American Journal of Plant Sciences
Vol.3 No.6(2012), Article ID:20026,9 pages DOI:10.4236/ajps.2012.36092
Differential Proteome Analysis of Chlamydomonas reinhardtii Response to Arsenic Exposure
![]()
1Department of Chemistry, Cleveland State University, Cleveland, USA; 2Department of Biological, Geological, and Environmental Sciences, Cleveland State University, Cleveland, USA; 3Lerner Research Institute, Cleveland Clinic, Cleveland, USA.
Email: h.vankeulen@csuohio.edu
Received February 28th, 2012; revised April 5th, 2012; accepted April 16th, 2012
Keywords: Arsenic Stress; Chlamydomonas reinhardtii; Heavy Metal; Oxygen Stress; Proteomics; 2-DE Mass Spectrometry
ABSTRACT
The fresh water unicellular green alga Chlamydomonas reinhardtii was used to explore whether it could function as a model system to identify proteins that are differentially expressed in response to arsenate exposure. Cells were treated with different concentrations of arsenate ranging from 100 - 400 mM. When exposed to 200 mM arsenate, the amount of live cells started to lessen on the second day and continued to diminish, indicating a toxic effect of arsenate. Proteomic analysis was used to investigate if these cells showed a specific response to arsenic-induced stress. Fifteen proteins were found that were over-expressed in the 200 mM arsenate-treated samples and two proteins were found to be very strongly over-expressed in samples treated with 400 µM. These were selected for identification using liquid chromatography coupled with tandem mass spectrometry. Oxidative stress and protein damage were the major effects as shown by the up-regulation of Mn-superoxide dismutase, an oxygen-evolving enhancer protein, a chaperonin-like protein and a heat shock protein.
1. Introduction
Arsenic (As) contamination of ground water and soil adversely affects human health and causes major environmental problems. Arsenic is well known as a ubiquitous metalloid in the environment, is the 20th most abundant element in the earth crust, but various anthropogenic activities (smelting, industrial waste release) and natural processes (volcanic eruptions, acid rain), have led to its accumulation over time in soil and water which has resulted in a worldwide problem, especially through contamination of groundwater [1-3].
Phytoextraction, one of the major processes in phytoremediation, has been utilized to clean up metal-contaminated soil and water [4]. It employs the potential of certain higher plants to uptake and concentrate toxic substances from the environment into its biomass. However, As is phytotoxic as evidenced by the inability of most plants to accumulate it [1,5]. Two common forms of inorganic arsenic are arsenate (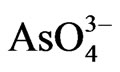 ) [As(V)], and arsenite (AsO2), [As(III)] which are interconvertible depending on the status of the environment they are in [6] and are more abundant than organic species [7]. Over 400 plants are hyperaccumulators of toxic heavy metals [8]. Tolerance to arsenic is well known for Pteris vittata
) [As(V)], and arsenite (AsO2), [As(III)] which are interconvertible depending on the status of the environment they are in [6] and are more abundant than organic species [7]. Over 400 plants are hyperaccumulators of toxic heavy metals [8]. Tolerance to arsenic is well known for Pteris vittata
[9], while other plants are less tolerant such as Zea mays [10] and Oriza sativa [11], the latter is known to be an arsenic accumulator and possibly a health risk as food when grown in arsenate contaminated soils.
Previous investigations using proteomic methods combined with mass spectrometry in our laboratory revealed that the Dwarf Sunflower, Helianthus annuus, is able to accumulate As and this accumulation results in the expression of stress-related proteins such as chitinase [12]. It can be assumed that the physiological and biochemical response towards As stress is highly complex since higher plants are organized as multicellular organisms with many types of tissues, which results in a complex proteome and stress-related responses that are at the cellular, tissue and organismal levels. Moreover, mass spectrometry relies heavily on the availability of a genomic database for the organism or cells under investigation, which is not available for certain plants such as H. annuus. Therefore, it was decided to investigate whether a single cell organism could be used to study stress response at the cellular level without the impediment of complex organ systems and one for which a genome database is available.
In this study, the unicellular green alga Chlamydomonas reinhardtii was chosen in order to perform proteomic analysis to investigate its response to arsenic. The alga’s complete genome is available [13] and research on its proteome and metabolome are being performed [14] and its sub-proteome [15] and chloroplast proteome have been studied in detail [16]. It is easy to grow, has a short-life cycle and a large number of experimental parameters can be incorporated into the study. In addition, data so far obtained for C. reinhardtii are similar to those obtained for higher plants, suggesting that it might be a good model organism. For example, recent work showed that phytochelatins (PC) are the major chelators induced upon cadmium (Cd) treatment and that these complexes sequestered up to 70% of the total Cd found in Cd-treated cells [17,18]. Other studies on C. reinhardtii discussed the fact that it might be a good model to study heavy metal homeostasis and tolerance [19], light stress [20] transition metal transport [21], and the role of PCs in Cd accumulation [22]. A proteomics study on the effect of Cd showed down-regulation of a number of proteins involved in photosynthesis, Calvin cycle and Chlorophyll biosynthesis, at the same time some typical stress-related proteins were up-regulated such as heat shock proteins, superoxide dismutase and glutathione-S-transferase to name a few of the proteins identified [23].
However, very little is known about arsenic toxicity in C. reinhardtii. Many have reported on arsenic toxicity and have described effects such as anti-oxidative response [24], changes in uptake kinetics and arsenic-phosphate interactions [9] mainly using P. vittata. Some studies using arsenic treatment have been reported using H. annuus [25] as well as C. reinhardtii [26,27]. But other than a few proteomic-level studies about arsenic accumulation such as in rice plants [28] and in the Dwarf Sunflower [12] and Pteris vittata [29], very little has been reported about proteome changes upon arsenic stress in Chlamydomonas. This study is based on the premise that C. reinhardtii might be useful to investigate the effects of As exposure and to search for differential expression of proteins in order to see 1) if the response also includes oxygen stress as found previously in the Dwarf Sunflower and 2) if other proteins such as metal-binding proteins could be identified.
