Open Journal of Veterinary Medicine
Vol.3 No.2(2013), Article ID:32697,13 pages DOI:10.4236/ojvm.2013.32023
Bifidobacteria Upregulate Expression of Toll-Like Receptor Negative Regulators Counteracting Enterotoxigenic Escherichia coli Mediated Inflammation in Bovine Intestinal Epitheliocytes
1Food and Feed Immunology Group, Laboratory of Animal Products Chemistry, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
2Laboratory of Clinical and Experimental Biochemistry, Reference Centre for Lactobacilli (CERELA-CONICET), Tucuman, Argentina
3Laboratory of Animal Breeding and Genetics, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
4Cell Biology Laboratory, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
5Department of Food, Agriculture and Environmental Science, Miyagi University, Sendai, Japan
6Food Science and Technology Institute, Morinaga Milk Industry Co. Ltd., Kanagawa, Japan
Email: #haruki@bios.tohoku.ac.jp
Copyright © 2013 Kozue Murata et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received February 12, 2013; revised March 23, 2013; accepted April 23, 2013
Keywords: Bovine Intestinal Epithelial Cells; Bifidobacteria; ETEC; TLRs Negative Regulators
ABSTRACT
We previously established a bovine intestinal epithelial cell line (BIE cells) and showed that BIE cells are useful in vitro model system for the study of interactions between pathogenic and beneficial microorganisms and bovine intestinal epithelial cells (IECs). In the present study, we aimed to select potential immunomodulatory bifidobacteria that may be used to beneficially modulate the inflammatory response in bovine IECs. We also aimed to gain insight into the molecular mechanisms involved in the anti-inflammatory effect of bifidobacteria by evaluating the role of Toll-like receptor (TLR)-2 and TLR negative regulators in the regulation of proinflammatory cytokines production and MAPK, NF-κB and PI3K pathways activation in BIE cells. Five bifidobacteria strains were evaluated in this study and according to their capacity to modulate the inflammatory response of BIE cells. Despite the unique effect of each strain, four common points were found when comparing the effect of the high and moderate anti-inflammatory strains: 1) Upregulation of TLR negative regulators and the intensity of that upregulation was related to the different immunomodulatory capacity of each bifidobacteria strain; 2) The balance between MAPK activation and MKP-1 upregulation affected the anti-inflammatory effect of bifidobacteria in BIE cells; 3) The inhibition of PI3K pathway was related to the anti-inflammatory effect of bifidobacteria; 4) The immunoregulatory effect of bifidobacteria in BIE cells is partially dependent on TLR2. This study shows that BIE cells can be used for the selection of immunoregulatory bifidobacteria and for studying the mechanisms involved in the protective activity of immunobiotics against TLR4-induced inflammatory damage. In addition, we have demonstrated that the anti-inflammatory effect of bifidobacteria was achieved by a complex interaction of multiple TLRs negative regulators as well as the inhibition/activation of multiple signaling pathways.
1. Introduction
Today, bifidobacteria are economically important because they are added in high numbers as live bacteria to numerous food preparations, with various health-related claims, especially those related to the beneficial modulation of the intestinal immune system to avoid unproductive inflammation [1-4]. In this sense, it was reported that Bifidobacterium animalis MB5 protect intestinal Caco-2 cells from the inflammation-associated response caused by enterotoxigenic Escherichia coli (ETEC) K88 by partly reducing pathogen adhesion and by counteracting neutrophil migration [1,2]. In addition, experiments in Caco- 2 cells showed that B. animalis MB5 counteracts ETECinduced upregulation of interleukin (IL)-1β and tumor necrosis factor α (TNF-α), and the downregulation of transforming growth factor β1 (TGF-β1) expression, and consequently to block the cytokine deregulation [1,2]. Others studies evaluating the effect of bifidobacteria in mice showed that the deliberate consumption of B. animalis AHC7 resulted in the attenuation of nuclear factor κB (NF-κB) activation within mice infected with a proinflammatory translocating microbe, Salmonella typhimurium. In addition, B. animalis AHC7 consumption was associated with modulation of cytokine signaling within the mucosa of healthy animals. In vitro, dendritic cells (DCs) bound B. animalis AHC7, secreted IL-10 and IL- 12p70, and DCs stimulated with B. animalis AHC7 enhanced Foxp3 expression in naïve T cells [5]. In addition, Hoarau et al. [6] demonstrated that the supernatant of Bifidobacterium breve C50 induces DCs maturation through TLR2, with a high IL-10 production. These and other studies clearly demonstrate that bifidobacteria are highly effective in inducing intestinal tolerance and regulating unproductive inflammation.
