutrition of field crops. 2nd Edition, Marcel Dekker, Inc., New York, 494.
- Sorrow, A. (2009) Georgia peanut yield may be record. http://www.southeastfarmpress.com/peanuts/Georgia-peanuts-1013/index