2. Materials and Methods
2.1. Cell Growth and Arsenic Treatment
C. reinhardtii wild type (+) was purchased from Carolina Biological Supply Co. (http://www.Carolina.com) and algae were purified from contaminating bacteria by serial dilution and plating out of cells on agar plates. The purified cells were grown at 25˚C in 250 ml foam-plugged (Jaece Industries, http://www.jaece.com), autoclaved Erlenmeyer flasks containing 100 ml growth medium. The growth medium was composed of Tris-acetate-phosphate
(TAP) as described by Dunford et al. (http://www.unbf.ca/vip/restools/TAP.htm). The flasks were shaken continuously (130 rpm) on an orbital shaker incubator using continuous illumination using 15 W “Plant & Aquarium” fluorescent lighting. Experiments were conducted to establish the toxicity ranges using dibasic sodium arsenate (Na2HAsO4∙7H2O). The concentration of arsenate used was 0, 100, 200, 300 and 400 μM. The flasks were inoculated with 1 × 105 cells ml–1. Cells were counted with a Levy-Hausser hemacytometer. Cell growth was monitored at least once a day until the experiment was terminated at the end of a six day period. The growth rate was determined by counting live cells in a hemacytometer and optical density at 750 nm. Cells were harvested when the density reached 0.6 - 0.7 optical density units for each concentration of arsenate. Each series of 0 - 400 µM arsenate was done twice, the first time in triplicate, the second time in duplicate. For the growth rate only live cells were counted. For proteomics the cells were collected by centrifugation at 6000 × g for 20 min at 20˚C. The cell pellets were stored at –80˚C or immediately processed for extraction of total proteins.
2.2. Sample Preparation for Gel Electrophoresis
After six days of incubation cells were harvested and pellets were ground to a fine powder in liquid N2. Approximately 1g of ground material was resuspended in 1 ml of 1× phosphate buffered saline (PBS) at pH 7.4 containing 1× nuclease solution (100 mM Tris-HCl, pH 7.0, 50 mM MgCl2, RNase (500 μg/ml) and DNase (1mg/ml)) and 1x plant protease inhibitor solution (G-Biosciences, http://www.gbiosciences.com). The suspension was vortexed briefly and 3 freeze-thaw cycles were performed in order to break the cell walls. The suspension was sonicated on ice six times for 10 s each with intermittent 1 min breaks using an ultrasonicator (Branson Ultrasonics, http://www.All-Spec.com/BransonUltrasonics). Samples were centrifuged in 3 cycles successively at 4˚C with the settings of 15,000 × g for 30 min; 20,000 × g for 30 min; and 20,000 × g for 15 min in order to obtain a clear protein extract without any debris. Protein precipitation was performed using freshly prepared ice-cold, saturated ammonium sulfate ((NH4)2SO4) solution. The first protein precipitation was performed by adding saturated ammonium sulfate drop-wise until the final concentration reached 40%. After 30 min incubation in an ice bath, the suspension was centrifuged at 15,000 × g for 20 min at 4˚C. The supernatant was subjected to a second precipitation by increasing the salt concentration up to 60% ammonium sulfate and a clean protein pellet was obtained. The protein pellets were resuspended in 1 × PBS buffer and dialyzed overnight at 4˚C against 1 × PBS buffer. Protein concentration in each fraction was determined using the Bradford assay. Five hundred micrograms (500
μg) of protein from the 60% ammonium sulfate fraction was precipitated with 10% Trichloroacetic acid (TCA)/ acetone with 0.07% dithiothreitol (DTT) at –20˚C overnight. After centrifugation (17,000 × g for 20 min at 4˚C), the protein pellet was washed twice with ice-cold acetone, centrifuged (17,000 × g for 20 min at 4˚C), vacuum dried and stored at –80˚C until use.
2.3. Two-Dimensional Protein Gel Electrophoresis
Protein pellets were resuspended in freshly prepared rehydration buffer (RB) (7M Urea, 2M thiourea, 1% CHAPS, 1% Triton X-100, 1% DTT and 0.2% Ampholytes and 1% BromoPhenolBlue). A total of 500 µg of protein was used for 2-DE analysis. Each pellet was resuspended in 200 μl of rehydration buffer at room temperature. Rehydration was performed by loading the total 200 µl RB on 11 cm pH 3 - 10 linear immobilized pH gradient (IPG) strips (BioRad, http://www.discover-bio-rad. com) into the tray. Active rehydration was performed at 5 V for 14 - 16 h at room temperature. The IPG strips were focused using a Protean IEF cell (Bio-Rad) according to the manufacturer’s instructions and the total volt-hours reached at the end of IEF was 40 kVh. After isoelectric focusing was completed, proteins were separated using a 12% SDS-PAGE gel (Bio-Rad) at a voltage of 200 V. The gels were fixed in a 100 ml fixing solution (50% ethanol, 10% acetic acid in water) for 30 min at room temperature with gentle shaking. After washing with water, the gels were stained overnight with GelCode Blue staining solution (Pierce, http://www.piercenet.com).
2.4. Image Analysis of Two-Dimensional Gels
Gels were scanned at 300 ppi on a Scan maker, 5900 scanner, (MicroTek Lab Inc, http://www.microtek.com) and the scanned images of protein spots were loaded into the PDQUEST gel analysis software (Version 6.2, BioRad). The data set was obtained from gels using proteins from control and arsenic-treated cells. The proper spot detection parameters were selected and adjusted using the Spot Detection Wizard in order to identify and count all the protein spots of interest in the gels. Match sets were created using all gels and up-regulated/over-expressed spots at different concentrations of arsenic exposure were determined using a Student’s t-test within PDQUEST with a significance level set to 95% (p < 0.05). Replicates from three independent extracts were used for analysis. Protein spots were finally visually inspected to validate the obtained results.
2.5. In-Gel Protein Digestion
Stained protein spots that appeared as up-regulated after As treatment were selected and excised manually. They were cut into smaller pieces and transferred to 1.5 ml microcentrifuge tubes. The excised gel fragments were washed twice for 1 h each with 175 μl of wash reagent (50% ethanol (95% USP grade), 5% acetic acid (99.9% Aldrich, http://www.sigmaaldrich.com) and water) to remove the dye that was bound to the protein. It was then dehydrated for 5 min using 175 μl of HPLC grade acetonitrile (Burdick and Jackson, (http://www.honeywell.com) and rehydrated for 5 min with 100 mM ammonium bicarbonate. It was again dehydrated for 5 min with acetonitrile and vacuum-dried in a SpeedVac for approximately 3 min. After this dehydration/rehydration procedure, the gel pieces were digested with trypsin by adding 10 - 15 μl of 10 ng/μl trypsin reagent (Promega sequenceing grade modified trypsin dissolved in 2 ml 50 mM ammonium bicarbonate, http://www.promega.com). The gel pieces were vortexed, centrifuged briefly and digested overnight at room temperature. The resulting peptides were extracted from the polyacrylamide in two aliquots of 30 μl of extraction reagent (50% acetonitrile, 5% ACS grade formic acid). These extracts were combined and evaporated to less than 10 μl in a SpeedVac and then resuspended in 1% acetic acid to make a final volume of approximately 30 μl for LC-MS/MS analysis.
