Creative Education
Vol.06 No.08(2015), Article ID:56499,10 pages
10.4236/ce.2015.68079
The Use of DNA Extraction for Molecular Biology and Biotechnology Training: A Practical and Alternative Approach
D. N. Lázaro-Silva1, J. C. P. De Mattos2, Helena C. Castro1, G. G. Alves1, L. M. F. Amorim1*
1Post-Graduation Program of Science and Biotechnology, Biology Institute, Fluminense Federal University, Niterói, Brazil
2Biophysics and Biometry Departament, Roberto Alcantara Gomes Biolology Institute, Rio de Janeiro State University, Rio de Janeiro, Brazil
Email: *amorim@vm.uff.br
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 27 October 2014; accepted 17 May 2015; published 21 May 2015
ABSTRACT
DNA extraction and polymerase chain reaction are molecular biology techniques widely used in research, diagnostics and biotechnology industry. In this work, we report a practical course designed for undergraduate students of health, biological sciences and biotechnology areas. The course is five days long and emphasizes basic procedures in practical activities. The students were challenged to use three different protocols for DNA isolation (one using commercial silica column and two, in-house) using no lab animals but themselves. They evaluated the DNA yield, and potential amplification of the extracted DNA by PCR using their own cells genetic material. Students were stimulated to adopt a critical view during the course to develop skills including critical analysis of primary results and possible errors, scientific report writing and a problem solving profile.
Keywords:
Molecular Biology, Practical Course, DNA Extraction, Whole Blood, Polymerase Chain Reaction

1. Introduction
Biological Sciences and Biotechnology are inherently complex and highly abstract. Therefore, professionals of health and biological sciences and biotechnology are challenged with techniques such blood collection and extraction of DNA, evaluation by spectrophotometry and agarose gel electrophoresis as well as polymerase chain reaction (PCR) amplification of the DNA. However, these complex topics are often presented to undergraduate students during theoretical classes or with practical activities involving classical protocols of DNA isolation from fruit (Miller, 1994, Tibell and Rundgren, 2010, Sitaraman, 2012) . Despite these strategies are very easy, low-cost and adequate to teach principles of molecular biology, they have limited resemblance to the actual professional activities on the environment of health science careers. In addition, some skills are necessary to perform these basic molecular procedures and should be obtained during graduation as endorsed by reviews of biology curricula like “BIO2010” (Slonczewsk, 2004) .
Isolating high-quality nucleic acids from blood is a part of the many diagnostics with forensic and/or scientific applications (Poh, 2014) . Despite the concern to use noninvasive sources of DNA (Hearn, 2010) , the use of peripheral blood still remains very common in research and diagnosis, due to its reliability as a source of genetic material (Martin, 1992). Furthermore, blood is the main target of study in clinical research and diagnosis to effectively detect pathogens (Schijman, 2011) as it allows to access the genetic material of pathogens such as Pneumococcal pneumonia (Marchese, 2011) , Trypanosoma cruzi (González, 2010) and several viruses (Mello, 2007) . Furthermore, regardless of the relative degree of invasiveness, blood collection can often be used to avoid techniques that are even more invasive. It allows prenatal diagnosis, and the detection of genetic disorders such as hemophilia in fetuses (without the use of amniotic fluid) (Tsui, 2011) or even sex determination (Mortarino, 2011) .
The use of blood from laboratory animals for in vivo experiments in education is still looking for alternatives (Cardoso and Almeida, 2014) . However, it is important that the students of health science careers be prepared for the issues involving this topic in their future professional lives (Badyal and Desai 2014) . This includes questions involving collecting, manipulating, evaluating and comparing human blood. Therefore, alternatives for the use of blood from lab animals should be developed for substitution purpose without loosing the applied perspective. In addition, besides approaching the technique, the alternative option should still allow the discussion in different levels, from research to the professional application with a dynamic view for every student.
