American Journal of Plant Sciences
Vol.5 No.1(2014), Article ID:41898,8 pages DOI:10.4236/ajps.2014.51009
Relative Response of Four Tomato Species to Rotylenchulus reniformis Infestation
1Department of Biological and Environmental Sciences, Alabama A&M University, Normal, USA; 2Department of Entomology and Plant Pathology, Auburn University, Auburn, USA; 3Department of Agronomy and Soils, Auburn University, Auburn, USA.
Email: *govind.sharma@aamu.edu
Copyright © 2014 Robert Ebow McEwan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is pro- perly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Robert Ebow McEwan et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received July 20th, 2013; revised November 25th, 2013; accepted December 15th, 2013
KEYWORDS
Nematode-Resistance; Rotylenchulus reniformis; Reniform Nematode; Solanum lycopersicum; S. chilense; S. peruvianum; S. pimpinellifolium; Tomato
ABSTRACT
The reniform nematode (Rotylenchulus reniformis) is among the most economically damaging plant pathogens in the United States. This nematode is mostly known for its damage to cotton but tomato is also well-within its vast host range that includes 314 plant species across 77 plant families. Nematode-resistant genotypes offer an effective, environmentally safe alternative to agro-chemicals for reniform nematode management. Resistance genes can be introgressed into cultivars through plant improvement efforts. Tomato is a diploid species which is more amenable to identification of resistance genes in contrast to cotton where cultivars are either tetraploid or hexaploid.This greenhouse study examined cultivated and wild Solanum species represented by 40 tomato accessions, to identify resistance and susceptibility responses to R. reniformis. Accessions were evaluated by using single plants in six replicates. Seeds were germinated in sterile soil and inoculated with mixed vermiform R. reniformis. After seven weeks, eggs and vermiform stages were extracted from the root system and counted. A susceptible control S. lycopersicum “Rutgers” (LA1090) was included. Seven putatively resistant tomato genotypes were identified. These genotypes in increasing order of resistance are S. chilense (LA1029), S. lycopersicum (LA1792), S. chilense (LA1932), S. peruvianum var. humifusum (LA0385) S. pimpinellifolium (LA2934), S. peruvianum f. glandulosum (LA1283) and S. pimpinellifolium (LA1579).
1. Introduction
Tomato (Solanum lycopersicum L. syn. Lycopersicon esculentum Mill.) offers an accessible model system from which to clone a nematode resistance gene [1]. Genes for resistance to the southern root-knot nematode (Meloidogyne incognita) and the cyst nematode (Globodera rostochiensis) have been successfully isolated and characterized from tomato species S. peruvianum L. [formerly L. peruvianum (L.) Mill.] and S. lycopersicum. Disease symptoms such as galling index cannot be relied on to assess reniform nematode resistance because such symptoms do not develop in infested plants. Roots infested with reniform nematodes may appear normal unless viewed under a microscope even when aboveground symptoms have been observed [2]. In a susceptible host, the female establishes a specialized feeding site or syncytium and develops to the egg-laying stage. Resistance occurs when the female fails to establish or maintain this feeding site [3,4]. As a result, most evaluation studies either measure egg production as an indicator of nematode reproduction, or vermiform count as an indicator of nematode feeding and survival or both. Often, these counts are expressed as a percentage of a susceptible or resistant control [5]. Tomato is a near perfect plant system for basic and applied plant research particularly for discovery of resistance genes, because of its photoperiod, insensitivity and high self-fertility, broad environmental adaptability, and wide difference in fruit size and short life cycle duration [6]. Nematode resistance assays require plants with mature seeds for the next generation, and tomato offers an easier and faster alternative to cotton because of shorter duration to attain maturity (60 - 95 days). Evaluation of 33 tomato genotypes for resistance to root-knot nematodes (RKN), showed tomato Mongal T-11 and tomato Beef Master to be highly resistant to Meloidogyne spp. and also with the lowest reproductive factors of 0.71 and 0.53, respectively [7].
Tomato and its wild relatives have good seed yield and ease of controlled pollination and hybridization. This plant is therefore amenable to asexual propagation and in vitro plant regeneration [8]. Tomato, a diploid species, has a moderate sized genome (~0.95 pg/1C, 950 Mbp) [9, 10] with minimal gene duplication. A wide array of mutants [11] and diverse genetic stocks are available including a diverse collection of wild species (http://tgrc.ucdavis.edu/; http://www.sgn.cornell.edu). The tomato genome encodes an estimated 35,000 genes [12]. The availability of high molecular weight insert genomic libraries, including both Yeast Artificial Chromosome (YAC) and Bacterial Artificial Chromosome (BAC) libraries [13,14], has facilitated map-based or positional cloning. Current genetic maps for tomato include at least 2200 Restriction Fragment Length Polymorphisms (RFLPs), Cleaved Amplified Polymorphic Sequences (CAPs), and Simple Sequence Repeats (SSRs), as well as emerging genetic resources that include a comparative map with Arabidopsis of over 500 Conserved Orthologous Set (COS) markers [15], http://www.sgn.cornell.edu. The populations used for generating these maps were derived from crosses between wild relatives of various Solanum species and cultivars. This has led to the discovery and introgression of novel alleles for disease resistance [16] and fruit traits into cultivated germplasm. Thus far, resistance to Meloidogyne incognita, Fusarium and Verticillium wilts, Phytophthora infestans, Globodera rostochiensis, and Pseudomonas syringae pv. tomato has been successfully introgressed into cultivated tomato germplasm. Very few economically important plant species are as well characterized as tomato and therefore strictly from identification of resistance genes to reniform nematode perspective tomato species are also being studied here. Tomato has been recognized as an excellent host of the reniform nematode and infection of tomato has been reported from Puerto Rico, Colombia, India and Pakistan [17]. The nematode causes substantial economic damage to tomato crops throughout tropical and subtropical regions. Systematic evaluation of tomato species and germplasm for reniform resistance has been reported in older studies and few of these have been undertaken in the past 25 years. Resistance in S. pimpinellifolium (PI375937) was first identified by earlier investigators working on tomato [17]. The progeny of S. pimpinellifolium (P1375937) × S. lycopersicum “Red Rock” cross was tested for resistance to R. reniformis, and the presence of one dominant gene in S. pimpinellifolium was suggested. Furthermore, in 22 tomato cultivars tested, resistance response to H. schachtii and reniform nematode was correlated [18]. In a greenhouse study that provided a 12-day exposure to reniform nematodes, three S. lycopersicum selections—Kalyanpur Sel I and III, LA121, and a yellow-fruited S. pimpinellifolium accession were categorized as being immune [19]. In this study, we present results from four species comprising 40 genotypes for their responses to reniform nematode infestation.