2.6. Mass Spectrometry Analysis and Protein Identification
The mass spectrometric analysis for the identified protein spots from the treated gels were performed using either LCQ-Deca or LTQ Linear ion trap mass spectrometer (ThermoScientific, http://www.thermoscientific.com) located in the Proteomics Core, Lerner Research Institute, the Cleveland Clinic, Cleveland. The HPLC column was a self-packed 10 cm × 75 µm Phenomenex Jupiter C18 reversed-phase capillary chromatography column. One to ten µl of the tryptic digests were injected and the peptides were eluted from the column by an acetonitrile/ 0.05 M acetic acid gradient at a flow rate of 0.2 µl/min. The nanoelectrospray source was operated at 2.5 kV and the mass spectrometer was operated in a data-dependent mode.
The Chlamydomonas protein database was obtained from the Chlamydomonas center JGI portal chlre4 genome browser at http://genome.jgi-psf.org/Chlre4/Chlre4. home.html. It was uploaded into the MASCOT search program in order to identify the proteins analyzed from the mass spectrometer. The LC-MS/MS data (from Xcalibur raw files) for each sample was searched against the above mentioned Chlamydomonas protein sequence database and additional searches were performed using the full NCBI non-redundant database (The Genetic sequence database at the National Center for Biotechnology Information). Protein versus protein alignment searches were performed for each protein sequence as well as for the peptide sequences obtained. The identity of these proteins was further proved by SEQUEST, tandem mass spectrometry database searching algorithm on Bioworks software (Thermo Finnigan, http://www.sisweb.com). All the MS/MS spectra were searched against the databases generated from identified protein sequences for each protein with filtering criteria having Xcorr scores ≥ 1.5 for singly charged, Xcorr scores ≥ 2.0 for doubly charged and Xcorr scores ≥ 2.5 for triply charged.
3. Results and Discussion
3.1. Arsenic Effect on Cell Growth
The first goal was to delineate the effect of arsenate on cellular growth as a response to arsenic stress. Arsenate was added to growth medium at the beginning of the assay and cell growth was monitored on a daily basis for 6 days. The results showed that at concentrations between 10 - 100 µM no significant growth inhibition was observed (not shown). However, the number of live cells, identified by moving flagella and a larger size than the dead cells, which formed clusters, began to decline at concentrations greater than l00 µM indicating that Chlamydomonas was unable to sustain growth in the presence of arsenate at doses above this level (Figure 1). The results are consistent with a previous study which showed that cell growth was stimulated in medium containing arsenic at a low concentration of 1.0 µM but was inhibited at higher concentrations [26], despite the fact that inorganic phosphate was present in the culture medium, which would act as a competitor of arsenate in transport

Figure 1. Effect of different arsenate concentrations on C. reinhardtii cell growth. The variation in the cell growth of C. reinhardtii upon exposure to increasing concentrations (0, 100, 200, 300 and 400 µM) of arsenate over 6 days of incubation at 25˚C temperature and continuous illumination was measured and expressed in live cell number per ml culture. Each assay was done in triplicate. The bar graph shows the standard deviation for each assay.
and binding to target molecules [30]. C. reinhardtii is thus incapable of detoxification of arsenate. It is possible that the inorganic form of arsenic is methylated to form monomethylor dimethyl arsenic such that the potential toxic effects of arsenic are increased, resulting in reactive oxygen species and formation of anti-oxidants [30,31].
3.2. Extraction of Soluble Proteins
The accuracy of 2-D gel based proteomic analysis depends on the yield and cleanliness of the protein extract which ultimately results in protein gels with a high resolution and maximum number of proteins visible. Initially, several commonly used methods for extraction of proteins from Chlamydomonas were applied such as described by Förster et al. [20] using acetone and 10% trichloroacetic acid (TCA) or as described by Gillet et al. [23] using streptomycin sulfate precipitation followed by a standard acetone—TCA precipitation, or polyethylene glycol (PEG) fractionation as modified used for sunflower proteins [32]. While all of these methods are commonly used to prepare proteins for 2-D gel electrophoresis, it was found that these procedures yielded gel patterns containing a considerable degree of heavy streaks with a fairly low amount of clearly visible protein spots (<100) (not shown). The low protein recovery and poor 2-D gels may be attributed to the presence of substances that coalesced soluble proteins, which was evident by the viscous nature of the final sample preparation and the color of which was always greenish. The heavy streaks could be due to the overabundance of proteins such as ribulose bisphosphate carboxylase oxygenase (RuBisCO). These streaks overshadow other lower abundant proteins with similar molecular mass and isoelectric points. Removal or reduction of the streaks would thus help to identify those proteins that are differentially expressed. Therefore, another protein precipitation technique employing ammonium sulfate precipitation was tested. Ammonium sulfate precipitation has long been used in protein purification but the approach is not considered a desirable method for extracting proteins from plants, most likely due to the inability to obviate the problems associated with high protease activities in plants and the presence of such compounds as polyphenols. The protocol consisted of treatment with two consecutive salt concentrations: 40% salt treatment, followed by an increase to 60%. The first salt treatment was effective in removing substances which contributed to both the greenish color and viscosity. In addition it was found, using gel electrophoresis, that RuBisCO was selectively removed, but not many other proteins (results not shown), in a similar manner as precipitation of Sunflower RuBisCO with polyethylene glycol [32]. An increase in salt concentration to 60% resulted in precipitation of proteins whose amount was 90% of the original protein concentration in crude extracts. The 2-D gel patterns obtained from the use of ammonium sulfate precipitated samples showed much improved protein resolution with significantly higher number of protein spots and a substantial reduction of streaks (Figure 2, top panel). Further, the 2-D gel patterns were highly reproducible.
