American Journal of Plant Sciences
Vol.07 No.10(2016), Article ID:69135,12 pages
10.4236/ajps.2016.710140
Phenolic Profiles and Polyphenol Oxidase (PPO) Gene Expression of Red Clover (Trifolium pratense) Selected for Decreased Postharvest Browning
Isabelle A. Kagan1*, Randy D. Dinkins1, Norman L. Taylor2#
1USDA-ARS, Forage-Animal Production Research Unit, Agricultural Science Center North, University of Kentucky, Lexington, KY, USA
2Department of Plant and Soil Sciences, University of Kentucky, Lexington, KY, USA

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 30 June 2016; accepted 24 July 2016; published 27 July 2016
ABSTRACT
Red clover (Trifolium pratense L.) is a legume forage abundant in phenolic compounds. It tends to brown when cut for hay, due to oxidation of phenolic compounds catalyzed by polyphenol oxidase (PPO), and subsequent binding to proteins. Selecting for a greener hay may provide information about the relationship of browning, PPO, and phenolics to each other. The red clover Kenland cultivar was selected over eight breeding cycles for decreased browning after being cut and dried 48 h in the field. Expression of PPO1 and PPO3, in Kenland and three of the eight cycles, was compared by real-time quantitative PCR. Phenolic compounds in Kenland and Cycle 8, collected 0, 24, and 48 h after cutting, were quantified by high-performance liquid chromatography (HPLC). Visual browning scores decreased 12% between Kenland and Cycle 8 (P = 0.02). PPO1 and PPO3 gene expression were not affected by selection. Clovamide decreased 26% in Cycle 8 relative to Kenland (P = 0.016). Sissotrin decreased 10% in Cycle 8 (P = 0.043). Neither total formononetin nor total biochanin A was affected by selection (P = 0.63 and 0.45, respectively). These results suggest that when selecting clover for decreased postharvest browning, a decrease occurs in a phenolic compound that can bind protein independently of PPO. However, PPO1 and PPO3 gene expression, and the major red clover isoflavones, are minimally affected.
Keywords:
Trifolium pratense L., Browning, PPO, Clovamide, Isoflavone, HPLC

1. Introduction
Red clover (Trifolium pratense L.) is a legume that is widely grown in the United States as a forage crop [1] . It contains a variety of soluble phenolic compounds, including phenolic acids, phenolic acid esters, flavonoids, and isoflavonoids [2] . Among about 20 red clover isoflavonoids [3] , the most abundant are biochanin A and formononetin (Figure 1(a)), with genistein and daidzein occurring at 5 to 10% of the concentrations of biochanin A and formononetin [4] . These latter two compounds are present primarily as malonylglucosides (Figure 1(b)) in planta [5] - [7] . Two dominant caffeic acid derivatives are phaselic acid and clovamide (Figure 1(c)) [8] - [10] . These are o-diphenols (compounds with hydroxy groups ortho to each other on the benzene ring).
These secondary metabolites have various effects on ruminant health. Formononetin has been linked to decreased fertility in sheep [11] . In ovine [12] and bovine [13] rumen fluid, it is converted into daidzein and then into equol, which may be the molecule primarily responsible for ovine reproductive disorders [14] . Equol and p-ethylphenol, a metabolite of biochanin A, have been linked to decreased bovine reproduction [15] . However, biochanin A may improve assimilation of protein, due to its ability to inhibit bovine [16] and caprine [17] ru-



Figure 1. Structures of selected phenolic compounds of red clover (Trifolium pratense L.).
minal hyper ammonia-producing bacteria (HAB) in vitro.
Phaselic acid and clovamide provide another mechanism for decreasing protein losses in the rumen. Studies have revealed a positive correlation between low rates of red clover proteolysis and the presence of both o-diphenols and polyphenol oxidase (PPO) [18] [19] . In intact plant cells, phenolic compounds are stored in the vacuole, while PPO is located in the plastid [20] . Wounding (e.g. mowing) or cell death disrupt cell walls and organelle membranes, leading to reactions between normally isolated molecules [20] . PPO catalyzes the oxidation of compounds with an o-diphenol moiety to o-quinones, which then react non-enzymatically with amino acids to form brown protein-phenolic complexes [21] . Phaselic acid is a PPO substrate, while clovamide can bind to proteins in the presence or absence of PPO [8] [9] .
The abovementioned browning that follows wounding or cell death, and that is often observed in harvested red clover [22] , is probably due in part to the brown protein-phenolic complexes formed upon tissue damage. Other contributing factors may be the color of oxidized phenolics alone, or of polymers of those oxidation products [23] [24] . The relationship between browning, concentration or activity of PPO, and amount of available soluble phenolic substrates is complex [22] , and selection for decreased post-harvest browning could clarify this relationship. Greater retention of green color after harvest could be due to decreased PPO activity, possibly with parallel decrease in gene expression. Postharvest retention of green color could also imply a decrease in phenolic PPO substrates, with lower overall concentrations or with a relative increase in the phenolic compounds lacking the o-diphenol moiety (e.g. the major red clover isoflavones). Jones et al. [22] found no relationship between browning and concentrations of total soluble phenolics but suggested that specific phenolic compounds might be implicated in the relationship. Browning of extracts of fresh tissue is linked to the expression of PPO and phenolic PPO substrates [25] , but browning due to curing in the field has not been linked to PPO and phenolics. Selection for decreased postharvest browning could have some commercial value because of the visual appeal of green hay to some animal producers. This appeal is not completely unfounded because in timothy, greener color has been correlated with greater protein content, although rumen degradability was not measured [26] . The following study was undertaken to analyze the phenolic profiles and PPO gene expression of Kenland red clover selected over eight breeding cycles for decreased browning after cutting, in a manner intended to simulate the browning that might occur in cut forage being cured for hay.