2. Materials and Methods
2.1. Growing and Stock Maintenance of the Tomato Genotypes
Seeds of the 40 accessions were obtained from the C.M. Rick Tomato Genetics Resource Center (TGRC), University of California, Davis, CA. These accessions carry LA designation followed by a four digit unique number. In the greenhouse, six seeds of each accession were germinated in trays using Metromix 200 Planting Mix (Sungro Horticulture, Bellevue, WA). At the first true leaf stage seedlings were transplanted into one-gallon pots at a single plant per pot. Transplants were watered as needed and fertilized weekly with Miracle Gro 15-30-15 (The Scotts Company, Marysville, OH). Upon maturation, fruits were collected and seeds were extracted and stored for future use.
2.2. Reniform Nematode Inocula
Reniform nematode population was collected from an infected field soil and confirmed morphometrically and increased on Delta PineLand 555 B2RF cotton (Gossypium hirsutum) in the greenhouse at Auburn University. The population of R. reniformis consisted of mixed vermiform stages obtained a day before inoculation. The nematodes were extracted from the cotton roots by gravity sieving followed by sucrose centrifugations. The nematode inoculum was quantified using the Nikon TSX 100 inverted microscope and standardized to apply 2000 vermiform life stages per 3 ml of water.
2.3. Reniform Nematode Bioassay in the Greenhouse
Twelve seeds of each tomato genotype were planted in plastic trays filled with Metromix 200 planting medium. At the first true leaf stage (10 - 14 days after planting), seedlings were transplanted one per pot into 150ml Ray Leach “Cone-tainers” (Stuewe and Sons, Corvallis, OR) filled with 60:40 (v/v) mixture of sand and clay. Tomatoes were screened for nematode response using a modified protocol [2]. The Cone-tainers with the six largest plants of each genotype were infested with nematodes by gently pipetting, the nematode suspension at the base of the tomato stem. Each Cone-tainer received two inoculations 7 days apart each with 2000 mixed vermiform R. reniformis. Plants were irrigated daily and fertilized weekly for seven weeks. Eggs were extracted from roots using 0.14 M NaOCl solution [20], followed by centrifugal flotation in 1 M sucrose solution. Vermiform stages were extracted and enumerated at 40× with the inverted microscope. A susceptible control S. lycopersicum “Rutgers” (LA 1090) was included. This genotype has been shown to support prolific reproduction of reniform nematodes [21]. In all, six randomized replications of 40 genotypes were tested against R. reniformis.
2.4. Statistical Analysis
Data were analyzed using generalized linear mixed models procedures as implemented in SAS® PROC GLIMMIX (SAS Institute, Cary, NC), employing a lognormal distribution function for counts. The experimental design was a randomized complete block design with six blocks; block was considered to be a random effect. Dunnett’s test was employed to compare accession means to the susceptible control cv. Rutgers. Mean counts and Upper and Lower confidence limits (UCL and LCL) on the log scale were back-transformed to the original scale.
3. Results and Discussion
Forty accessions were screened for nematode resistance. All accessions tested supported development and reproduction of reniform nematode. Female nematodes exhibited a preference for penetration of young tender roots generally near root tips (Figure 1). As shown in Tables 1-3, total count nematodes (eggs + vermiform) extracted ranged from a high of 2522/125 mL of soil on S. lycopersicum “Moboline” (LA3152) to a low of 122 on S. pimpinellifolium (LA1579). Across all accessions, count per gram of fresh root varied from 48 to 1645 on LA1579 and LA3318, respectively. Nematode egg counts were greater than vermiform numbers for majority of the accessions evaluated (Figure 2). However, there was not sufficient evidence to suggest a direct relationship between the two measurements. A strong correlation existed between egg and vermiform counts for the 14 wild species examined (r = 0.88, P= 0.00002), the two estimates were somewhat loosely correlated for the 8 background cultivars (r = 0.65, P= 0.04) and 18 mutants
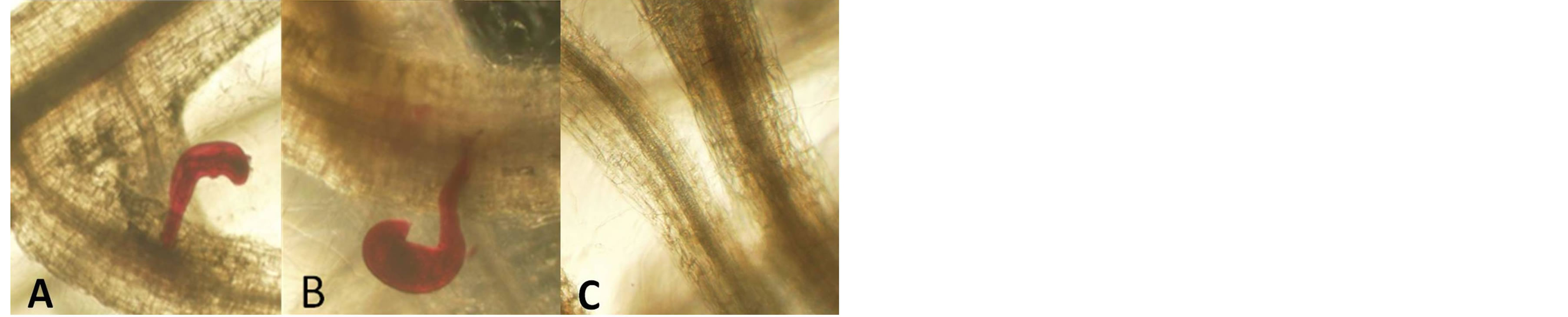
Figure 1. Reniform nematode infested roots seven weeks post inoculation (wpi). A = Roots of susceptible wild tomato S. lycopersicum var. cerasiforme (LA 2070). B = Roots of susceptible tomato cultivar S. lycopersicum cv. Micro-tom (LA 3911). C = Roots of putatively resistant tomato cultivar S. lycopersicum (LA1792).