3.3. Proteomics
Protein extracts prepared from control and arsenic-treated cells were compared to determine differential responses to As-stress. Proteins were extracted from the algae at the end of the 6th day of the incubation during which measurable decline in cell density began to occur. In the gel
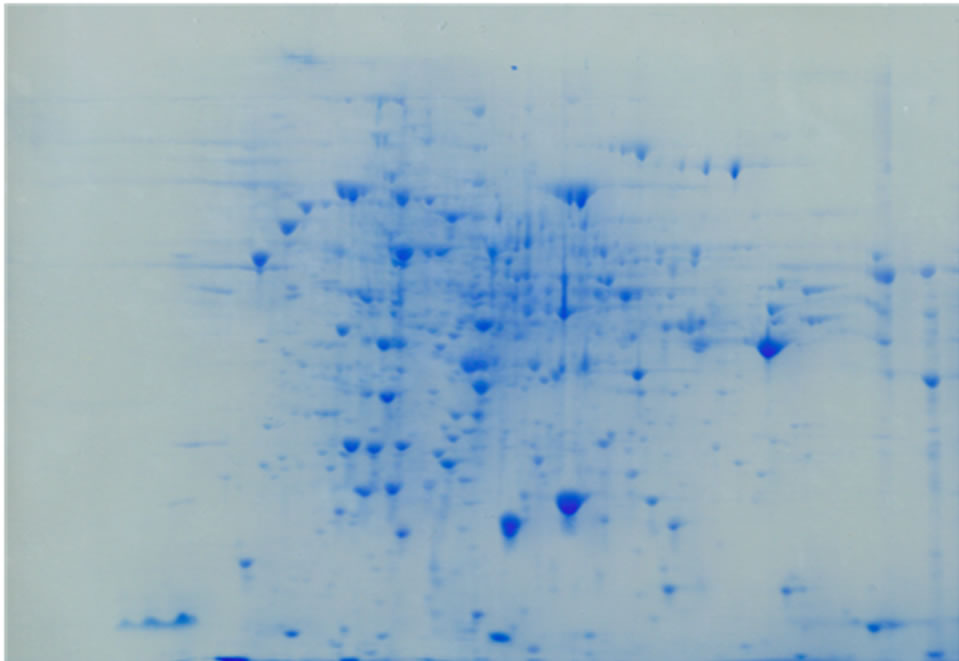
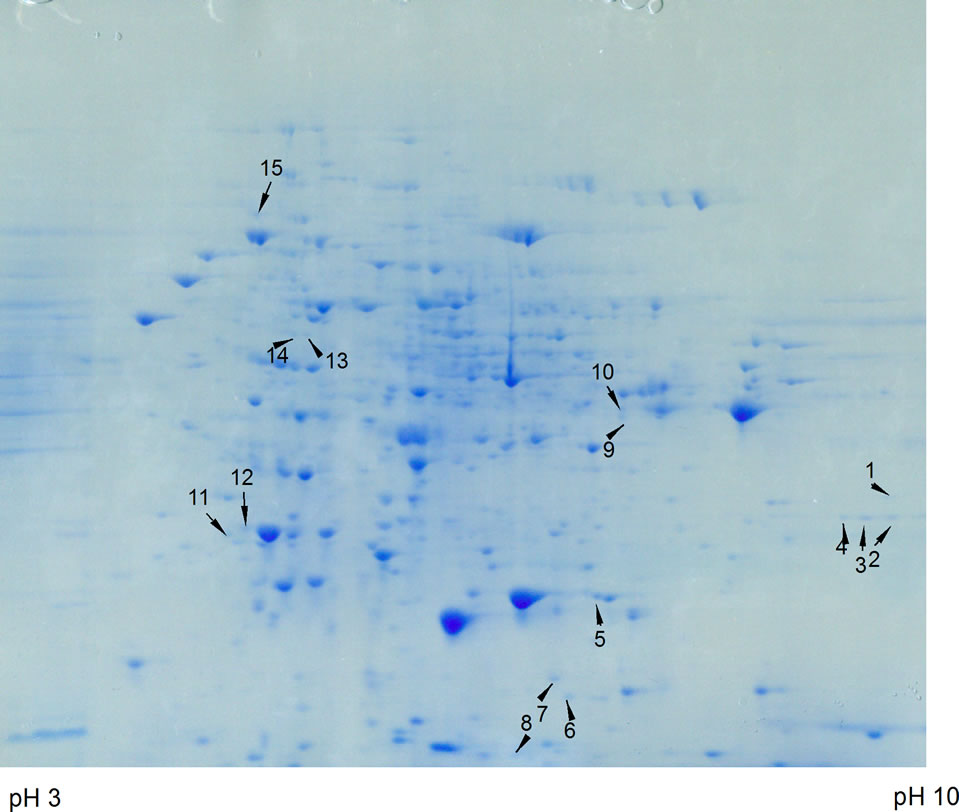
Figure 2. Top panel. 2-D gel of ammonium sulfate precipitated proteins from control C. reinhardtii. Proteins were separated in the first dimension on an IPG strip pH 3 - 10 (from left to right in the figure) and in the second dimension on a 12% polyacrylamide SDS gel. A total of 500 µg protein from a 6 day culture was used. The gel was stained with GelCode Blue. Bottom Panel. 2-D gel of ammonium sulfate precipitated proteins from 200 µM arsenate treated C. reinhardtii. The samples was treated in the same way as the control. Fifteen protein spots were differentially expressed and identified using LC-MS/MS, shown by arrows.
electrophoresis step, separation of proteins was achieved by isoelectric focusing wherein proteins were distributed in the pI range of 3 - 10. In the second dimension, SDSpolyacrylamide gel electrophoresis was performed and the gels were stained with GelCode Blue (Figure 2). The spots that disappeared or whose intensity decreased by the arsenic treatment were not considered for identification, because they could be down-regulated proteins or represent loss of proteins resulting from degradation by cell death. Fifteen spots (1 - 15) represented over-expressed proteins when cells were grown in the presence of 200 µM arsenate (Figure 2, bottom panel). When cells were grown in 300 or 400 µM arsenate more protein spots disappeared probably due to the high degree of cell death (not shown), therefore, 400µM arsenate was used as the maximal concentration. Usually 100, 200 and 300 µM arsenate is used in experiments with the arsenic hyperaccumulator Pteris vittata (24), thus 400 µM arsenate was tested as the maximal possible concentration. However, two proteins were strongly up-regulated after treatment with 400 µM arsenate and extracted for analysis. These proteins are marked extra 1 and 2 in Table 1.
The up-regulated proteins can be categorized on the basis of their putative functions. Of the proteins with known functions, most are associated with the removal of damaged proteins, oxidative stress, increased energy demand, protein synthesis and protein folding. The remaining proteins were annotated in the C. reinhardtii database as unknown or hypothetical without specific functions.
One of the up-regulated proteins is the a subunit D of the 26S proteasome (spot 1), which is part of the 20S core complex [33]. The 26S proteasome is the proteolytic complex in eukaryotes responsible for the removal of short-lived or abnormal intracellular proteins [34]. The molecular organization of the 26S proteosome from the plant Arabidopsis thaliana has been described in some detail [34]. It is structurally and functionally conserved among eukaryotes, suggesting that the subunit arrangement of the 26S proteasome in C. reinhardii is probably similar to those determined for higher plants. The function of the proteasome in plants appears to be affected by external stress resulting in an increase in the core 20S proteasome at the cost of the 26S proteasome, which has a number of additional proteins. The increase of the 20S core unit appears to result in an increase in protein degradation of oxidized proteins and so enhances tolerance towards oxidative stress [35]. The expression of this 20S proteasome subunit might be increased in the As-treated cells in order to remove proteins that are damaged by exposure to arsenate. However, no other proteasome proteins were up-regulated, so the biosynthesis of proteasome proteins might be the result of a loss of a controlling mechanism. Proteasome assembly can be impaired when one of the lid proteins is absent as was found in fission yeast [33].