2. Materials and Methods
2.1. Selection for Decreased Browning and Evaluation of Multiple Cycles
The selection process was directed from 1995 to 2010. About 1500 to 3000 spaced red clover plants were grown at the University of Kentucky Spindletop Research Farm Lexington, KY, USA (38˚10'N, 84˚49'W). In 1995, selections were made from the Kenland variety. A few shoots of each plant were cut near the base. Plants whose cuttings appeared less brown than others after 48 to 72 h of curing (5% to 8% of the total number of spaced plants) were retained and allowed to intercross. The seed from this selection cycle was sown the following year, and the process of cutting, marking the least brown plants after 48 to 72 h, and saving seed was repeated. At the time of this study, eight selection cycles had taken place. The final number of cycles in 2010 was thirteen, due to losses in a couple of years.
For comparison of browning, PPO expression, and phenolic profiles among the eight selection cycles, seed from those cycles, plus the original Kenland cultivar, were planted in a randomized block design with four replicates. Each replicate consisted of nine rows, ~2 m long, for Kenland and the eight cycles. Clover seed was sown on 16 April 2004, and the same stands were used for two consecutive years. Plants were cut three times in 2004 (13 July, 13 August, and 20 September), and twice in 2005 (16 May and 21 June). Each cutting date was at the early flowering stage. The difference in the first cutting date between the two years was due to the longer time needed in 2004 for growth of a stand started from seed. In August and September 2004, the mean temperatures near the harvest site were 22 and 20˚C, respectively, while in May and June 2005, mean temperatures were 16 and 24˚C, respectively. No precipitation occurred during these curing periods.
Immediately prior to cutting the clover stands in 2004, leaves were collected for RNA analysis (one leaf from each of five plants per selection cycle in each of two replicate blocks) and promptly frozen in liquid nitrogen. Within each row, all plants were trimmed (usually around mid-morning) to 5 cm above the soil line with hand scythes. Stems and leaves from three to six plants per row were collected immediately after cutting in order to obtain phenolic profiles prior to curing. Plants were allowed to dry in the field for 48 to 72 h in order to simulate curing of hay. Tissue was collected 24 and 48 h after cutting, in order to analyze changes in phenolic profiles over time. Tissue was transported on dry ice to storage at −20˚C, and freeze-dried (Model 18DX48SA freeze dryer; Botanique Preservation Equipment, Peoria, AZ, USA). The freeze-dried material was ground to pass a 1-mm mesh in either a Wiley mill (Thomas Scientific, Swedesboro, NJ, USA) or a cyclone mill (Foss, Eden Prairie, MN; harvest 2 of 2004 and a few samples from harvest 1 of 2004) and stored at −20˚C until extracted.
2.2. Assessment of Changes in Browning
Cut clover was scored for browning on a scale of 1 to 9 (1 = green, and 9 = very dark brown/black). Visual assessments were made 24, 48, and 72 h after cutting, although color changes at 24 h were sometimes not pronounced enough to observe any differences among selections. In 2005, the 24- and 48-h scores from the June harvest were evaluated by analysis of variance (ANOVA) using the GLM procedure from SAS (Cary, NC, USA).
2.3. PPO RNA Extraction and Gene Expression Analysis
Total RNA from Kenland and selection cycles 1, 7, and 8 was isolated from 100 mg of leaf sample in 1mL of TRIzolâ reagent (Invitrogen™, Life Technologies, Carlsbad, CA). The total RNA isolated was purified by running the samples through Qiagen columns (RNeasy Plant Mini Kit, Qiagen Inc., Valencia, CA) combined with an on-column DNase digestion (RNase-Free DNase set, Qiagen Inc., Valencia, CA) to ensure DNA-free RNA preparations. RNA was extracted from two biological replicates of each cycle in this manner. Expression levels of the clover PPO1 and PPO3 genes, which are expressed in leaves [25] , were monitored by quantitative real- time reverse transcription PCR (qRT-PCR) using SYBR Green I [27] . Real-time PCR quantification was performed by designing specific oligonucleotide primers for the following genes, using PrimerQuestSM software (Integrated DNA Technologies, Coralville, IA).
PPO1 (Forward primer: 5’-GAACAAAAAGCATCACCTAGAAG-3’; Reverse primer: 5’-ACTGATTGGAGGGTCTATATTTT-3’),
PPO3 (Forward primer: 5’ GAACAAAAAGCATCACCTAGAAG-3’; Reverse primer: 5’-TGGACAACAATTGATGTTCACGG-3’)
a-Tubulin (Forward primer: 5’-AGGTTTGTGTAAGTGGGACGCTCA-3’; Reverse primer: 5’-ACACACTGATGTCGCTGTTCTCCT-3’).