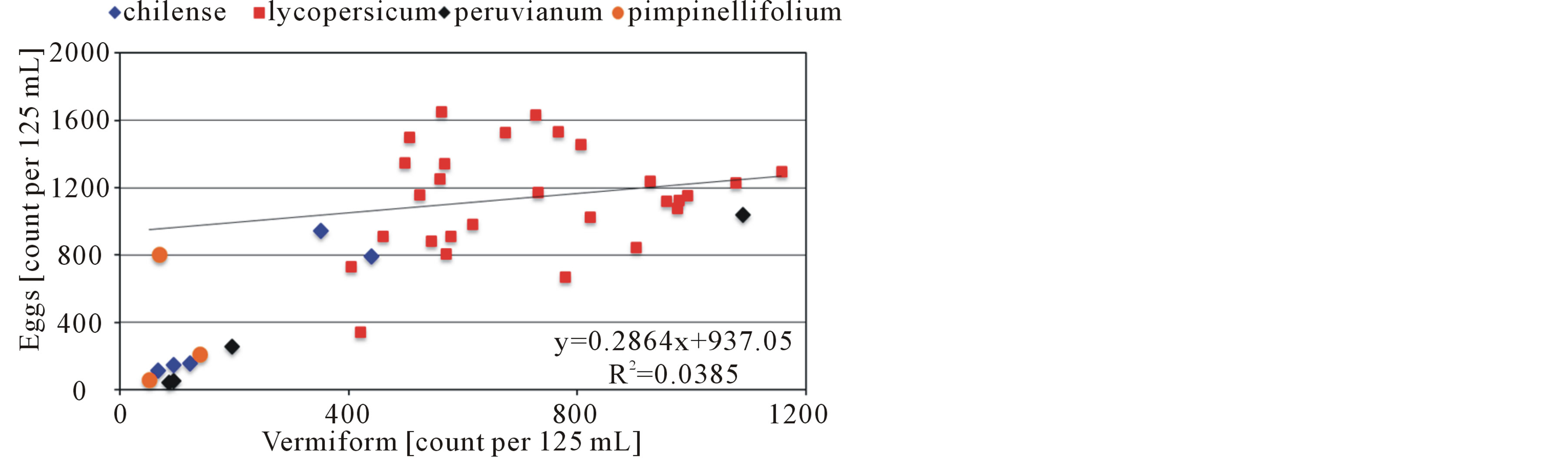
Figure 2. Relationship between vermiform and egg ratios of reniform nematode for 40 accessions belonging to four cultivated and wild species of tomato (Solanum).
tested (r = 0.35, P = 0.08).
Solanum lycopersicum “Rutgers” (LA1090), used in this study as a susceptible check supported abundant nematode reproduction. The average count (including vermiform and eggs) recorded was 1235/125 ml soil. Thirtytwo additional accessions registered mean counts comparable to LA1090 and were thus classified susceptible. Seven accessions (~18%) suppressed nematode reproduction significantly (at P ≤ 0.05) when compared to LA1090 in both counts per 125 mL of soil and gram root fresh weight. These were classified as “putatively resistant”. The egg counts observed on the putatively resistant genotypes were more than 10% but less than 30% of LA1090. Mean counts were not significantly different from the control LA1090 (Table 1). In contrast, 6 of the 7 (~86%) putatively resistant accessions were from the wild species collection with varying geographical origins. These had levels of nematode reproduction that were significantly lower than LA1090 (Table 2). Solanum pimpinellifolium (LA2934), S. chilense (LA1932), S. chilense (LA1029), S. peruvianum var. humifusum (LA0385), S. pimpinellifolium (LA1579), and S. peruvianum var. glandulosum (LA1283) all supported nematode populations that were significantly smaller than those of the susceptible control. Among the background
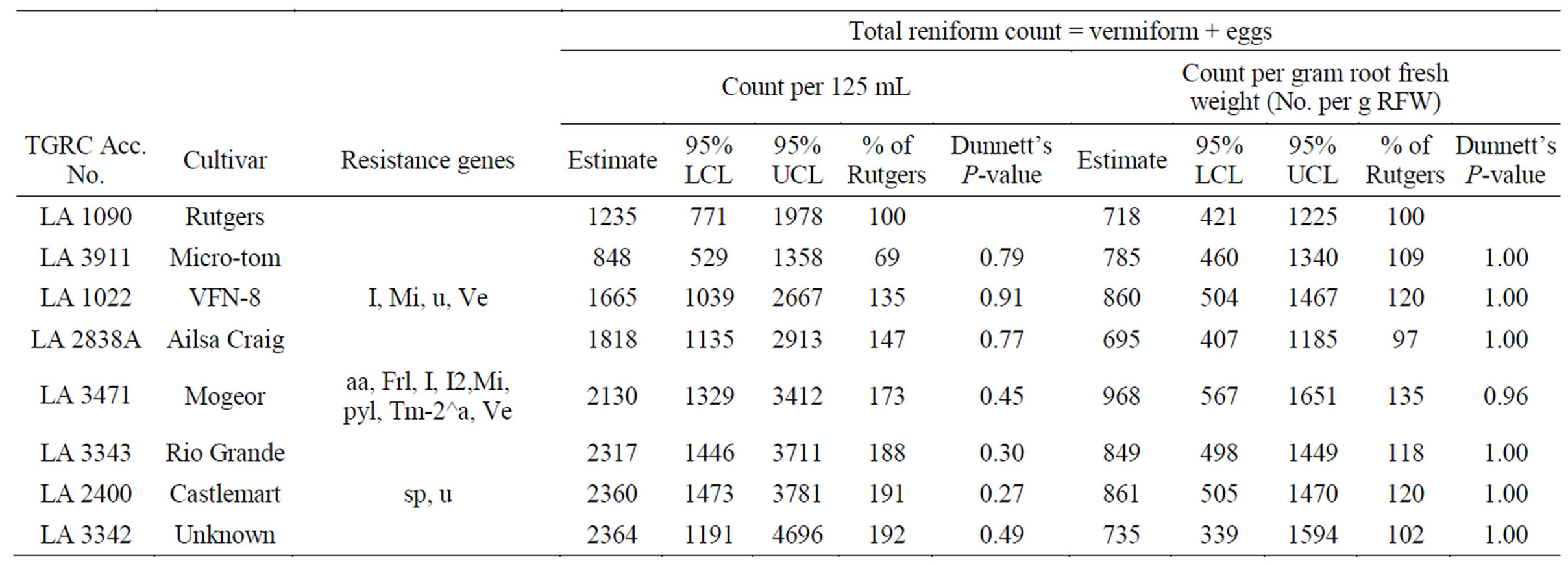
Table 1. Relative resistance to reniform nematode (RN) by Solanum lycopersicum genotypes as compared to susceptible control cv. Rutgers. Data average of six replications.