Thus, the aim of this work is to propose a practical summer course to undergraduate students of biomedicine, biotechnology and biological sciences, containing basic procedures of using human blood DNA extraction, including spectrophotometry, agarose gels and Biosafety on blood manipulation. Therefore, the students are challenged to evaluate their own blood by using three different DNA extraction techniques (one from commercial sources and two, in-house), getting first-hand contact with the equipments and procedures of a real molecular biology laboratory. The course requires and works several skills of these students, including: data interpretation, problem solving, experimental design, scientific report writing and collaborative work, with critical analysis of primary results and possible errors.
2. Methodology
2.1. Subjects
A group of ten undergraduate students of both genders (7 females and 3 males), age 21.4 ± 1.43 (20 to 24 years), 100% single, 100% student occupation, 100% auto-declared white skin color, from São Paulo (10%), Minas Gerais (10%) and Rio de Janeiro (80%) on the fourth period (2nd year) of Biology (n = 6) or Biomedicine (n = 4) courses from the Fluminense Federal University (UFF), Rio de Janeiro―Brazil, were invited to participate in the molecular biology summer course offered at the Molecular Oncology Laboratory of Biology Institute of UFF. All students signed the informed consent to collect blood, perform DNA extractions and donate DNA to the laboratory’s DNA collection (Antônio Pedro Hospital Ethics Committee CONEP 02509 No. 0012.0258000-09, 2009).
2.2. Summer Course Design
The laboratory classes were divided into five meetings with a maximum of 7 hours (Table 1). The DNA extraction was distributed into the first three days (Figure 1). The basic procedures executed were:
1) Preparation of solutions, dilutions and sterilization;
2) Preparation and use of agarose gels to separate genomic DNA and PCR products;
3) Interpretation and trouble shooting of agarose gel electrophoresis;
4) PCR reaction, the importance of controls; interpretation and troubleshooting of PCR reaction;
5) The use of the laboratory equipments as: autoclave, micropipetters, centrifuges, balance, horizontal electro- phoresis system, UV transilluminator, termocycler and photodocumentation system.
Table 1. Schedule of activities of the molecular biology summer course.
On the last day of the course they were asked to write a final text, including graphics, to describe and illustrate the obtained results.
2.3. BioSafety
Students were instructed about laboratory safety, such as the use of individual protection equipments. They wore laboratory coat, gloves and safety glasses all time when in the laboratory and handling blood samples. Any residual blood or contaminated vial was disposed in a Biohazard waste container that was autoclaved prior to discard. Students used ultraviolet protective glasses and gloves when viewing and handling gels with ethidium bromide (EtBr) on the UV transilluminator. To minimize exposure to EtBr, only the dilute staining solution was available to them. Besides, a restricted area of the laboratory was reserved to gel staining, and students were instructed to leave all EtBr-contaminated materials in this area only. All solutions and remaining buffer were treated as hazardous waste and disposed in adequate containers to be discarded.