Polymerase chain reactions were carried out in an iCyclerTM iQ detection system (Bio-Rad Laboratories, Hercules, CA) using SYBR Green I to monitor dsDNA synthesis. For negative controls, the cDNA samples of the same treatments and DNase treated minus RT controls were used. Real-time PCR amplification was performed in a total volume of 20 µl reaction mixture containing 1 µl of cDNA gene specific primers, SYBR Green I (Molecular Probes, Eugene, OR) and Platinum Taq DNA polymerase (Invitrogen, Life technologies, Carlsbad, CA). Each sample was loaded in triplicate and the experiments were repeated twice using the following thermal cycling program conditions: initial denaturation for 2 min at 95˚C, 95˚C 30 s, 55˚C 30 s and 72˚C 30 s for 35 cycles followed by 5 min extension at 72˚C. In order to compare the transcript levels of a gene from different samples, the values were normalized relative to transcripts of a-tubulin, which is assumed to be at a constant level in all the samples. Melt curve analysis was done to characterize the gene-specific dsDNA product by slowly raising the temperature (0.2˚C/10s) from 60˚C to 95˚C with fluorescence data collected at 0.2˚C intervals [28] . A sample of the PCR products was sequenced to verify the correct amplification.
2.4. Reagents and Chemicals
Methanol (HPLC grade) was purchased from EMD Chemicals (Gibbstown, NJ, USA). Ethyl ether, caffeic acid, chlorogenic acid, and catechol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ethyl acetate, acetic acid, hydrochloric acid, and magnesium sulfate were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Formononetin and biochanin A were purchased from both Sigma-Aldrich and Indofine Chemical (Hillsboro, NJ, USA). Sissotrin (biochanin A 7-O-glucoside), ononin (formononetin 7-O-glucoside), genistein, and daidzein were purchased from Indofine. Clovamide (caffeoyl-DOPA, or N-[3’,4’-dihydroxy-(E)-cinna- moyl]-3-hydroxy-L-tyrosine) was purchased from Santa Cruz Biotechnology Company (Santa Cruz, CA, USA). Phaselic acid and p-coumaroyl malate were a kind gift from Drs. Wayne Zeller and Paul Schatz (USDA-ARS Dairy Forage Research Center, Madison, WI, USA). The synthesis of phaselic acid has been described by Zeller [29] , and the synthesis of p-coumaroyl malate has been described by Sullivan and Zarnowski [30] .
2.5. Extractions of Phenolic Compounds from Red Clover Tissue
Samples harvested in August 2004, September 2004, May 2005, and June 2005 were analyzed in this study. The original Kenland cultivar and selection cycle 8 (hereafter referred to as Cycle 0 and Cycle 8, respectively) were analyzed from each harvest because the greatest differences were expected to be between the initial and final cycles. Soluble phenolics were extracted from freeze-dried, ground clover tissue (125 mg) in methanol-acetic acid-water (85:0.5:14.5, v/v/v), and partitioned with ethyl acetate-ethyl ether (1:1, v/v), according to the method of Kagan and Flythe [31] .
2.6. Separation of Phenolic Compounds by High-Performance Liquid Chromatography (HPLC)
Phenolic extracts were resuspended in 1.0 mL MeOH, and an aliquot was diluted 5-fold in a MeOH-acetic acid solution so that the final solvent ratio was MeOH-water-acetic acid (60:39.6:0.35, v/v/v). Dilutions (15 µL) were injected onto a 4.6 × 250 mm C18 column (LiChrospher RP-18 adsorbent from EMD Chemicals; packed by Grace Davison Discovery Sciences, Deerfield, IL, USA) on an Agilent (Santa Clara, CA) model 1100 HPLC. Separation was done at 28˚C and a flow of 0.8 mL/min, using the method of Kagan and Flythe [31] . Absorbance, on a single-wavelength detector, was monitored at 270 nm.
2.7. Identification and Quantification of Phenolic Compounds
Phaselic acid, clovamide, and p-coumaroyl malate were identified initially by spiking extracts with standards of those compounds and verifying the coelution of sample peaks with the standards. The same was done to identify formononetin, ononin, biochanin A, and sissotrin. Subsequently, these compounds were identified by comparing retention times of peaks in extracts to the retention times of standards. Consistency of retention times was verified by injecting standards of clovamide, biochanin A, genistein, daidzein, and formononetin with every HPLC run. Phaselic acid, sissotrin, and ononin, due to scarcity, were not injected regularly, but peaks were identifiable because of minimal shifts in retention time (standard deviation of 0.1 to 0.4 min for 11 compounds identified in 96 injections). Formononetin glucoside malonate (peak G, Figure 2) and biochanin A glucoside malonate (peak I, Figure 2) were identified by hydrolyzing extracts under conditions based on the methods of de Rijke et al. [6] . Extracts (0.7 mL), diluted in the proportions of MeOH, water, and acetic acid used for HPLC, were heated 75 min at 80˚C - 87˚C. Increases in the concentrations of sissotrin and ononin were compared to decreases in the concentrations of the corresponding malonylglucoside peaks, to verify that changes in the glucosides and malonylglucosides were of a similar magnitude. Genistein and daidzein were not found in extracts when standards were coinjected in a preliminary study.