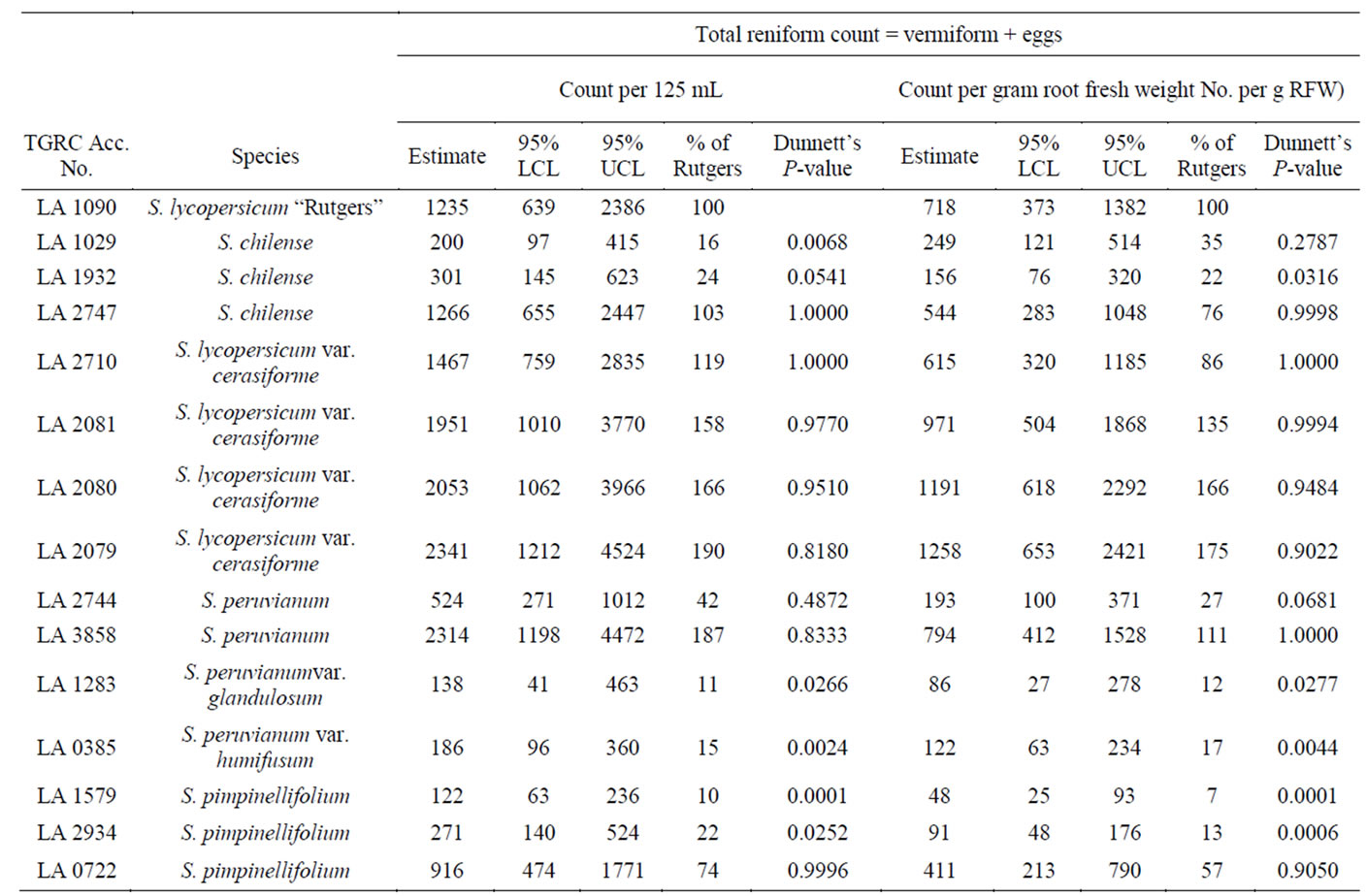
Table 2. Relative resistance to reniform nematode (RN) in wild tomato species compared to susceptible control Solanum lycopersicum “Rutgers”. Data average of six replications.
genotypes tested and examined, only S. lycopersicum (LA1792) was able to meaningfully stifle nematode reproduction (Table 3).