2.4. DNA Extraction Protocols
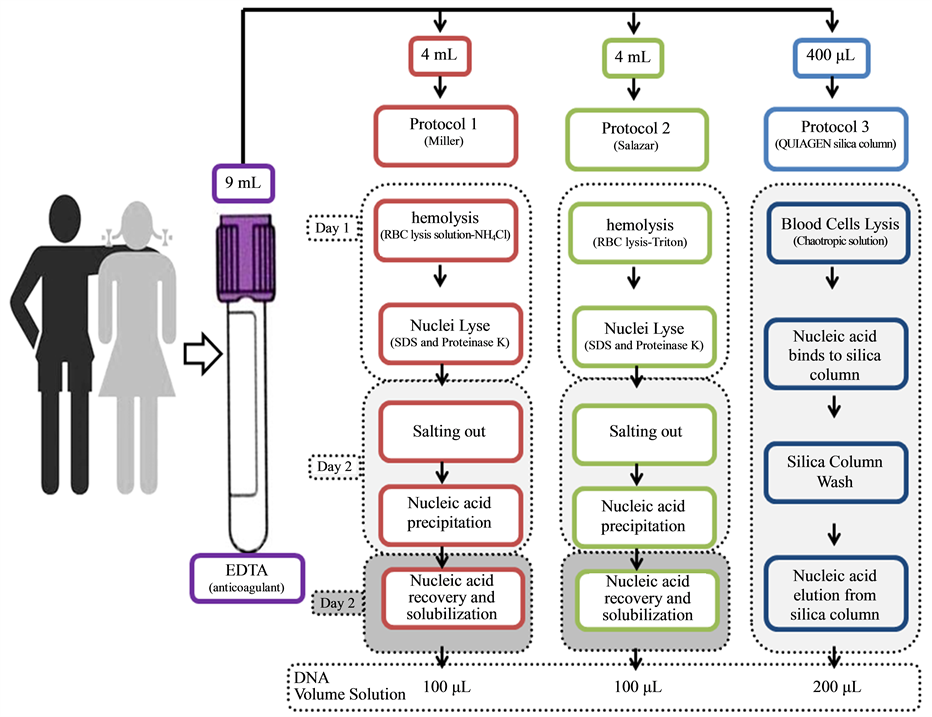
Nine milliliters of blood were collected from the each student by venous puncture with Ethylenediaminetetraacetic acid (EDTA)―containing tubes, and aliquoted to be submitted to three different DNA extraction protocols (Figure 1). For safety reasons, students manipulated only their own blood samples.
2.4.1. Protocol 1― (MILLER, 1988)
Red blood cells in 4 mL of blood were lysed with 8 mL of of haemolysis buffer (155 mM NH4Cl, 10 mM KHCO3, 1 mM EDTA) in an ice bath per thirty minutes. After centrifugation (500 ×g/10 min) the leukocytes nuclei pellet was washed with 8 mL of hemolytic buffer until disappearance of red color, indicating the absence
Figure 1. Detailed schedule of the procedures used in the practical summer course applied to undergraduate students of biomedicine and biological sciences at Universidade Federal Fluminense, containing basic procedures using human blood, and three different protocols.
of hemoglobin. Leukocytes were lysed with 3 mL of nuclei lysis buffer (10 mM Tris-HCl, 400 mM NaCl and 2 mM Na2EDTA, pH 8.2) containing 200 μL of 20% SDS and 20 μL of proteinase K (25 mg/mL) at 56˚C overnight. On the next day, the digestion was centrifuged (500 ×g/10 min) and the supernatant passed to another tube. Proteins were precipitated with 1 mL of NaCl (6 M) and after centrifugation (12,000 ×g/10 min), the aqueous phase was transferred to a new tube. Two volumes of ice cold ethanol were added and DNA precipitation visualized. DNA was collected (14,000 ×g/10 min) and washed with 1 mL of 70% ethanol to remove excess of salt. This step must be repeated until the precipitate becomes translucent. Dried samples were eluted with 100 μL of TE (10 mM Tris-HCl, 0.2 mM Na2EDTA, pH 7.5) for 2 hours at 37˚C and stored at ?20˚C.
2.4.2. Protocol 2― SALAZAR (1998)
Red blood cells in 4 mL of blood were lysed with 8 mL of TM1-TRITON (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, pH 8.0, and 25 mL/L Triton X-100) for 15 minutes at room temperature. After centrifugation (500 ×g/10 min), the pellet was washed with 6 mL of TM1 (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, pH 8.0) until hemoglobin removal. The nuclei of leukocytes was lysed with 500 µL of TM2 (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, pH 8.0, 0.4 M NaCl) and 50 µL of SDS 10% at 37˚C overnight. The proteins were removed by saline precipitation with 80 µL of NaCl (5 M) and centrifugation (10,000 ×g/5 min). The supernatant was transferred to a new tube, DNA precipitated with 2 volumes of ice cold ethanol and washed, for salt removing, with ethanol 70%. The elution was made in 100 µL of TE (10 mM Tris-HCl, 0.2 mM Na2EDTA, pH 7.5) in a water bath for 15 minutes at 60˚C. The DNA was stored at ?20˚C until further use.