Because of the beneficial immunoregulatory properties of probiotics, there is increasing interest in the use of these microorganisms for decreasing incidence of infections and ameliorating inflammatory gastrointestinal diseases in animals. In this sense, several studies have been conducted in vivo utilizing different probiotic strains to evaluate the effect of immunobiotics against intestinal infections and unproductive inflammation, however the majority of these studies were performed in swine and only a few in the cattle [7,8]. The use of probiotics in cattle could be improved by the development of in vitro systems specifically for cattle that allow the simple and efficient assess of the immunomodulatory activity of the potential probiotic strains.
We have recently established a bovine intestinal epithetlial cell line (BIE cells) [9]. When BIE cells are cultured they assume monolayer cobblestone and epithelial-like morphology with close contact between the cells. Moreover, scanning electron microscopy examination of BIE cell revealed that 3-days old cells have irregular and slender microvilli-like structures on their surface and that these structures increase in complexity as the cells grow [9]. We have also showed that BIE cells are useful in vitro model system for the study of interactions between pathogen-associated molecular patterns (PAMPs) and bovine intestinal epithelial cells (IECs) [10]. In fact, our findings indicate that BIE cells are useful cell line for studying inflammatory responses via Toll-like receptor (TLR) activation in vitro. Considering that inflammatory responses induced by PAMPs derived from intestinal pathogens can lead to dysregulation of IECs signaling, disruption of membrane barrier integrity, enhancement of pathogen translocation and disease [11], BIE cells can be used to evaluate therapies designed for preventing inflammatory damage caused by these PAMPs during infections in the gut [10].
In the present study, we aimed to select potential immunomodulatory bifidobacteria that may be used to beneficially modulate the inflammatory response in bovine IECs. Over the past decade it has become clear that probiotic and commensal organisms can interact with IECs lining the mucosa to modulate specific functions of the mucosal immune system and that interaction involves the innate pattern recognition receptors (PRRs) such as TLRs. Therefore, we also aimed to gain insight into the molecular mechanisms involved in the anti-inflammatory effect of bifidobacteria by evaluating the role of TLR2 and TLR negative regulators in the regulation of proinflammatory cytokines production and mitogen-activated protein kinases (MAPK), NF-κB and phosphatidylinositol 3-kinase (PI3K) pathways activation in BIE cells.
2. Materials and Methods
2.1. BIE Cells
The bovine intestinal epithelial cell line (BIE cells) was originally derived from fetal bovine intestinal epitheliocytes, and then established as a SV40 large T antigentransformed intestinal cell line as previously described [9]. BIE cells were maintained in Dulbecco’s modified Eagle medium (DMEM; GIBCO, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum (FBS) and penicillin-streptomycin until it they were used for further studies. For the passage, BIE cells were treated with a sucrose/EDTA buffer (0.1 M Na2HPO4/12H2O, 0.45 M Sucrose, 0.36% EDTA/4Na, BSA) for 4 min, detached using 0.04% trypsin in phosphate-buffered saline (PBS, pH 7.2). Then, BIE cells were plated in collagen type I-coated culture dishes (Sumilon, Tokyo, Japan) at 1.5 × 104 cells/cm2 and cultured at 37˚C in an atmosphere of 5% CO2 in Dulbecco’s Modified Eagle media (DMEM) (10% FCS, 1% streptomycin/penicillin, 100 U/ml streptomycin, high glucose, L-glutamine, 0.11 mg/ml sodium pyruvate; GIBCO).