Resistance has been identified by measuring eggs per gram of root [22], vermiform nematodes recovered from soil [5], or a combination of number of eggs and vermiform stages extracted [23,24]. Combining both life stages to evaluate resistance provides information on nematode reproduction, feeding and survival ability and thus serves as an indicator of the resistance and susceptibility within a host genotype. In this study, our approach involved a combination of eggs and vermiform life stages extracted from roots of forty cotton accessions for resistance evaluation. Near isogenic lines (NIL) are vital genetic stocks
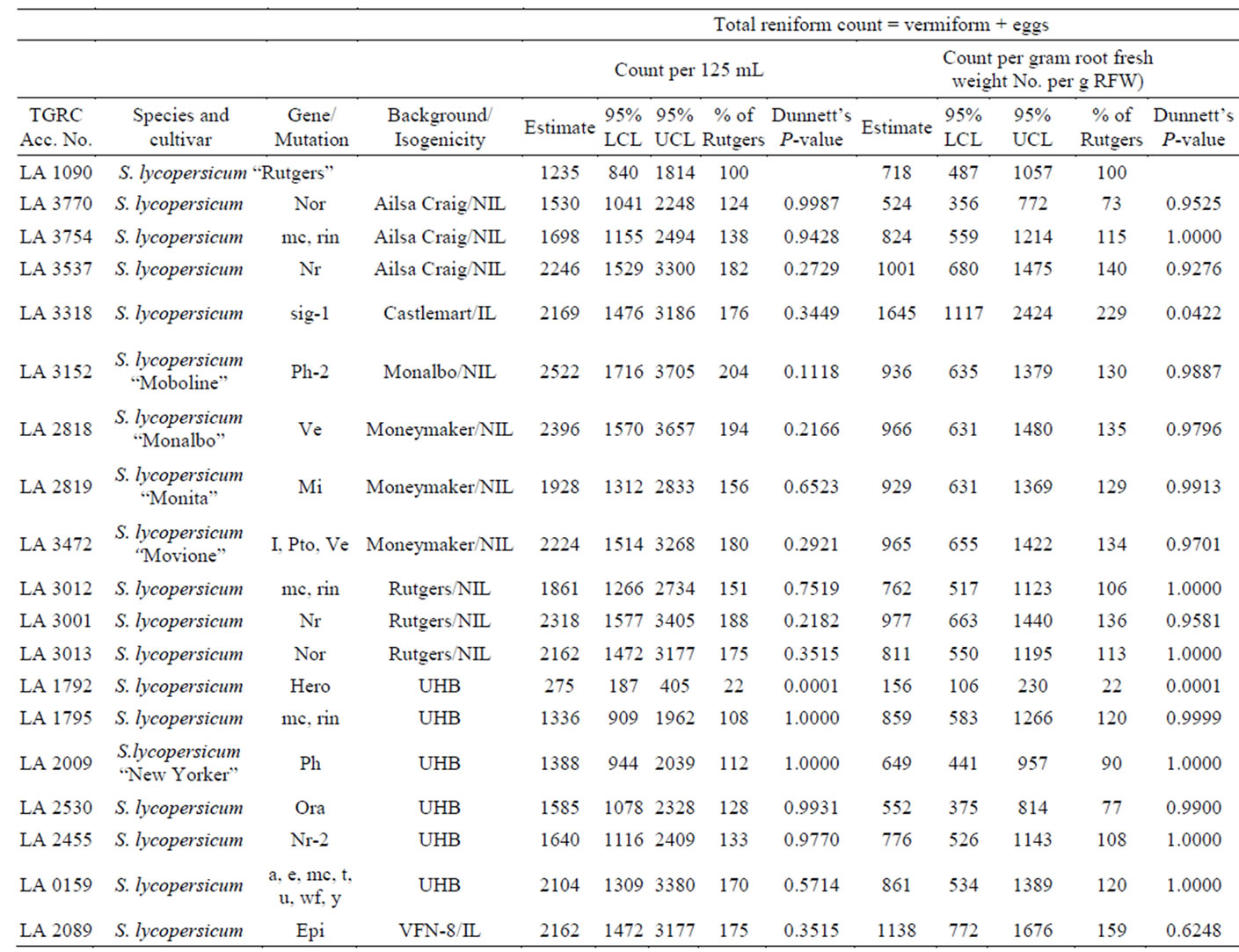
Table 3. Relative resistance to reniform nematode (RN) in tomato cultivars, isogenic lines (IL), near isogenic lines (NIL) and accessions of unknown hybrid background (UHB) compared to susceptible control Rutgers. Data average of six replications.
for investigating the function and regulation of single genes [25], and aid in isolation of genes [26]. Tomato genotypes evaluated in this study were carefully selected to include parental lines, NILs, and S. lycopersicum cultivars with considerable common genetic background. Narrow genetic base of cultivars make it imperative to look for novel genes in the wild relatives. The majority of these wild species can be crossed directly to the cultivated tomato, making resistance genes readily transferable.
Seven tomato genotypes supported lower populations of R. reniformis than the control LA1090: S. chilense (LA1029), S. lycopersicum (LA1792), S. chilense (LA1932), S. peruvianum var. humifusum (LA0385), S. pimpinellifolium (LA2934), S. peruvianum f. glandulosum (LA1283), and S. pimpinellifolium (LA1579). Solanum peruvianum, one of the most distant relatives of the cultivated tomato, originated in central Peru and northern Chile, has been the source of many major resistance genes [27]. An example is the tomato yellow leaf curl virus (TYLCV) that has been introgressed into the tomato breeding line TY172. The TYLCV resistance in TY172 is controlled by a previously unknown major QTL (Ty-5, mapped to chromosome 4), originating from the resistant line, and four additional minor QTLs (mapped to chromosomes 1, 7, 9 and 11) [28]. Several accessions of S. peruvianum have been identified with resistance to root-knot nematode controlled by Mi gene located on chromosome six [29]. This gene confers resistance to tomato against several Meloidogyne species M. incognita, M. javanica, and M. arenaria [30]. One out of the three R gene homologues at the Mi locus, (Mi 1.2) was found to confer resistance to both a nematode and an aphid [31]. Our results however did not reveal any resistance to the reniform nematode in the cultivated tomato accessions—S. lycopersicum “Monita” (LA2819), S. lycopersicum “VFN-8” (LA1022), and S. lycopersicum “Mogeor” (LA3471) which possess the introgressed Mi gene. This suggests a different resistance mechanism for RN than Mi gene identified for Meloidogyne species.
Plants lack a diversity of receptors associated with their immune system as found in vertebrates, and therefore rely on only a relatively small set of innate immune receptors against pathogenic attacks. Recently, S. pimpinellifolium has been observed to possess a plant immune receptor protein Cf-2, providing a dual resistance to fungi and nematode. The Cf-2 protein, previously was identified as an immune receptor for the leaf mold fungus Cladosporium fulvum. However, this protein also mediates disease resistance to the root parasitic nematode G. rostochiensis pathotype Ro1-Mierenbos [32].Two S. pimpinellifolium accessions, LA2934 and LA1579 suppressed reniform nematode numbers significantly which may be associated with the Cf-2 protein. There are also earlier reports of low reproduction rate of R. reniformis on certain accessions of this species [17,19]. The disease resistance gene Pto was introgressed into the cultivated tomato species from S. pimpinellifolium [33]. The Pto gene confers resistance to the bacterial pathogen, P. syringae pv tomato (Pst), the causal agent for bacterial speck disease. Pst-infected leaf tissue and fruits develop black specks surrounded by chlorotic halos leading to a reduction in yield [34]. Pto, one of the first R-genes to be cloned and sequenced, encodes a serine-threonine protein kinase and is 963 bp long with no introns. This gene belongs to a family of six genes clustered in a 60-kb region of chromosome five of tomato [35]. The S. lycopersicum “Movione” (LA3472), with the introgressed Pto and othergenes did not demonstrate any resistance to the reniform nematode in this study.