2.4.3. Protocol 3―QIAamp DNA Blood Mini Kit Silica Column
The extraction using QIAamp DNA Blood Mini Kit (Qiagen, Germany) was performed following manufacturer’s instructions. Briefly, 30 μL of proteinase K (100 mg/mL), 400 μL of blood and 400 μL of AL buffer were added, in this order, to a microcentrifuge tube and mixed by inversion. After incubation at 56˚C for 10 minutes, 200 μL of absolute ethanol was added and the tube mixed again by pulse-vortexing for 15 seconds. The mixture was transferred to the spin column and centrifuged (6000 ×g/1 min). The column was then placed in a clean 2 mL tube, 500 μL of AW1 buffer was added and after centrifugation (6000 ×g/1 min) the column was placed into a new collection tube. The AW2 buffer (500 μL) was added, and the column was centrifuged (20,000 ×g/3 min). The column was then placed in a new 1.5 mL microcentrifuge tube, 200 μL double-distilled water was added, incubated at room temperature for 5 minutes and finally centrifuged (8000 ×g/3 min) to recover DNA. The eluted DNA was stored at ?20˚C until further use.
2.5. Genomic DNA Quantification and Purity Measurements
The concentration and purity of genomic DNA were determined by measuring the absorbance at 260 nm and A260:A280 absorbance ratios respectively, using a spectrophotometer NanoDrop ND-1000.
2.6. Genomic DNA Integrity and PCR Product Verification
Agarose gel electrophoresis was used to determine the integrity of the DNA. Briefly, 5 µL of genomic DNA mixed with 1 µl of loading buffer (50% of Glycerol, 0.125% of Bromophenol blue, 0.125% of Xylene Cyanol, TE pH 8.0) was loaded onto 1% agarose gel with ethidium bromide (0.5 µg/mL) and electrophoresed for 45 minutes at 80 mV followed by visualization under UV light (Sambrook, 1998).
The size of the product (197 bp) was confirmed by electrophoresis on a 2% agarose gel containing 8 µL of the PCR products mixed with 1 µL of loading buffer and using a 100 base pair marker as a reference (Fermentas Life Sciences, Lithuania).
2.7. PCR Reaction
In order to evaluate DNA quality for amplification by PCR-RFLP, DNA samples of each protocol were assayed as previously described by Nomura (2007) with modifications. Briefly, the PCR reaction was carried out in a 50 µL mix containing 100 ng genomic DNA, Taq Buffer (Fermentas, MD, USA), 0.2 mM of dNTPs (Life Tech., Brazil), 0.2 µM of each primer (Forward: 5’-CTCCTCCTCCTCTGCTCCTC-3’; Reverse: 5’-GGGGCTAGCT- CGGGACTC-3’-Operon, USA), 5% of DMSO and 1.25 U Taq DNA Polymerase (Fermentas, MD, USA). Cycling conditions comprised an initial denaturation step for 10 minutes at 95˚C, followed by 35 cycles of three steps: 95˚C for 30 seconds, 60˚C for 30 seconds, and 72˚C for 1 minute. A final extension step was performed at 72˚C for 10 minutes.
2.8. Assessment of Student Acceptance
A survey with 5 questions regarding the students’ opinions about the summer course was applied on the last day for the 10 participants. The questions involved the evaluation of the class dynamics and how the course reached their expectation, as detailed in Table 2.
In order to assess if there is a public interest in attending the summer course, which justifies the assembly of it, ninety undergraduate students of biology and medicine were asked if they would like to perform a summer curse of molecular biology. They were previously informed about the content of the course (DNA extraction from blood, electrophoresis and PCR) and answered this question after a practical class of DNA extraction from fruits in the discipline of biochemistry.
3. Results and Discussion
According to Voet et al. (2003) , research experience is an essential part of any molecular biology curriculum, and may be obtained through well planned laboratory courses. Current literature reports about the lack of disciplines that not only get the themes in a practical perspective, but also involves data interpretation, problem solving, experimental design, scientific report writing and collaborative work, with critical analysis of primary results and possible errors (Coil, 2010; Azer, 2013) .
Table 2. Survey of students’ opinions about the summer course.