2.2. Microorganisms
Bifidobacteria were provided by Morinaga Milk Industry Co. Ltd. (Zama, Japan). Five different strains were used in the experiments: B. longum BB536, B. adolescentis ATCC15705, B. adolescentis MCC-75, B. breve MCC- 117 and B. infantis MCC-1021 [12].
Bifidobacteria were grown in Man-Rogosa-Sharpe broth and agar (Difco, Detroit, MI, USA) supplemented with 0.05% (w/v) cysteine (Sigma, Tokyo, Japan), and incubated at 37˚C for 16 h under anerobic conditions (AnaeroGen; Oxoid, Basingstoke, UK). Cultures were then centrifuged at 3000 rpm for 10 minutes and bifidobacteria were washed with PBS, and heat killed (60˚C, 30 min). These bacterial samples were resuspended in DMEM, enumerated using a Petroff-Hausser counting chamber, and stored at −80˚C until use.
Enterotoxigenic Escherichia coli (ETEC) strain 987 (O9: H−: 987P+: STa+) was kindly provided by Dr. M. Nakazawa, National Institute of Animal Health (Tsukuba, Japan) [13,14]. ETEC cells were grown in blood agar (5% sheep blood) for 2 hours at 37˚C and then transferred to tryptic soy broth (TSB; Becton, Dickinson and Company, USA) for 5 days at 37˚C without shaking. ETEC cell were collected and transferred to 1L TSB and cultured 20 hours at 37˚C with shaking. After incubation, the subcultures of bacteria were centrifuged at 5000 × g for 10 min at 4˚C and washed with PBS (pH 7.2). Finally, ETEC cell were heat killed at 100˚C for 15 minutes and then washed with PBS. Heat-killed ETEC were suspended in DMEM for use.
2.3. Anti-Inflammatory Assay in BIE Cells
Bifidobacteria were re-suspended in DMEM (10% FSC, 1% SP), enumerated in a microscope using a PetroffHausser counting chamber, and stored at –80˚C until use. BIE cells were plated at 3 × 104 cells/well of a 12-well ptype I collagen-coated plates (Iwaki, Tokyo, Japan), and cultured for three days. After changing medium, bifidobacteria (5 × 107 cells/ml) were added and 48 hours later, each well was washed vigorously with medium at least 3 times to eliminate all the stimulants. BIE cells were treated with heat-killed ETEC (final concentration: 5 × 107 cells/ml) for indicated time and the expression of cytokines, chemokines and TLRs negative regulators were studied by using real time PCR as de scribed below. In addition, activation of p38, c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase (ERK) MAPKs, PI3K and NF-kB pathways were studied by using western blotting as described below. In some experiments, the synthetic TLR2 agonist tripalmitoylated lipopeptide Pam3CysSerLys4 (Pam3CSK4) was also used. BIE cells were stimulated with Pam3CSK4 (final concentration: 200 ng/ml) for the indicated time same as the other stimuli.
In blocking experiments, unlabeled anti-human TLR2- mouse IgG (Clone #TL2.1, Cat. #309710, Biolegend, San Diego, CA) and its isotype control antibody (MOPC-173, Cat. #400224, Biolegend) was used. Cultured PIE cells were incubated with the unlabeled anti-TLR2 or isotype control antibody for 12 h before stimulation with bacteria.