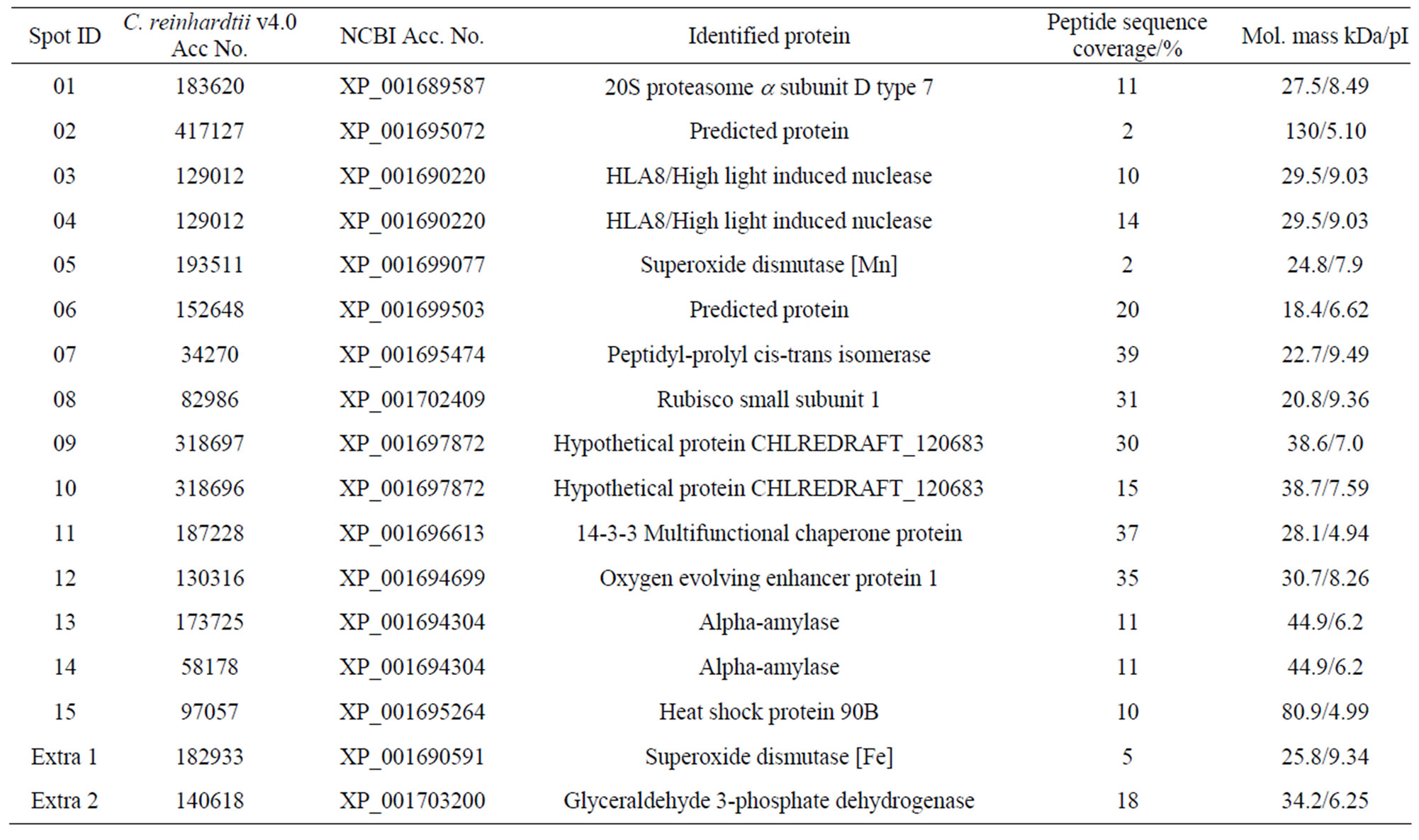
Table 1. Overview of identified proteins resulting from arsenate stress.
It is also possible that the other core proteins were not detected in the 2-D gels. Another protein identified in these experiments is Mn-dependent superoxide dismutase (SOD) (spot 5), which catalyzes the reduction of superoxide anions to hydrogen peroxide. The increase of this enzyme might be to reduce oxidative stress, which results in production of reactive oxygen species, an observation that also was made using the Dwarf Sunflower [36], a similar observation was made when Chlamydomonas was exposed to Cd [23]. When cells were exposed to 400 µM arsenate a different SOD was identified namely a Fe-dependent SOD (spot extra 1). Studies on maize, for instance, have indicated that an increase in antioxidant capacity improved tolerance to arsenic [24]. Another protein expressed under arsenic stress is peptidyl-prolyl cis-trans isomerase (spot 7), which catalyzes the interconversion of peptidyl-prolyl imide bonds in peptides and proteins. It is also expressed in higher plants in response to oxidative stress [37]. In addition, prolyl isomerases act together with chaperonins, and protein disulfide isomerases that are involved in protein folding [38]. The chaperonin proteins are expressed in hyperaccumulator plants when exposed to arsenic and other toxic metals [39]. In the present study, one spot was identified as a chaperonin having multifunctional activities (spot 11). Chaperonins appear to have diverse functions including protein synthesis, folding, and posttranslational modification. During metal stress they are able to prevent irreversible protein denatuaration or help channel proteins for proteolytic degradation [40]. Recently, this protein has been identified and characterized in higher plants but its precise role in plant cells remains unclear. It may be that the expression of this protein plays an essential role in the removal of damaged or abnormally folded proteins. Heat shock protein 90B that was expressed as spot 15 is also a chaperonin protein and it has been reported to be expressed under metal stress [41,42]. Previously it was shown that a heat shock protein was up-regulated in Dwarf Sunflowers upon exposure to lead [36].
Ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) (spot 8) is a major enzyme in green plants that catalyzes carbon assimilation, hence controls photosynthetic rate and thus has a major effect on plant growth. The increased expression of the RuBisCO small subunit could be to compensate for damage to the photosynthesis system. Degradation of RuBisCO in metal non-tolerant plants has been reported in response to redox-reaction associated heavy metals such as copper and cadmium and other metals including mercury, cobalt, manganese and zinc [43-45]. A down-regulation of RuBisCO subunits, on the other hand, was observed in hyperaccumulator plants following exposure to metal stress [46] and also was shown in a previous investigation using Asexposed Dwarf Sunflowers [12], suggesting a decrease in net photosynthesis under metal stress. The large subunit of RuBisCO was not detected because it was removed by the ammonium sulfate precipitation. Treatment of Chlamydomonas with Cd resulted in a down-regulation of the large subunit; the small subunit was not identified as upor down-regulated in this particular study [23].