In addition to injections of standards to verify retention times, calibration curves of biochanin A and caffeic acid were run during each period of regular sample injection in order to quantify compounds. Phaselic acid, p-coumaroyl malate, and clovamide were quantified as equivalents of caffeic acid, and isoflavonoids were quantified as equivalents of biochanin A. Over a 13-month period, three standard curves of biochanin A and caffeic acid had slopes with relative standard deviations of 2.1% for biochanin A, and 4.3% for caffeic acid.
Besides the isoflavonoids identified by malonylglucoside hydrolysis or comparison of retention times to those of standards, two unidentified peaks (D and E, Figure 2) were quantified as isoflavones based on the spectra generated by a photodiode array detector (model PDA-100) on a Summit HPLC (Dionex, Bannockburn, IL, USA). Isoflavone spectra are characterized by an absorbance maximum at 240 - 280 nm, and a minor absorbance maximum at 300 - 380 nm [32] .
2.8. Statistical Analysis of Differences in Phenolic Profiles
Peaks on the 96 HPLC chromatograms were aligned with the peaks having the closest retention time, shape, and most similar neighboring peaks. Eleven compounds (indicated by uppercase letters on Figure 2) were chosen for analysis based on size, resolution, and known relationship to browning, PPO, or phytoestrogenic activity. Minor shoulder peaks, or the smallest unidentifiable peaks, were not analyzed because very small or poorly-
Figure 2. Chromatogram of an extract of Kenland red clover, collected immediately after cutting on 16 May 2005. The compounds indicated by lowercase letters are those analyzed for differences between cycles 0 and 8 of the selections for decreased browning. A, phaselic acid; B, clovamide; C, p-coumaroyl malate; D and E, unidentified isoflavones; F, formononetin glucoside (ononin); G, formononetin malonylglucoside; H, biochanin A glucoside (sissotrin); I, biochanin A malonylglucoside; J, formononetin; K, biochanin A.
resolved peaks, or shoulder peaks, were less likely to be reproducibly quantifiable. Concentrations of the 11 selected compounds were analyzed with JMP version 10 (SAS Institute, Cary, NC, USA), using analysis of variance (ANOVA) and a full factorial analysis of the effects of year (2004 or 2005), selection cycle (0 or 8), and collection time (0, 24, or 48 hours after cutting). Where significant (P < 0.05) differences were found, Student’s t-tests were used to compare phenolic concentrations.
3. Results
3.1. Visual Browning Scores of Selection Cycles
Differences among selections were generally difficult to see, but the analysis of selection cycle vs. browning score indicated a slight overall decrease in browning. Least squares means of the ratings for each selection indicated a fairly steady decrease in browning score between Kenland (6.5) and cycle 3 (5.4). However, cycles 4 and 5 appeared slightly more brown (scores of 5.5 and 6.1, respectively); and after a decrease in browning in cycles 6 and 7 (4.8 and 4.6, respectively), a slight increase was observed in cycle 8 (score of 5.4). The overall trend in rating was towards a decrease (P = 0.023) in a manner modeled by Equation (1):
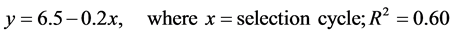 (1)
(1)
According to this equation, a shift towards a low browning score (3 to 4) would require 12 to 18 selection cycles.
3.2. Effects of Selection on PPO Gene Transcript Abundance in Kenland and Three Selection Cycles
Comparisons in gene expression were done between the early selection cycles (cycle 0, or Kenland, and cycle 1) and later cycles (cycle 7 and cycle 8). No significant differences were observed in the expression of the clover PPO genes from early to later selection cycles (Figure 3).
3.3. Effect of Selection on Red Clover Phenolic Profiles and Concentrations
The fingerprint of phenolic compounds did not change among samples. Concentrations changed, but no appearance or disappearance of compounds was observed. A main effect of selection for decreased browning was observed for the concentrations of clovamide (P = 0.016) and sissotrin (P = 0.043). When averaged across all collection times and harvests, clovamide concentrations were 27% less in Cycle 8 (4.7 ± 0.4 micromole per gram dry weight, µmol/gdw) than in Cycle 0 (6.4 ± 0.4 µmol/gdw), and biochanin A glucoside (sissotrin) concentrations were 10% less in Cycle 8 (1.8 ± 0.1 µmol/gdw) than in Cycle 0 (2.0 ± 0.1 µmol/gdw). No effects of selection cycle were observed for concentrations of total phenolics (P = 0.72, determined by summing concentrations of the 11 analyzed phenolic compounds), total formononetin as the sum of the aglycone and glycosides (P = 0.63), or total biochanin A as the sum of the aglycone and glycosides (P = 0.45).
3.4. Effects of Year or Curing Time on Red Clover Phenolic Profiles and Concentrations
In 2004, pronounced changes in the concentrations of some phenolic compounds were observable over a 48-h drying/curing period (Figure 2). These changes included a decrease in phaselic acid and clovamide (peaks A and B, respectively; Figure 2). Decreases in the malonylglucosides of formononetin (peak G) and biochanin A (peak I), and sharp increases in the aglycones of formononetin (peak J) and biochanin A (peak K), were also observable.