Solanum lycopersicum (LA1792) is a tomato line with the introgressed Hero gene family shown to confer high level (95%) of resistance to all pathotypes of a potato cyst nematode (PCN), G. rostochiensis [36], and partial resistance to G. pallida [37]. Resistance to PCN in two tomato accessions S. pimpinellifolium B6173 and S. peruvianum B6001 has been confirmed [38]. The level of resistance in S. pimpinellifolium was noted to be greater than that in S. peruvianum. As a result of this greater resistance, coupled with ease of hybridization with tomato cultivars, S. pimpinellifolium was chosen as a donor parent for imparting PCN resistance into commercial tomato varieties. The resistance of S. pimpinellifolium B6173 to the Wren isolate of G. rostochiensis was shown to be controlled by a single dominant gene for which the symbol Hero was proposed. The Hero gene was introgressed into tomato cultivar LA1792 from wild species S. pimpinellifolium LA121 [38]. Map-based cloning and structural characterization of the Hero gene from tomato and its genomic organization has been reported on extensively [39]. Hero gene encodes a protein with a nucleotide-binding site (NBS) and a leucine-rich-repeat (LRR) domain. This gene is a member of a family of 14 homologous genes located in a 118 kb region on chromosome four. A great majority of resistance (R) genes are organized in gene clusters [40], and the Hero gene is no exception. In our study, S. lycopersicum (LA1792) was among the seven genotypes with the highest resistance to reniform nematode. Further studies are needed to ascertain which of the 14 R gene homologues at the Hero gene cluster is most responsible for reniform nematode resistance. In cotton, where resistance to reniform nematode is a critical economic necessity, tomato germplasm identified here provides a simpler approach for identification of resistance genes. Arduous efforts in cotton have shown the introgression and back crossing of resistance from G. longicalyx to G. hirsutum [41,42] and in other Gossypium species [22] including G. aridum [43] and from G. arboreum and a G. hirsutum/G. aridum bridging line [44]. Genetic engineering of resistance utilizing oryzacystatin gene for R. reniformis has been demonstrated in Arabidopsis thaliana [45], in a species with a well-characterized genome. The nature of RN resistance mechanism in tomato at present is unknown.
4. Conclusion
This study provides a comprehensive comparative evaluation of resistance in four Solanum spp. to R. reniformis. Among these NILs, near NILs, and parental checks within which, specific accessions and cultivars were identified with resistance to R. reniformis. Furthermore, a protocol for undertaking rapid detection of RN in vivo in tomato field soils is currently being pursued. In the root samples of resistant Solanum cultivars and accessions, the female nematodes failed to penetrate and therefore were unable to develop any further while the relatively less resistant cultivars facilitated greater penetration and subsequent development of the RN.
Acknowledgements
The authors extend their sincerest gratitude to the C.M. Rick Tomato Genetics Resource Center (TGRC), University of California, Davis, CA, for supplying seed and information. Ms. Sarah Beth Cseke provided laboratory support and for stimulating discussions. This is journal article # 658 of Alabama A & M Agricultural Experiment Station. This work was supported by the Experiment Station and USDA/NIFA Evans-Allen grants and by the National Science Foundation/PGRP award DBI 703470. The co-authors are dedicating this study to the late Dr. Ramesh Kantety.
REFERENCES
- J. Ho, R. Weide, H. M. Mai, M. F. van Wordragen, K. N. Lambed, M. Koornneef, P. Zabe and V. M. Williamson, “The Root-Knot Nematode Resistance Gene (Mi) in Tomato: Construction of a Molecular Linkage Map and Identification of Dominant cDNA Markers in Resistant Genotypes,” The Plant Journal, Vol. 2, 1992, pp. 971- 982.
- A. F. Robinson, “Cotton Nematodes,” In: C. W. Smith and J. T. Cothren, Eds., Cotton: Origin, History, Technology, and Production, John Wiley & Sons, New York, 1999, pp. 595-615.
- J. E. Erpelding and S. R. Stetina, “Genetics of Reniform Nematode Resistance in Gossypium arboreum Germplasm Line PI 529728,” World Journal of Agricultural Research, Vol. 1, 2013, pp. 48-53.
- P. Agudelo, R. T. Robbins, K. S. Kim and J. M. Stewart, “Histological Changes in Gossypium hirsutum Associated with Reduced Reproduction of Rotylenchulus reniformis,” Journal of Nematology, Vol. 37, 2005, pp. 185-189.
- D. B. Weaver, K. S. Lawrence and E. van Santen, “Reniform Nematode Resistance in Upland Cotton Germplasm,” Crop Science, Vol. 47, No. 1, 2007, pp. 19-24. http://dx.doi.org/10.2135/cropsci2006.02.0130
- C. M. Rick and J. I. Yoder, “Classical and Molecular Genetics of Tomato: Highlights and Perspectives,” Annual Review of Genetics, Vol. 22, 1988, pp. 281-300. http://dx.doi.org/10.1146/annurev.ge.22.120188.001433
- F. Jaiteh, C. Kwoseh and R. Akromah, “Evaluation of Tomato Genotypes for Resistance to Root-Knot Nematodes,” African Crop Science Journal, Vol. 20, 2012, pp. 41-49.
- J. J. Fillatti, J. Kiser, R. Rose and L. Comai, “Efficient Transfer of Glyphosate Tolerance Gene into Tomato Using Binary Agrobacterium tumefaciens Vector,” Nature Biotechnology, Vol. 5, 1987, pp. 726-730. http://dx.doi.org/10.1038/nbt0787-726
- K. Arumuganathan and E. D. Earle, “Nuclear DNA Content of Some Important Plant Species,” Plant Molecular Biology Reporter, Vol. 9, No. 3, 1991, pp. 208-218. http://dx.doi.org/10.1007/BF02672069
- D. G. Peterson, W. R. Pearson and S. M. Stack, “Characterization of the Tomato (Lycopersicon esculentum) Genome Using in Vitro and in Situ DNA Reassociation,” Genome, Vol. 41, 1998, pp. 346-356.