The increase of expression of α-amylase (spot 13 and 14) is indicative of an increase in energy requirement, which maybe the result of damage to the alga’s photosynthetic capacity. It must be noted that the cells were growing in TAP medium, which contains acetate that can be used as a carbon source. Under similar conditions using Cd, it was found that enzymes in the glyoxylate pathway (isocitrate lyase) and in glycolysis/gluconeo-genesis (phosphoglycerate mutase) were up-regulated [23]. Treatment with As had a different effect, but at 400 µM As, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (spot extra 2) was strongly up-regulated, supporting the possible increase in gluconeogenesis. However, it has been shown that GAPDH is a pleiotropic protein involved in various stresses, programmed cell death, DNA damage repair and DNA replication [47]. Differential expression of GAPDH in response to metal stress has been observed in several proteomic studies [45,48,49], and might not be related to energy metabolism but to any of the above mentioned effects.
Oxygen evolving enhancer protein 1 (OEE) (spot 12) is another protein expressed under stress (oxygen radicals and heat). It plays a major catalytic role in photosynthetic oxygen evolution in green plants, and is associated with the photosystem II complex, the site of oxygen evolution in all higher plants. The OEE of photosystem II also was shown to exhibit thioredoxin activity in C. reinhardtii [50,51] and it has been reported that these genes of C. reinhardtii are expressed upon exposure of cells to high light and low CO2 concentrations [52]. The dual role of OEE 1 as a thioredoxin in metabolism and a function in light-driven electron flow [50] is supported by the increase of its synthesis during arsenate-induced stress and the overall importance of maintenance of the proper redox state.
In conclusion, the response of C. reinhardtii is similar as found for Sunflowers and includes mainly a response to protein and oxidative damage. No specific As-binding proteins were found and it appears that C. reinhardtii might not be a good model to study the response to As uptake.
4. Acknowledgements
The authors thank the Ohio Plant Biotechnology Consortium through The Ohio State University, Ohio Agricultural Research and Development Center for funding. In addition, funds were provided from the Cleveland State University Research Council in the form of a Faculty Research Development Program award, and a Doctoral Dissertation Research Expense Award. Also the authors would like to thank Dr. Ling Li and Dr. Dongmei Zhang for training and advice with the mass spectrometry at the Proteomics Core at the Lerner Research Institute, Cleveland Clinic Foundation.
REFERENCES
- M. I. S. Gonzaga, J. A. G. Santos and L. Q. Ma, “Arsenic Chemistry in the Rhizosphere of Pteris vittata L. and Nephrolepis exaltata L.,” Environmental Pollution, Vol. 143, No. 2, 2006, pp. 254-260. doi:10.1016/j.envpol.2005.11.037
- M. I. S. Gonzaga, J. A. G. Santos and L. Q. Ma, “Arsenic Phytoextraction and Hyperaccumulation by Fern Species,” The Journal of Agricultural Science, Vol. 63, No. 1, 2006, pp. 90-101. doi:10.1590/S0103-90162006000100015
- B. Kumar Mandal and K. T. Suzuki, “Arsenic: Round the World: A Review,” Talanta, Vol. 58, No. 1, 2002, pp. 201-235. doi:10.1016/S0039-9140(02)00268-0
- C. Tu, L. Q. Ma and B. Bondada, “Arsenic Accumulation in the Hyperaccumulator Chinese Brake (Pteris vittata L.) and Its Utilization Potential for Phytoremediation,” Journal of Environmental Quality, Vol. 31, No. 5, 2002, pp. 1671-1675. doi:10.2134/jeq2002.1671
- A. A. Carbonell, M. A. Aarabi, R. D. DeLaune, R. P. Gambrell and W. H. Patrick Jr., “Arsenic in Wetland Vegetation: Availability, Phytotoxicity, Uptake and Effects on Plant Growth and Nutrition,” Science of the Total Environment, Vol. 217, No. 3, 1998, pp. 189-199. doi:10.1016/S0048-9697(98)00195-8
- R. D. Tripathi, S. Srivastava, S. Mishra, N. Singh, R. Tuli, D. K. Gupta and F. J. M. Maathuis, “Arsenic Hazards: Strategies for Tolerance and Remediation by Plants,” Trends in Biotechnology, Vol. 25, No. 4, 2007, pp. 158-165. doi:10.1016/j.tibtech.2007.02.003
- T. Takamatsu, H. Aoki and T. Yoshida, “Determination of Arsenate, Arsenite, Monomethylarsonate and Dimethylarsinite in Soil Polluted with Arsenic,” Soil Science, Vol. 133, No. 4, 1982, pp. 239-246. doi:10.1097/00010694-198204000-00007
- A. J. M. Baker, S. P. McGrath, R. D. Reeves and J. A. C. Smith, “Metal Hyperaccumulator Plants, a Review of the Ecology and Physiology of a Biological Resource for Phytoremediation of Metal Polluted Soils,” In: N. Terry and G. Baelos, Eds., Phytoremediation of Contaminated Soil and Water, Lewis Publishers, Boca Raton, 2000, pp. 85-107.
- J. Wang, F.-J. Zhao, A. A. Meharg, A. Raab, J. Feldmann and S. P. McGrath, “Mechanisms of Arsenic Hyperaccumulation in Pteris vittata. Uptake Kinetics, Interactions with Phosphate, and Arsenic Speciation,” Plant Physiology, Vol. 130, No. 3, 2002, pp. 1552-1561. doi:10.1104/pp.008185
- R. Requejo and M. Tena, “Maize Response to Acute Arsenic Toxicity as Revealed by Proteome Analysis of Plant Shoots,” Proteomics, Vol. 6, No. S1, 2006, pp. S156-S162. doi:10.1002/pmic.200500381
- M. J. Abedin, J. Feldmann and A. A. Meharg, “Uptake Kinetics of Arsenic Species in Rice Plants,” Plant Physiology, Vol. 128, No. 3, 2002, pp. 1120-1128. doi:10.1104/pp.010733
- H. van Keulen, R. Wei and T. J. Cutright, “ArsenateInduced Expression of a Class III Chitinase in the Dwarf Sunflower Helianthus annuus,” Environmental and Experimental Botany, Vol. 63, No. 1-3, 2008, pp. 281-288. doi:10.1016/j.envexpbot.2007.11.012
- S. S. Merchant, S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz, et al., “The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions,” Science, Vol. 318, No. 5848, 2007, pp. 245-250. doi:10.1126/science.1143609
- P. May, S. Wienkoop, S. Kempa, B. Usadel, N. Christian, J. Rupprecht, J. Weiss, L. Recuenco-Munoz, O. Ebenhöh, W. Weckwerth and D. Walther, “Metabolomicsand Proteomics-Assisted Genome Annotation and Analysis of the Draft Metabolic Network of Chlamydomonas reinhardtii,” Genetics, Vol. 179, No. 1, 2008, pp. 157-166. doi:10.1534/genetics.108.088336
- V. Wagner, J. Boesger and M. Mittag, “Sub-Proteome Analysis in the Green Flagellate Alga Chlamydomonas reinhardtii,” Journal of Basic Microbiology, Vol. 49, No. 1, 2009, pp. 32-41. doi:10.1002/jobm.200800292
- M. Terashima, M. Specht and M. Hippler, “The Chloroplast Proteome, a Survey from the Chlamydomonas reinhardtii Perspective with a Focus on Distinctive Features,” Current Genetics, Vol. 57, No. 3, 2011, pp. 151-168. doi:10.1007/s00294-011-0339-1
- J. M. Collard and R. F. Matagne, “Isolation and Genetic Analysis of Chlamydomonas reinhardtii Strains Resistant to Cadmium,” Applied and Environmental Microbiology, Vol. 56, No. 7, 1990, pp. 2051-2055.