A main effect of curing time (Table 1) was observed for p-coumaroyl malate, isoflavones D and E, sissotrin
Figure 3. Relative expression (means ± standard deviations of two independent PCR experiments, with four replicates per experiment) of the red clover PPO1 and PPO3 genes compared to the control a-tubulin gene for four selection cycles (Cycle 0 is the original Kenland variety from which selections were made). The a-tubulin gene expression was set to 100%.
Table 1. Effect of sampling time (number of hours after cutting) on concentrations of red clover phenolic compounds, in micromole per gram of dry weight (µmol/gdw, least squares means ± standard error). Within a row, means with a common letter are not significantly different (P < 0.05). SE, standard error; p-coum-Mal, p-coumaroyl malate; fn, formononetin; biA, biochanin A.
(biochanin A glucoside), total phenolics, and total formononetin and biochanin A (sum of the aglycone, glucoside, and malonylglucoside). All decreased from cutting time to 48 h later.
A main effect of harvest year was observed for p-coumaroyl malate and sissotrin. The former increased (P < 0.0001) by 70% in 2005 (from 0.48 to 0.83 µmol/gdw), and the latter increased (P < 0.001) by 40% in 2005 (from 1.6 to 2.2 µmol/gdw).
An interaction of harvest year with curing time was observed for phaselic acid, clovamide, ononin, formononetin, biochanin A, and the malonylglucosides of formononetin and biochanin A (Table 2). In 2005, no change in concentration over time was measured in formononetin, biochanin A, clovamide, or ononin. In contrast, in 2004, concentrations of these compounds changed between 0 and 24 h after cutting, and in clovamide and ononin, changes also were measured between 24 and 48 h after cutting (Table 2). Phaselic acid, formononetin malonylglucoside, and biochanin A malonylglucoside decreased between 0 and 24 h after cutting in 2004, but in 2005, a decrease occurred only between 24 and 48 h after cutting.
4. Discussion
Analysis of visual browning scores (Equation (1)) demonstrated that the eight cycles of selection had led to a red clover line that browned slightly less than the original Kenland during a 48-h curing period, and that another four to eight selection cycles would be necessary to obtain clover with a browning score less than intermediate. These results demonstrate the difficulty of selecting for decreased browning entirely by visual evaluation.
The small magnitude of change in browning over eight selection cycles may partially explain why no changes in PPO1 or PPO3 gene expression were observed among cycles (Figure 3). It is also possible that measurement of enzyme activity would have revealed changes not revealed by gene expression analysis. However, monitoring expression of PPO1 and PPO3 may not have adequately captured PPO variation among selection cycles because it is now known that red clover has about six PPO genes [33] . The PPO4 gene, which was identified by Winters et al. [33] , was expressed more strongly than PPO1 in mature leaves of red clover [10] . Given the maturity of the red clover at harvest, it is possible that PPO4 expression would have been stronger than PPO1 or PPO3 expression, and more responsive to selection for decreased browning.
The small magnitude of change in browning may also partially explain why only clovamide and sissotrin concentrations were affected by selection. However, given the lack of change in PPO1 gene expression, the observed decrease in clovamide, which can bind to proteins in the presence or absence of PPO1 [9] , may indicate that decreased browning has the greatest effect on concentrations of phenolics that can bind protein independently of PPO1 activity. Dependence on PPO4 activity cannot be ruled out because PPO4 can utilize clovamide as a substrate [10] . Potential effects of a decrease in sissotrin (biochanin A glucoside) are difficult to assess because total biochanin A did not decrease in Cycle 8.
The effects of the 48-h curing period on phenolic concentrations, averaged across cycles 0 and 8, reflect the processes of oxidation, proteolysis, and loss of cellular integrity expected in wounded and dying plant tissue.
Table 2. Effects of sampling time and year on concentrations in micromole per gram of dry weight (µmol/gdw, least squares means ± standard error) of selected phenolic compounds in red clover. Within a row, means with a common letter are not significantly different (P < 0.05). SE, standard error; fn-MalGlc, formononetin malonylglucoside; biA-MalGlc, biochanin A malonylglucoside; fn, formononetin, biA, biochanin A.
The decrease in p-coumaroyl malate over time (Table 1) could be due to binding to proteins, because p-coumaroyl malate can inhibit proteolysis in alfalfa extracts, although the mechanism does not involve PPO or oxidation [9] . The observed decreases in isoflavones D, E, sissotrin, and total formononetin and biochanin A, may also indicate non-PPO-mediated binding to protein because formononetin and biochanin A lack an o-diphenol moiety, as do the majority of the red clover isoflavones [3] . The decreases observed over 48 h in phaselic acid and clovamide in 2004 (Table 2) suggest binding of PPO-oxidized diphenols to proteins. A feeding study utilizing freshly cut and conditioned (chopped and collected after 1 h) red clover demonstrated that protein-bound phenolics increased in conditioned red clover, concomitantly with a 10-fold decrease in phaselic acid and disappearance of clovamide [34] . In our study, the trend towards decreased phaselic acid and clovamide over time were affected by sampling year, with no change in clovamide in 2005, and a more gradual decrease in phaselic acid in 2005 than in 2004 (Table 2). This difference between years may be due to variations in mean temperatures. Lower temperatures during the May 2005 harvest might have led to slower disintegration of cells and consequently a decrease in the concentrations of o-diphenols available to react with PPO.