- N. Menda, Y. Semel, D. Peled, Y. Eshed and D. Zamir, “In Silico Screening of a Saturated Mutation Library of Tomato,” Plant Journal, Vol. 38, No. 5, 2004, pp. 861- 872. http://dx.doi.org/10.1111/j.1365-313X.2004.02088.x
- R. Van der Hoeven, C. Ronning, J. Giovannoni, G. Martin and S. Tanksley, “Deductions about the Number, Organization, and Evolution of Genes in the Tomato Genome Based on Analysis of a Large Expressed Sequence Tag Collection and Selective Genomic Sequencing,” Plant Cell, Vol. 14, No. 7, 2002, pp. 1441-1456. http://dx.doi.org/10.1105/tpc.010478
- M. A. Budiman, L. Mao, T. C. Wood and R. A. Wing, “A Deep-Coverage Tomato BAC Library and Prospects Toward Development of an STC Framework for Genome Sequencing,” Genome Research, Vol. 10, 2000, pp. 129- 136.
- G. Bonnema, J. Hontelez and R. Verkerk, “An Improved Method of Partially Digesting Plant Megabase DNA Suitable for YAC Cloning: Application to the Construction of a 5.5 Genome Equivalent YAC Library of Tomato,” The Plant Journal, Vol. 9, 1996, pp. 125-133. http://dx.doi.org/10.1046/j.1365-313X.1996.09010125.x
- S. D. Tanksley, M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale, P. Broun, T. M. Fulton, J. J. Giovanonni, S. Grandillo, G. B. Martin, R. Messeguer, J. C. Miller, L. Miller, A. H. Paterson, O. Pineda, M. Roder, R. A.Wing, W. Wu and N. D. Young, “High Density Molecular Linkage Maps of the Tomato and Potato Genomes,” Genetics, Vol. 132, 1992, pp. 1141-1160.
- E. Kabelka, W. C. Yang and D. Francis, “Improved Tomato Fruit Color within an Inbred Backcross Line Derived from Lycopersicon esculentum and L. hirsutum Involves the Interaction of Loci,” Journal of the American Society for Horticultural Science, Vol. 129, 2004, pp. 250-257.
- R. V. Rebois, B. J. Eldridge, J. M. Good and A. K. Stoner, “Tomato Resistance and Susceptibility to the Reniform Nematode,” Plant Disease Reporter, Vol. 57, 1973, pp. 169-172.
- R. V. Rebois, A. E. Steele, A. K. Stoner and B. J. Eldridge, “A Gene for Resistance to Rotylenchulus reniformis in Tomato, and a Possible Correlation with Resistance to Heterodera schachtii,” Journal of Nematology, Vol. 9, 1977, pp. 280-281.
- P. Balasubramanian and C. Ramakrishnan, “Resistance to the Reniform Nematode, Rotylenchulus reniformis in Tomato,” Nematologia Mediterranea, Vol. 11, 1983, pp. 203-204.
- R. S. Hussey and K. R. Barker, “A Comparison of Methods of Collecting Inocula of Meloidogyne Species, Including a New Technique,” Plant Disease Reporter, Vol. 57, 1973, pp. 1025-1028.
- A. F. Robinson, C. G. Cook and A. E. Percival, “Resistance to Rotylenchulus reniformis and Meloidogyne incognita Race 3 in the Major Cotton Cultivars Planted Since 1950,” Crop Science, Vol. 39, 1999, pp. 850-858. http://dx.doi.org/10.2135/cropsci1999.0011183X003900030039x
- C. Yik and B. Wray, “Resistant Germplasm in Gossypium Species and Related Plants to Rotylenchulus reniformis,” Journal of Nematology, Vol. 16, 1984, pp. 146-153.
- A. F. Robinson and A. E. Percival, “Resistance to Meloidogyne incognita Race 3 and Rotylenchulus reniformis in Wild Accessions of Gossypium hirsutum and G. barbadense from Mexico,” Journal of Nematology, Vol. 29, 1997, pp. 746-755.
- R. T. Robbins, L. Rakes, L. E. Jackson and D. G. Dombek, “Reniform Nematode Resistance in Selected Soybean Cultivars,” Journal of Nematology, Vol. 31, 1999, pp. 667-677.
- H. Tsujimoto, “Production of Near-Isogenic Lines and Marked Monosomic Lines in Common Wheat (Triticum aestivum) cv Chinese Spring,” The American Genetic Association, Vol. 92, 2001, pp. 254-259.
- T. Kojima, H. Tsujimoto and Y. Ogihara, “High-Resolution RFLP Mapping of the Fertility Restoration (Rf3) Gene against Triticum timopheevi Cytoplasm Located on Chromosome 1BS of Common Wheat,” Genes and Genetic Systems, Vol. 72, 1997, pp. 353-359. http://dx.doi.org/10.1266/ggs.72.353
- G. Kalloo, “Interspecific and Intergeneric Hybridization in Tomato,” In: Kalloo, Ed., Genetic Improvement of Tomato, Springer, New York, 1991, pp. 73-82.
- I. Anbinder, M. Reuveni, R. Azari, I. Paran, S. Nahon, H. Shlomo, L. Chen, M. Lapido and I. Levin, “Molecular Dissection of Tomato Leaf Curl Virus Resistance in Tomato Line TY172 Derived from Solanum peruvianum,” Theoretical and Applied Genetics, Vol. 119, 2009, pp. 519-530. http://dx.doi.org/10.1007/s00122-009-1060-z
- W. S. Barham and N. N. Winstead, “Inheritance of Resistance to Root-Knot Nematodes in Tomatoes,” Proceedings of the American Society for Horticultural Science, Vol. 69, 1957, pp. 372-377.