In the present work, the proposed Summer Course in Molecular Biology challenged the students from biomedicine and biology undergraduate courses to isolate and manipulate DNA from human blood samples. Thus, these future specialists could get first-hand contact with the professional laboratory environment, techniques and equipment. Herein we report the results of the students’ experience and reports about the comparison of three DNA extraction techniques performed during this summer course.
3.1. DNA Yield and Purity
DNA extraction for DNA collection samples is a critical procedure as it is time-consuming and may involve ethical aspects. The three protocols proposed here included two in-house procedures of low cost (Aidar, 2007) and low-toxicity as it does not use phenol-chloroform extraction (Lum, 1998; Rogers, 2007 ).
According to the students’ assay reports, the concentration of DNA samples were 260.1 ± 44.9 ng/µL (26.3 ? 529.6 ng/µL), 268.3 ± 43.7 ng/µL (131.4 - 463.5 ng/µL) and 33.8 ± 3.4 ng/µL (16.5 - 46.4 ng/µL), with a total DNA mass of 26.0, 26.8 and 6.8 µg to Protocols 1, 2 and 3 respectively (Figure 2(a) and Figure 2(b)). The two in-house procedures extracted 4 more times DNA than the commercial silica column (p < 0.001). However, considering the DNA mass corrected per blood volume used on extractions (6.503 ± 1.122 ng, 6.707 ± 1.092 ng and 33.75 ± 3.319 ng/ml of blood to Protocols 1, 2 and 3 respectively) commercial silica column was the most profitable protocol (p < 0.001; Figure 2(c)).
The 260/280 ratio is used to assess the purity of DNA and RNA. A ratio of ~1.8 is generally accepted as “pure” for DNA (Sambrook, 1989) . The OD 260/280 ratio was 1.838 ± 0.014 (1.74 - 1.87), 1.831 ± 0.028 (1.60 - 1.89) and 1.779 ± 0.047 (1.55 ? 2.02) to Protocols 1, 2 and 3 respectively. The student data showed that the in-house procedures provided DNA with good purity with no statistical difference of the commercial silica column (p > 0.05, Figure 2(d)).
Although silica columns involved a four step procedure: 1) blood cells lysis, 2) DNA bind to the silica, 3) silica wash and 4) DNA elution from silica (Figure 1(a)), they were performed in only one day. The students’ results showed that the in-house methods were able to extract large quantities of DNA in three days (Figure 1(a)). The comparison of the students revealed that the salt-precipitation method is economically more viable while the technique described by Salazar (1998) can be performed in a smaller number of days since nuclei lysis could be carried out at 65˚C for 30 minutes. The students detected these protocols advantages and disadvantages and informed in their reports.
3.2. DNA Integrity
The student assays reports described all DNA extraction procedures (Figure 2). They described that some samples obtained by using protocol 2 presented a smearing, indicating degradation (Figure 2(e)). It is also possible to see a pattern of bands, characteristic of apoptosis, in samples 1S and 3S. According to Takayama (2001) , it is common to observe apoptotic DNA fragments in samples stored at room temperature or stored for up to 2 days at 4˚C, with no implications for the PCR. The blood samples were collected at the beginning of classes and were kept at least 3 hours at room temperature prior to be extracted. To avoid this, the students suggested that in the



Figure 2. DNA concentration (a), purity (b), mass (c) and yield per milliliters of blood (d) obtained on each protocol of DNA extraction assay performed by the students (***p < 0.0001). (e) Agarose gel electrophoresis of genomic DNA obtained by three different DNA extractions. Samples (5 µL) were separated in 1% agarose gel for 45 minutes at 80 mA and stained with ethidium bromide. The numbers refer to the students DNAs and the letters to the extraction techniques used (M―Protocol 1- Miller, S―Protocol 2-Salazar, C―Protocol 3-silica column). (f) Agarose gel electrophoresis of PCR products. Samples (8 µL) were separated in 2% agarose gel for 45 minutes at 80 mA and stained with ethidium bromide. The numbers refer to the students DNAs and the letters to the extraction techniques used (PM―molecular weight, M―Protocol 1-Miller, S―Protocol 2-Salazar, C―Protocol 3-Silica column).
next course, the blood collection be carried out after the preparation of DNA extraction solutions (Table 1). The Protocol 2 seems to promote precipitation of fragmented DNA as it was detected apoptotic DNA fragments from apoptotic leukocytes (Figure 2(e)).