Decreases in the malonylglucosides of formononetin and biochanin A were observed over the 48-h curing period (Table 2). The malonylglucosides and glucosides of formononetin and biochanin A are readily hydrolyzed to the aglycones, due to the presence of glucosidases in plant tissue [7] . A decrease in formononetin glucoside (ononin), and increases in formononetin and biochanin A aglycones over 48 h, presumably due to hydrolysis of glycosides, were observed in 2004, but not in 2005 (Table 2). This discrepancy may be due to the temperature differences between the two years, as mentioned for phaselic acid and clovamide.
A key attribute of red clover in ruminant nutrition is a relatively low rate of protein degradation. Red clover lacks tannins, high-molecular-weight polymers of flavonoids or phenolic acids that bind protein and other macromolecules [35] . However, more protein escaped in vitro ruminal degradation in red clover than in other non- tannin-containing legumes like alfalfa (Medicago sativa) and cicer milkvetch (Astragalus cicer), and more than in two low-tannin cultivars of birdsfoot trefoil (Lotus corniculatus) [36] . Some feeding trials have demonstrated superior nitrogen use efficiency [37] or average daily gains [38] on red clover compared to other legume forages like alfalfa, although results have been variable [39] . The extent of ruminal red clover protein degradation may depend partly on environment [40] or genetics [41] . It is uncertain if a decrease in clovamide concentrations would significantly increase the rumen degradability of protein in the Cycle 8 selection. Phaselic acid was unchanged, and because total biochanin A was not affected by selection for decreased browning, protection from ruminal HAB would still be effective. In terms of other ruminant health aspects, a lack of change in total biochanin A and total formononetin indicates that estrogenic effects would be unlikely to increase in the Cycle 8 selection. Future work may involve analysis of later selection cycles to determine if those cycles reveal more pronounced changes in PPO gene expression and phenolic concentrations.
Acknowledgements
This manuscript is dedicated to Dr. Norman L. Taylor, who did the field selections and was responsible for the initial ideas on phenolic content in relation to the browning phenotype. Dr. Taylor passed away on 25 October 2010. The study was funded by the US Department of Agriculture. Mention of trade names or commercial products in the article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Cite this paper
Isabelle A. Kagan,Randy D. Dinkins,Norman L. Taylor, (2016) Phenolic Profiles and Polyphenol Oxidase (PPO) Gene Expression of Red Clover (Trifolium pratense) Selected for Decreased Postharvest Browning. American Journal of Plant Sciences,07,1478-1489. doi: 10.4236/ajps.2016.710140
References
- 1. Taylor, N.L. (2008) A Century of Clover Breeding Developments in the United States. Crop Science, 48, 1-13.
http://dx.doi.org/10.2135/cropsci2007.08.0446 - 2. Oleszek, W., Stochmal, A. and Janda, B. (2007) Concentration of Isoflavones and Other Phenolics in the Aerial Parts of Trifolium Species. Journal of Agricultural and Food Chemistry, 55, 8095-8100.
http://dx.doi.org/10.1021/jf072024w - 3. Lin, L.-Z., He, X.-G., Lindenmaier, M., Yang, J., Cleary, M., Qiu, S.-X. and Cordell, G.A. (2000) LC-ESI-MS Study of the Flavonoid Glycoside Malonates of Red Clover (Trifolium pratense). Journal of Agricultural and Food Chemistry, 48, 354-365.
http://dx.doi.org/10.1021/jf991002+ - 4. Krenn, L., Unterrieder, I. and Ruprechter, R. (2002) Quantification of Isoflavones in Red Clover by High-Performance Liquid Chromatography. Journal of Chromatography B, 777, 123-128.
http://dx.doi.org/10.1016/S1570-0232(02)00079-X - 5. Beck, A.B. and Knox, J.R. (1971) The Acylated Isoflavone Glycosides from Subterranean Clover and Red Clover. Australian Journal of Chemistry, 24, 1509-1518.
http://dx.doi.org/10.1071/CH9711509 - 6. De Rijke, E., Zafra-Gómez, A., Ariese, F., Brinkman, U.A.Th. and Gooijer, C. (2001) Determination of Isoflavone Glucoside Malonates in Trifolium pratense L. (Red Clover) Extracts: Quantification and Stability Studies. Journal of Chromatography A, 932, 55-64.
http://dx.doi.org/10.1016/S0021-9673(01)01231-6 - 7. Toebes, A.H.W., de Boer, V., Verkleij, J.A.C, Lingeman, H. and Ernst, W.H.O. (2005) Extraction of Isoflavone Malonylglucosides from Trifolium pratense L. Journal of Agricultural and Food Chemistry, 53, 4660-4666.
http://dx.doi.org/10.1021/jf047995f - 8. Lee, M.R.F., Tweed, J.K.S. and Sullivan, M.L. (2013) Oxidation of ortho-Diphenols in Red Clover With and Without Polyphenol Oxidase (PPO) Activity and Their Role in PPO Activation and Inactivation. Grass and Forage Science, 68, 83-92.
http://dx.doi.org/10.1111/j.1365-2494.2012.00873.x - 9. Sullivan, M.L. and Zeller, W.E. (2013) Efficacy of Various Naturally Occurring Caffeic Acid Derivatives in Preventing Post-Harvest Protein Losses in Forages. Journal of the Science of Food and Agriculture, 93, 219-226.