- S. Hu, K. W. K. Lau and M. Wu, “Cadmium Sequestration in Chlamydomonas reinhardtii,” Plant Science, Vol. 161, No. 5, 2001, pp. 987-996. doi:10.1016/S0168-9452(01)00501-5
- M. Hanikenne, “Chlamydomonas reinhardtii as a Eukaryotic Photosynthetic Model for Studies of Heavy Metal Homeostasis and Tolerance,” New Phytologist, Vol. 159, No. 2, 2003, pp. 331-340. doi:10.1046/j.1469-8137.2003.00788.x
- B. Förster, U. Mathesius and B. J. Pogson, “Comparative Proteomics of High Light Stress in the Model Alga Chlamydomonas reinhardtii,” Proteomics, Vol. 6, No. 15, 2006, pp. 4309-4320. doi:10.1002/pmic.200500907
- A. Rosakis and W. Köster, “Transition Metal Transport in the Green Microalga, Chlamydomonas reinhardtii—Genomic Sequence Analysis,” Research in Microbiology, Vol. 155, No. 3, 2004, pp. 201-210. doi:10.1016/j.resmic.2003.12.004
- K. Nishikawa, A. Onodera and N. Tominaga, “Phytochelatins Do Not Correlate with the Level of Cd Accumulation in Chlamydomonas spp.,” Chemosphere, Vol. 63, No. 9, 2006, pp. 1553-1559. doi:10.1016/j.chemosphere.2005.09.056
- S. Gillet, P. Decottignies, S. Chardonnet and P. Le Maré- chal, “Cadmium Response and Redoxin Targets in Chlamydomonas reinhardtii: A Proteomics Approach,” Photosynthesis Research, Vol. 89, No. 2-3, 2006, pp. 201-211. doi:10.1007/s11120-006-9108-2
- M. Srivastava, L. Q. Ma, N. Singh and S. Singh, “Antioxidant Responses in Hyper-Accumulator and Sensitive Fern Species to Arsenic,” Journal of Experimental Botany, Vol. 56, No. 415, 2005, pp. 1335-1342. doi:10.1093/jxb/eri134
- A. Raab, H. Schat, A. A. Meharg and J. Feldmann, “Uptake, Translocation and Transformation of Arsenate and Arsenite in Sunflower (Helianthus annuus): Formation of Arsenic-Phytochelatin Complexes during Exposure to High Arsenic Concentrations,” New Phytologist, Vol. 168, No. 3, 2005, pp. 551-558. doi:10.1111/j.1469-8137.2005.01519.x
- T. Kaise, S. Fujiwara, M. Tsuzuki, T. Sakurai, T. Saitoh and C. Mastubara, “Accumulation of Arsenic in a Unicellular Alga Chlamydomonas reinhardtii,” Applied Organometallic Chemistry, Vol. 13, No. 2, 1999, pp. 107-111. doi:10.1002/(SICI)1099-0739(199902)13:2<107::AID-AOC824>3.0.CO;2-9
- I. Kobayashi, S. Fugiwara, H. Saegusa, M. Inouhe, H. Matsumoto and M. Tsuzuki, “Relief of Arsenate Toxicity by Cd-Stimulated Phytochelatin Synthesis in the Green Alga Chlamydomonas reinhardtii,” Marine Biotechnology, Vol. 8, 2005, pp. 94-101. doi:10.1007/s10126-005-5092-3
- N. Ahsan, D.-G. Lee, K.-H. Kim, I. Alam, S.-H. Lee, K.-W. Lee, H. Lee and B.-H. Lee, “Analysis of Arsenic StressInduced Differentially Expressed Proteins in Rice Leaves by Two-Dimensional Gel Electrophoresis Coupled with Mass Spectrometry,” Chemosphere, Vol. 78, No. 3, 2010, pp. 224-231. doi:10.1016/j.chemosphere.2009.11.004
- E. Bona, C. Cattaneo, P. Cesaro, F. Marsano, G. Lingua, M. cavaletto and G. Berta, “Proteomic Analysis of Pteris vittata Fronds, Two Arbuscular Mycorrhizal Fungi Differentially Modulate Protein Expression under Arsenic Contamination,” Proteomics, Vol. 10, No. 21, 2010, pp. 3811-3834. doi:10.1002/pmic.200900436
- A. A. Meharg and J. Hartley-Whitaker, “Arsenic Uptake and Metabolism in Arsenic Resistant and Nonresistant Plant Species,” New Phytologist, Vol. 154, No. 1, 2002, pp. 29-43. doi:10.1046/j.1469-8137.2002.00363.x
- X. Cao, L. Q. Ma and C. Tu, “Antioxidative Responses to Arsenic in the Arsenic-Hyperaccumulator Chinese Brake Fern (Pteris vittata L.),” Environmental Pollution, Vol. 128, No. 3, 2004, pp. 317-325. doi:10.1016/j.envpol.2003.09.018
- C. Walliwalagedara, H. van Keulen, T. J. Cutright and R. Wei, “Comparison of Sample Preparation Methods for the Resolution of Metal-Regulated Proteins in Helianthus annuus by 2-Dimensional Gel Electrophoresis,” The Open Proteomics Journal, Vol. 3, 2010, pp. 20-25.
- F. A. Salomons, K. Ács and N. P. Dantuma, “Illuminating the Ubiquitin/Proteasome System,” Experimental Cell Research, Vol. 316, No. 8, 2010, pp. 1289-1295. doi:10.1016/j.yexcr.2010.02.003
- H. Fu, J. H. Doelling, C. S. Arendt, M. Hochstrasser and R. D. Vierstra, “Molecular Organization of the 20S Proteasome Gene Family from Arabidopsis thaliana,” Genetics, Vol. 149, No. 2, 1998, pp. 677-692.