3.3. PCR Amplification
Polymerase chain reaction (PCR) is an important tool in molecular biology, genetics, forensics, and other fields. This technique is used to manipulate DNA for cloning and mutagenesis, and it can be used to detect genetic disorders, DNA fingerprinting, and identification of criminals from small samples of bodily fluids and hair (Sambrook, 1989) . The presence of contaminants, DNA degradation or low quantity of the DNA could inhibit PCR reaction (Gryson, 2010) .
The students of biomedicine and biological sciences were able to analyze their own blood in this course. PCR reactions using DNA samples obtained from protocol 2 and commercial kit were successfully performed on all samples, and in 80% of the samples extracted by protocol 1 (Figure 2(f)). According to the students’ observation, the failure in amplifying the two DNA samples from protocol 1 was due to the students handling error since samples 5 M and 9 M were successfully amplified in a second reaction.
3.4. Assessment of Student Learning, Acceptance and Demand
Student learning was assessed by: instructor observation of the student laboratory work and a questionnaire presented in Table 2. The results of the answers of the 10 participants are presented in Figure 3.



Figure 3. Evaluation of the summer course by the students of biomedicine and biological sciences. (a) Perception of Learning as inferred from the answers to the question 1 (Q1) of the questionnaire (n = 9 answers). (b) Mean Rating on a Lickert 1 - 5 Scale (1 = completely disagree, 5 = completely agree) to the understanding of content, dynamics of the course, reaching initial expectations and quality of course (results as mean ± SD, n = 10 answers). (c) Evaluation of the features that the students liked the most (blue bars), or would change (red bars) on the Summer Course (n = 9 answers).
According to the answers to the first question (Q1), most students felt that they learned molecular biology-related content (e.g. PCR, DNA isolation and amplification), indicating that the course somehow reached its main objective. In addition, some answers also stated that they learned practices pertaining to the daily routine of a laboratory (Figure 3(a)). This kind of statement reveals the potential of this dynamic as a practical course involving real equipment and laboratory techniques approximating the undergraduate students to their future professional environment.
The rates of the course quality and dynamics (1 - 5 Likert scale) determined by the students were all similar (~4) to both understanding of content and dynamics, reaching the initial expectations and course quality (Figure 3(b)). Overall, these results indicate a very good acceptance of the Course format, even though indicating that some changes could be performed to be totally approved by students, according to Q6.
Figure 3(c) shows what the students liked the most, or would prefer to change on the Course. The fact that they were able to learn the techniques by hands-on activities led most of them to enjoy the course, while at least one statement reported the simulation of the researchers’ activities and group interactions. Once again, students enjoyed and identified the importance of the practical activities where they were in first-hand contact with professional “researcher laboratory” activities.
Regarding to the suggestions of changes (red bars on Figure 3), students asked for improvements on the infrastructure (3 students), more interactive activities (2 students) and more molecular biology content (1 student). In fact, one important concern in practical courses is the demand for adequate infrastructure, especially concerning the availability of individual equipments, to avoid adopting a “demonstrative” profile for these activities.
Nevertheless, in agreement with the good acceptance revealed in Figure 3(b), all students stated that they would like to attend other editions of the Summer Course, when answering the Question 7. When asking other 90 students from medicine and biology courses (47% biology and 53% medicine) that had had the class using fruit as DNA source about attending a summer course regarding molecular biology, many of them answered positively (62%) about this possibility (48% medicine and 52% biology), which pointed to the need of knowing more about this subject and possibly for more applied classes.