http://dx.doi.org/10.1002/jsfa.5781 - 10. Webb, K.J., Cookson, A., Allison, G., Sullivan, M.L. and Winters, A.L. (2013) Gene Expression Patterns, Localization and Substrates of Polyphenol Oxidase in Red Clover (Trifolium pratense L.). Journal of Agricultural and Food Chemistry, 61, 7421-7430.
http://dx.doi.org/10.1021/jf401122d - 11. Braden, A.W.H., Hart, N.K. and Lamberton, J.A. (1967) The Oestrogenic Activity and Metabolism of Certain Isoflavones in Sheep. Australian Journal of Agricultural Research, 18, 335-348.
http://dx.doi.org/10.1071/AR9670355 - 12. Nilsson, A., Hill, J.L. and Davies, H.L. (1967) An in Vitro Study of Formononetin and Biochanin A Metabolism in Rumen Fluid From Sheep. Biochimica et Biophysica Acta, 148, 92-98.
http://dx.doi.org/10.1016/0304-4165(67)90282-6 - 13. Dickinson, J.M., Smith, G.R., Randel, R.D. and Pemberton, I.J. (1988) In Vitro Metabolism of Formononetin and Biochanin A in Bovine Rumen Fluid. Journal of Animal Science, 66, 1969-1973.
- 14. Lundh, T.J.-O., Pettersson, H.I. and Martinsson, K.A. (1990) Comparative Levels of Free and Conjugated Plant Estrogens in Blood Plasma of Sheep and Cattle fed Estrogenic Silage. Journal of Agricultural and Food Chemistry, 38, 1530-1534.
http://dx.doi.org/10.1021/jf00097a022 - 15. Woclawek-Potocka, I., Bah, M.M., Korzekwa, A., Piskula, M.K., Wiczkowski, W., Depta, A. and Skarzynski, D.J. (2005) Soybean-Derived Phytoestrogens Regulate Prostaglandin Secretion in Endometrium During Cattle Estrous Cycle and Early Pregnancy. Experimental Biology and Medicine, 230, 189-199.
- 16. Flythe, M.D. and Kagan, I.A. (2010) Antimicrobial Effect of Red Clover (Trifolium pratense) Extract on the Ruminal Hyper-Ammonia Producing Bacterium, Clostridium sticklandii. Current Microbiology, 61, 125-131.
http://dx.doi.org/10.1007/s00284-010-9586-5 - 17. Flythe, M.D., Harrison, B., Kagan, I.A., Klotz, J.L., Gellin, G.L., Goff, B. and Aiken, G.E. (2013) Antimicrobial Activity of Red Clover (Trifolium pratense L.) Extract on Caprine Hyper-Ammonia-Producing Bacteria. Agriculture, Food and Analytical Bacteriology, 3, 176-185.
- 18. Sullivan, M.L. and Hatfield, R.D. (2006) Polyphenol Oxidase and o-Diphenols Inhibit Postharvest Proteolysis in Red Clover and Alfalfa. Crop Science, 46, 662-670.
http://dx.doi.org/10.2135/cropsci2005.06-0132 - 19. Winters, A.L., Minchin, F.R., Michaelson-Yeates, T.P.T., Lee, M.R.F. and Morris, P. (2008) Latent and Active Polyphenol Oxidase (PPO) in Red Clover (Trifolium pratense) and Use of a Low PPO Mutant to Study the Role of PPO in Proteolysis Reduction. Journal of Agricultural and Food Chemistry, 56, 2817-2824.
http://dx.doi.org/10.1021/jf0726177 - 20. Vaughn, K.C. and Duke, S.O. (1984) Function of Polyphenol Oxidase in Higher Plants. Physiologia Plantarum, 60, 106-112.
http://dx.doi.org/10.1111/j.1399-3054.1984.tb04258.x - 21. Bittner, S. (2006) When Quinones Meet Amino Acids: Chemical, Physical and Biological Consequences. Amino Acids, 30, 205-224.
http://dx.doi.org/10.1007/s00726-005-0298-2 - 22. Jones, B.A., Hatfield, R.D. and Muck, R.E. (1995) Screening Legume Forages for Soluble Phenols, Polyphenol Oxidase and Extract Browning. Journal of the Science of Food and Agriculture, 67, 109-112.
http://dx.doi.org/10.1002/jsfa.2740670117 - 23. Goodenough, P.W., Kessell, S., Lea, A.G.H. and Loeffler, T. (1983) Mono- and Diphenolase Activity from Fruit of Malus pumila. Phytochemistry, 22, 359-363.
http://dx.doi.org/10.1016/0031-9422(83)83005-2 - 24. Rouet-Mayer, M.A., Ralambosoa, J. and Philippon, J. (1990) Roles of o-Quinones and their Polymers in the Enzymic Browning of Apples. Phytochemistry, 29, 435-440.
http://dx.doi.org/10.1016/0031-9422(90)85092-T - 25. Sullivan, M.L., Hatfield, R.D., Thoma, S.L. and Samac, D.A. (2004) Cloning and Characterization of Red Clover Polyphenol Oxidase cDNAs and Expression of Active Protein in Escherichia coli and Transgenic Alfalfa. Plant Physiology, 136, 3234-3244.