- J. Kurepa, S. Wang, Y. Li and J. Smalle, “Proteasome Regulation, Plant Growth and Stress Tolerance,” Plant Signaling & Behavior, Vol. 4, No. 10, 2009, pp. 924-927. doi:10.4161/psb.4.10.9469
- C. Walliwalagedara, I. Atkinson, H. van Keulen, T. Cutright and R. Wei, “Differential Expression of Proteins Induced by Lead in the Dwarf Sunflower Helianthus annuus,” Phytochemistry, Vol. 71, No. 13, 2010, pp. 1460- 1465. doi:10.1016/j.phytochem.2010.05.018
- A. Shapiguzov, A. Edvardsson and A. V. Vener, “Profound Redox Sensitivity of Peptidyl-Prolyl Isomerase Activity in Arabidopsis Thylakoid Lumen,” FEBS Letters, Vol. 580, No. 15, 2006, pp. 3671-3676. doi:10.1016/j.febslet.2006.05.054
- S. F. Göthel and M. A. Marahiel, “Peptidyl-Prolyl CisTrans Isomerases, a Superfamily of Ubiquitous Folding Catalysis,” Cellular and Molecular Life Sciences, Vol. 55, No. 3. 1999, pp. 423-436. doi:10.1007/s000180050299
- N. Ahsan, J. Renaut and S. Komatsu, “Recent Developments in the Application of Proteomics to the Analysis of Plant Responses to Heavy Metals,” Proteomics, Vol. 9, No. 10, 2009, pp. 2602-2621. doi:10.1002/pmic.200800935
- J. E. Sarry, L. Kuhn, C. Ducruix, A. Lafaye, C. Junot, V. Hugouvieux, A. Jourdain, O. Bastein, J. B. Fievet, D. Vailhen, B. Amekaraz, C. Moulin, E. Ezan, J. Garinand and J. Bourguignon, “The Early Responses of Arabidopsis thaliana Cells to Cadmium Exposure Explored by Protein and Metabolic Profiling Analysis,” Proteomics, Vol. 6, No. 7, 2006, pp. 2180-2198. doi:10.1002/pmic.200500543
- K. B. Aly, J. L. Pipkin, W. G. Hinson, R. J. Feuers, P. H. Duffy, L. Lyn-Cook and R. W. Hart, “Chronic Caloric Restriction Induces Stress Proteins in the Hypothalamus of Rats,” Mechanisms of Ageing and Development, Vol. 76, No. 1, 1994, pp. 11-23. doi:10.1016/0047-6374(94)90003-5
- M. E. Feder and G. E. Hofmann, “Heat-Shock Proteins, Molecular Chaperones, and the Stress Response, Evolutionary and Ecological Physiology,” Annual Review of Physiology, Vol. 61, 1999, pp. 243-282. doi:10.1146/annurev.physiol.61.1.243
- H. Führs, M. Hartwig, L. E. B. Molina, D. Heintz, A. V. van Dorsselaer, H. P. Braun and W. J. Horst, “Early Manganese-Toxicity Response in Vigna unguiculata L.—A Proteomic and Transcriptomic Study,” Proteomics, Vol. 8, No. 1, 2008, pp. 149-159. doi:10.1002/pmic.200700478
- M. Hajduch, R. Rakwal, G. K. Agrawal, M. Yonekura and A. Pretova, “High Resolution Two-Dimensional Electrophoresis Separation of Proteins from Metal Stressed Rice (Oryza sativa L.) Leaves: Drastic Reductions/Fragmentation of Ribulose-1,5-bisphosphate Carboxylase/Oxygenase and Induction of Stress-Related Proteins,” Electrophoresis, Vol. 22, No. 13, 2001, pp. 2824-2831. doi:10.1002/1522-2683(200108)22:13<2824::AID-ELPS2824>3.0.CO;2-C
- P. Kieffer, P. Schröder, J. Dommes, L. Hoffmann, J. Renaut and J. F. Hausman, “Proteomic and enzymatic Response of Poplar to Cadmium Stress,” Journal of Proteomics, Vol. 72, No. 3, 2009, pp. 379-396. doi:10.1016/j.jprot.2009.01.014
- M. H. Tuomainen, N. Nunan, S. J. Lehesranta, A. I. Tervahauta, V. H. Hassinen, H. Schat, K. M. Koistinen, S. Auriola, J. McNicol and S. O. Kärenlampi,” Multivariate Analysis of Protein Profiles of Metal Hyperaccumulator Thlaspi caerulescens Accessions,” Proteomics, Vol. 6, No. 12, 2006, pp. 3696-3706. doi:10.1002/pmic.200501357
- J.-W. Kim and C. V. Dang, “Multifaceted Roles of Glycolytic Enzymes,” Trends in Biochemical Sciences, Vol. 30, No. 3, 2005, pp. 142-150. doi:10.1016/j.tibs.2005.01.005
- N. Ahsan, D-G. Lee, I. Alam, P. J. Kim, J. J. Lee, Y. O. Ahn, S. S. Kwak, I. J. Lee, J. D. Bahk, K. Y. Kang, J. Renaut, S. Komatsu and B-H. Lee, “Comparative Proteomic Study of Arsenic-Induced Differentially Expressed Proteins in Rice Roots Reveals Glutathione Plays a Central Role during As Stress,” Proteomics, Vol. 8, No. 17, 2008, pp. 3561-3576. doi:10.1002/pmic.200701189
- T. Fukuda, A. Saito, J. Wasaki, T. Shinano and M. Osaki, “Metabolic Alterations Proposed by Proteome in Rice Roots Grown under Low P and High Al Concentration under Low pH,” Plant Science, Vol. 172, No. 6, 2007, pp. 1157-1165. doi:10.1016/j.plantsci.2007.02.020
- H. Heide, H. M. Kalisz and H. Follmann, “The Oxygen Evolving Enhancer Protein 1, (OEE) of Photosystem II in Green Algae Exhibits Thioredoxin Activity,” Journal of Plant Physiology, Vol. 161, No. 2, 2004, pp. 139-149. doi:10.1078/0176-1617-01033
- N. Rolland, A. Atteia, P. Decottignies, J. Garin, M. Hippler, G. Kreimer, S. D. Lemaire, M. Mittag and V. Wagner, “Chlamydomonas Proteomics,” Current Opinion in Microbiology, Vol. 12, No. 3, 2009, pp. 285-291. doi:10.1016/j.mib.2009.04.001
- C. S. Im and A. R. Grossman, “Identification and Regulation of High Light-Induced Genes in Chlamydomonas reinhardtii,” The Plant Journal, Vol. 30, No. 3, 2001, pp. 301-313. doi:10.1046/j.1365-313X.2001.01287.x.