4. Final Considerations
According to different authors, there is an urgent need for different approaches with an applied perspective for teaching biotechnology and molecular biology. In this work, we create a dynamic and practical course that receives positive evaluation by the students of biological sciences and biomedicine. The students’ reports reveal that the DNA extraction protocols evaluated here are simple and suitable for using PCR. The use of in-house procedures that are lower-cost methods and that can be performed in just one day (e.g. Protocol 2) allows the inclusion of this practical class in the disciplines of undergraduate courses of Health and Biological Sciences areas.
Moreover, another important feature of the parallel evaluation of three different methods during the summer course includes that the students are challenged with 3 different options, and they have to consider other parameters such as costs, time-consumption, availability of infrastructure, evaluation and comparison of results, quality assessment and other advanced issues usually not covered in classical courses. In this regard, students are able to conclude that even though the use of commercial kit shortens the experimental time, it is much more expensive than the in-house version, even though with similar or even better results. Such kind of stimuli may contribute to the potential development of critical thinking and problem-solving skills, both highly praised on cell and molecular biology curricula (Voet et al., 2003) , and extremely related to the reality of the laboratory/work environment of these professionals.
Acknowledgements
We thank to FAPERJ, UFF-FOPESQ, CAPES, CNPq and CNPq/PIBIC for the fellowships and financial support.
References
- Aidar, M., & Line, S. R. (2007). A Simple and Cost-Effective Protocol for DNA Isolation from Buccal Epithelial Cells. Brazilian Dental Journal, 18, 148-152.
- Azer, S. A., Guerrero, A. P., & Walsh, A. (2013). Enhancing Learning Approaches: Practical Tips for Students and Teachers. Medical Teacher, 35, 433-443.
- Badyal, D. K., & Desai, C. (2014). Animal Use in Pharmacology Education and Research: The Changing Scenario. Indian Journal of Pharmacology, 46, 257-265.
- Cardoso, C. V. P., & Almeida, A. E. C. C. (2014). Laboratory Animal: Biological Reagent or Living Being? Brazilian Journal of Medical and Biological Research, 47, 19-23.
- Coil, D., Wenderoth, M. P., Cunningham, M., & Dirks, C. (2010). Teaching the Process of Science: Faculty Perceptions and an Effective Methodology. CBE-Life Sciences Education, 9, 524-535.
- González, C. I., Ortiz, S., & Solari Colombian, A. (2010). Trypanosoma Cruzi Major Genotypes Circulating in Patients: Minicircle Homologies by Cross-Hybridization Analysis. International Journal for Parasitology, 40, 1685-1692.
- Gryson N. (2010). Effect of Food Processing on plant DNA Degradation and PCR-Based GMO Analysis: A Review. Analytical and Bioanalytical Chemistry, 396, 2003-2022.
- Hearn, R. P., & Arblaster, K. E. (2010). DNA Extraction Techniques for Use in Education. Biochemistry and Molecular Biology Education, 3, 161-166.
- Marchese, A., Esposito, S., Coppo, E., Rossi, G. A., Tozzi, A., Romano, M., Da Dalt, L., Schito, G. C., & Principi, N. (2011). Detection of Streptococcus pneumoniae and Identification of Pneumococcal Serotypes by Real-Time Polymerase Chain Reaction Using Blood Samples from Italian Children ≤5 Years of Age with Community-Acquired Pneumonia. Microbial Drug Resistance, 20, 1-6.
- Mello, F. C., Souto, F. J., Nabuco, L. C., Villela-Nogueira, C. A., Coelho, H. S., Franz, H. C., Saraiva, J. C., Virgolino, H. A., Motta-Castro, A. R., Melo, M. M., Martins, R. M., & Gomes, S. A. (2007). Hepatitis B Virus Genotypes Circulating in Brazil: Molecular Characterization of Genotype F Isolates. BMC Microbiology, 23, 103.