http://dx.doi.org/10.1104/pp.104.047449 - 26. Evans, M.W. and Ely, J.E. (1936) Timothy Selection for Improvement in Quality of Hay. Journal of the American Society of Agronomy, 28, 941-947.
http://dx.doi.org/10.2134/agronj1936.00021962002800110008x - 27. Dinkins, R.D., Tavva, V.S., Palli, S.R. and Collins, G.B. (2012) Mutant and Over-Expression Analysis of a C2H2 Single Zinc Finger Gene of Arabidopsis. Plant Molecular Biology Reports, 30, 99-110.
http://dx.doi.org/10.1007/s11105-011-0320-7 - 28. Ririe, K.M., Rasmussen, R.P. and Wittwer, C.T. (1997) Product Differentiation by Analysis of DNA Melting Curves During the Polymerase Chain Reaction. Analytical Biochemistry, 245, 154-160.
http://dx.doi.org/10.1006/abio.1996.9916 - 29. Zeller, W.E. (2013) Synthesis of (+)- and (-) Phaselic Acid. Synthetic Communications, 43, 1345-1350.
http://dx.doi.org/10.1080/00397911.2011.633252 - 30. Sullivan, M.L. and Zarnowski, R. (2011) Red clover HCT2, a Hydroxycinnamoyl-Coenzyme A: Malate Hydroxycinnamoyl Transferase, Plays a Crucial Role in Biosynthesis of Phaselic Acid and Other Hydroxycinnamoyl-Malate Esters in Vivo. Plant Physiology, 155, 1060-1067.
http://dx.doi.org/10.1104/pp.110.166793 - 31. Kagan, I.A. and Flythe, M.D. (2012) Factors Affecting the Separation and Bioactivity of Red Clover (Trifolium pratense) Extracts Assayed against Clostridium sticklandii, a Ruminal Hyper Ammonia-Producing Bacterium. Natural Product Communications, 7, 1605-1608.
- 32. Mabry, T.J., Markham, K.R. and Thomas, M.B. (1970) The Systematic Identification of the Flavonoids. Springer-Verlag, New York.
http://dx.doi.org/10.1007/978-3-642-88458-0 - 33. Winters, A., Heywood, S., Farrar, K., Donnison, I., Thomas, A. and Webb, K.J. (2009) Identification of an Extensive Gene Cluster Among a Family of PPOs in Trifolium pratense L. (Red Clover) Using a Large Insert BAC Library. BMC Plant Biology, 9, 94.
http://dx.doi.org/10.1186/1471-2229-9-94 - 34. Lee, M.R.F., Theobald, V.J., Tweed, J.K.S., Winters, A.L. and Scollan, N.D. (2008) Effect of Feeding Fresh or Conditioned Red Clover on Milk Fatty Acids and Nitrogen Utilization in Lactating Dairy Cows. Journal of Dairy Science, 92, 1136-1147.
http://dx.doi.org/10.3168/jds.2008-1692 - 35. Reed, J.D. (1995) Nutritional Toxicology of Tannins and Related Polyphenols in Forage Legumes. Journal of Animal Science, 73, 1516-1528.
- 36. Broderick, G.A. and Albrecht, K.A. (1997) Ruminal in Vitro Degradation of Protein in Tannin-Free and Tannin-Containing Forage Legume Species. Crop Science, 37, 1884-1891.
http://dx.doi.org/10.2135/cropsci1997.0011183X003700060037x - 37. Broderick, G.A., Walgenbach, R.P. and Maignan, S. (2001) Production of Lactating Dairy Cows Fed Alfalfa or Red Clover Silage at Equal Dry Matter or Crude Protein Contents in the Diet. Journal of Dairy Science, 84, 1728-1737.
http://dx.doi.org/10.3168/jds.S0022-0302(01)74608-5 - 38. Fraser, M.D., Speijers, M.H.M., Theobald, V.J., Fychan, R. and Jones, R. (2004) Production Performance and Meat Quality of Grazing Lambs Finished on Red Clover, Lucerne or Perennial Ryegrass Swards. Grass and Forage Science, 59, 345-356.
http://dx.doi.org/10.1111/j.1365-2494.2004.00436.x - 39. Marten, G.C., Jordan, R.M. and Ristau, E.A. (1990) Performance and Adverse Response of Sheep during Grazing of Four Legumes. Crop Science, 30, 860-866.
http://dx.doi.org/10.2135/cropsci1990.0011183X003000040019x - 40. Cassida, K.A., Griffin, T.S., Rodriguez, J., Patching, S.C., Herterman, O.B. and Rust, S.R. (2000) Protein Degradability and Forage Quality in Maturing Alfalfa, Red Clover, and Birdsfoot Trefoil. Crop Science, 40, 209-215.
http://dx.doi.org/10.2135/cropsci2000.401209x - 41. Broderick, G.A., Albrecht, K.A., Owens, V.N. and Smith, R.R. (2004) Genetic Variation in Red Clover for Rumen Protein Degradability. Animal Feed Science and Technology, 113, 157-167.
http://dx.doi.org/10.1016/j.anifeedsci.2003.12.004
NOTES
*Corresponding author.
#Deceased.