- B. P. Corbett, L. Jia, R. J. Sayler, L. M. Arevalo-Soliz and F. Goggin, “The Effects of Root-knot Nematode Infection and Mi-Mediated Nematode Resistance in Tomato on Plant Fitness,” Journal of Nematology, Vol. 43, 2011, pp. 82-89.
- M. Rossi, F. L. Goggin, S. B. Milligan, I. Kaloshian, D. E. Ullman and V. M. Williamson, “The Nematode Resistance Gene Mi of Tomato Confers Resistance against the Potato Aphid,” Proceedings of the National Academy of Sciences, Vol. 95, 1998, pp. 9750-9754. http://dx.doi.org/10.1073/pnas.95.17.9750
- J. L. Lozano-Torres, R. H. P. Wilbers, P. Gawronski, J. C. Boshoven, A. Finkers-Tomczak, J. H. G. Cordewener, A. H. P. America, H. A. Overmars, J. W. Van’t Klooster, L. Baranowski, M. Sobczak, M. Ilyas, R. A. L. van der Hoorn, A. Schots, P. J. G. M. de Wit, J. Bakker, A. Goverse and G. Smanta, “Dual Disease Resistance Mediated by the Immune Receptor Cf-2 in Tomato Requires a Common Virulence Target of a Fungus and a Nematode,” Proceedings of the National Academy of Sciences of USA, Vol. 109, 2012, pp. 10119-10124. http://dx.doi.org/10.1073/pnas.1202867109
- M. Pilowsky and D. Zutra, “Screening Wild Tomatoes for Resistance to Bacterial Speck Pathogen (Pseudomonas tomato),” Plant Disease, Vol. 66, 1982, pp. 46-47. http://dx.doi.org/10.1094/PD-66-46
- L. E. Rose, R. W. Michelmore and C. H. Langley, “Natural Variation in the Pto Disease Resistance Gene within Species of Wild Tomato (Lycopersicon). II. Population Genetics of Pto,” Genetics, Vol. 175, 2006, pp. 1307- 1319. http://dx.doi.org/10.1534/genetics.106.063602
- G. B. Martin, S. H. Brommonschenkel and J. Chunwongse, “Map-Based Cloning of a Protein Kinase Gene Conferring Disease Resistance in Tomato,” Science, Vol. 262, 1993, pp. 1432-1436. http://dx.doi.org/10.1126/science.7902614
- M. W. Ganal, R. Simon, S. Brommonschenkel, M. Arndt, M. S. Phillips, S. D. Tanksley and A. Kumar, “Genetic Mapping of a Wide Spectrum Nematode Resistance Gene (Hero) against Globodera rostochiensis in Tomato,” Molecular Plant-Microbe Interactions, Vol. 8, 1995, pp. 886-891. http://dx.doi.org/10.1094/MPMI-8-0886
- M. Sobczak, A. Avrova, J. Jupowicz, M. S. Phillips, K. Ernst and A. Kumar, “Characterization of Susceptibility and Resistance Responses to Potato Cyst Nematode (Globodera Spp.) Infection of Tomato Lines in the Absence and Presence of the Broad-Spectrum Nematode Resistance Hero Gene,” Molecular Plant-Microbe Interactions, Vol. 18, 2005, pp.158-168. http://dx.doi.org/10.1094/MPMI-18-0158
- P. R. Ellis and J. W. Maxon-Smith, “Inheritance of Resistance to Potato Cyst Eelworm (Heterodera rostochiensis Woll.) in the Genus Lycopersicon,” Euphytica, Vol. 20, 1971, pp. 93-101. http://dx.doi.org/10.1007/BF00146779
- K. Ernst, A. Kumar, D. Kriseleit, D. U. Kloos, M. S. Phillips and M. W. Ganal, “The Broad-Spectrum Potato Cyst Nematode Resistance Gene (Hero) from Tomato Is the Only Member of a Large Gene Family of NBSLRR Genes with an Unusual Amino Acid Repeat in the LRR Region,” The Plant Journal, Vol. 31, 2002, pp. 127-136. http://dx.doi.org/10.1046/j.1365-313X.2002.01341.x
- R. W. Michelmore and B. C. Meyers, “Clusters of Resistance Genes in Plants Evolve by Divergent Selection and a Birth-and-Death Process,” Genome Research, Vol. 8, 1998, pp. 1113-1130.
- A. F. Robinson, A. A. Bell, N. Dighe, M. A. Menz, R. L. Nichols and D. M. Stelly, “Introgression of Resistance to Nematode Rotylenchulus reniformis into Upland Cotton (Gossypium hirsutum) from G. longicalyx,” Crop Science, Vol. 47, 2007, pp. 1865-1877. http://dx.doi.org/10.2135/cropsci2006.12.0776
- N. D. Dighe, A. F. Robinson, A. A. Bell, M. A. Menz, R. G. Cantrell and D. M. Stelly, “Linkage Mapping of Resistance to Reniform Nematode in Cotton following Introgression from Gossypium longicalyx (Hutch. & Lee),” Crop Science, Vol. 49, 2009, pp. 1151-1164. http://dx.doi.org/10.2135/cropsci2008.03.0129
- G. B. Romano, E. J. Sacks, S. R. Stetina, A. F. Robinson, D. D. Fang, O. A. Gutierrez and J. A. Scheffler, “Reniform Nematode (Rotylenchulus reniformis) Resistance Locus from Gossypium aridum Identified and Introgressed into Upland Cotton (G. hirsutum),” Journal of Nematology, Vol. 41, 2009, pp. 375-376.
- E. J. Sacks and A. F. Robinson, “Introgression of Resistance to Reniform Nematode (Rotylenchulus reniformis) into Upland Cotton (Gossypium hirsutum) from G. arboreum and a G. hirsutum/G. aridum Bridging Line,” Field Crops Research, Vol. 112, 2009, pp. 1-6. http://dx.doi.org/10.1016/j.fcr.2009.01.006
- P. E. Urwin, A. Levesley, M. J. McPherson and H. J. Atkinson, “Transgenic Resistance to the Nematode Rotylenchulus reniformis Conferred by Arabidopsis thaliana Plants Expressing Proteinase Inhibitors,” Molecular Breeding, Vol. 6, 2000, pp. 257-264. http://dx.doi.org/10.1023/A:1009669325944
NOTES
*Corresponding author.