- Miller, S. A., Dykes, D. D., & Polesky, H. F. (1988). A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Research, 16, 1215. http://dx.doi.org/10.1093/nar/16.3.1215
- Miller, M. B. (1994). Practical DNA Technology in School. Journal of Biological Education, 28, 203-211. http://dx.doi.org/10.1080/00219266.1994.9655393
- Mortarino, M., Garagiola, I., Lotta, L. A., Siboni, S. M., Semprini, A. E., & Peyvandi, F. (2011). Non-Invasive Tool for Foetal Sex Determination in Early Gestational Age. Haemophilia, 17, 952-956. http://dx.doi.org/10.1111/j.1365-2516.2011.02537.x
- Nomura, M., Shigematsu, H., Li, L., Suzuki, M., Takahashi, T. et al. (2007). Polymorphisms, Mutations, and Amplification of the EGFR Gene in Non-Small Cell Lung Cancers. PLoS Medicine, 4, e125. http://dx.doi.org/10.1371/journal.pmed.0040125
- Poh, J. J., & Gan, S. K. (2014). Comparison of Customized Spin-Column and Salt-Precipitation Finger-Prick Blood DNA Extraction. Bioscience Reports, 34, 629-634. http://dx.doi.org/10.1042/BSR20140105
- Rogers, N. L., Cole, S. A., Lan, H. C., Crossa, A., & Demerath, E. W. (2007). New Saliva DNA Collection Method Compared to Buccal Cell Collection Techniques for Epidemiological Studies. American Journal of Human Biology,19, 319- 326. http://dx.doi.org/10.1002/ajhb.20586
- Salazar, L. A., Hirata, M. H., Cavalli, S. A., Machado, M. O. et al. (1998). Optimized Procedure for DNA Isolation from Fresh and Cryopreserved Clotted Human Blood Useful in Clinical Molecular Testing. Clinical Chemistry, 44, 1748-1450.
- Sambrook, J., Fritsch, E. R., & Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual (2nd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schijman, A. G., Bisio, M., Orellana, L., Sued, M., Duffy, T., Mejia Jaramillo, A. M. et al. (2011). International Study to Evaluate PCR Methods for Detection of Trypanosoma cruzi DNA in Blood Samples from Chagas Disease Patients. PLoS Neglected Tropical Diseases, 5, e931. http://dx.doi.org/10.1371/journal.pntd.0000931
- Slonczewski, J. L., & Marusak, R. (2004). A Response to BIO 2010: Transforming Undergraduate Education for Future Research Biologists, from the Perspective of the Biochemistry and Molecular Biology Major Program at Kenyon College. Biochemistry and Molecular Biology Education, 32, 151-155. http://dx.doi.org/10.1002/bmb.2004.494032030342
- Sitaraman, R. (2012). From Bedside to Blackboard: The Benefits of Teaching Molecular Biology within a Medical Context. Perspectives in Biology and Medicine, 55, 461-466. http://dx.doi.org/10.1353/pbm.2012.0030
- Takayama, T., Yamada, S., Watanabe, Y., Hirata, K., Nagai, A., Nakamura, I., Bunai, Y., & Ohya, I. (2001). Origin of DNA in Human Serum and Usefulness of Serum as a Material for DNA Typing. Legal Medicine, 3, 109-113. http://dx.doi.org/10.1016/S1344-6223(01)00018-9
- Tsui, N. B. Y., Kadir, R. A., Chan, K. C. A., Chi, C., Mellars, G., Tuddenham, E. G., Leung, T. Y., Lau, T. K., Chiu, R. W. K., & Dennis Lo, Y. M. (2011). PCR Analysis of Maternal Plasma DNA Noninvasive Prenatal Diagnosis of Hemophilia by Microfluidics Digital. Blood, 117, 3684-3691. http://dx.doi.org/10.1182/blood-2010-10-310789
- Tibell, L. A., & Rundgren, C. J. (2010). Educational Challenges of Molecular Life Science: Characteristics and Implications for Education and Research. CBE Life Sciences Education, 9, 25-33.
- Voet, J. G., Bell, E., Boyer, R., Boyle, J., O’Leary, M., & Zimmerman, J. K. (2003). Recommended Curriculum for a Program in Biochemistry and Molecular Biology. Biochemistry and Molecular Biology Education, 31, 161-162. http://dx.doi.org/10.1002/bmb.2003.494031030223
NOTES

*Corresponding author.