2.4. Quantitative Expression Analysis by PCR in BIE Cells
Two-step real-time quantitative PCR was used to characterize the expression of cytokines, chemokines and TLRs negative regulators mRNAs in BIE cells. Total RNA from each sample was isolated from the BIE cells using TRIzol reagent (Invitrogen). All cDNAs were synthesized from 5 μg of total RNA using a Quantitect Reverse Transcription kit (Qiagen, Tokyo, Japan) according to the manufacturer’s recommendations. Real-time quantitative PCR was carried out using a 7300 Real-time PCR System (Applied Biosystems, Warrington, UK) using Platinum SYBR Green qPCR SuperMix UDG with ROX (Invitrogen). The primers for cytokines, chemokines and TLRs negative regulators used in this study are described in Table 1. The PCR cycling conditions were 5 min at 50˚C; followed by 5 min at 95˚C; then 40 cycles of 15 sec at 95˚C, 30 sec at 60˚C and 30 sec at 72˚C. The reaction mixture contained 5 μl of the sample cDNA and 15 μl of the master mix including the sense and antisense primers. Expression of β-actin was used to normalize cDNA levels for differences in total cDNA levels in the samples. TLRs mRNA levels in BIE cells were calibrated by the bovine β-actin level, and normalized by common logarithmic transformation in comparison to the each control (as 1.00).
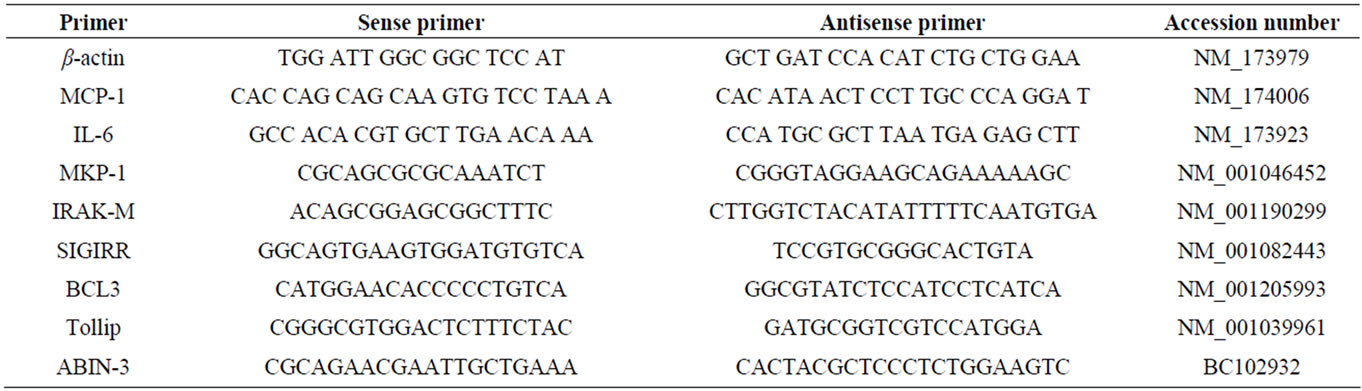
Table 1. Primer sequences used in this study.
2.5. Western Blotting
BIE cells cultured in 1.8 × 105 cells/60mm dishes were simulated with bifidobacteria or Pam3CSK4 with same time schedule and equivalent amount as mentioned above. BIE cells were then washed and stimulated with heatkilled ETEC for indicated time. After stimulation, BIE cells were washed three times with PBS and resuspended in 200 μl of CelLytic M Cell Lysis Reagent (Sigma-Aldrich, St. Louis, MO, USA) including protease and phosphates inhibitors (complete Mini, PhosSTOP: Roche, Mannheim, Germany). Protein concentration was measured with BCA protein assay kit (Pierce, Rockford, IL, USA). Extracts (120 μl) were transferred into Eppendorf tubes and were added with 40 μl of Sample Buffer Solution (2ME+) (×4) (Wako), and boiled for 5 min at 95˚C. Equal amounts of extracted proteins (2 μg) were loaded on 10% SDS-polyacrylamide gel electrophoresis (SDSPAGE). Separated proteins were transferred electrophoretically to a PVDF membrane. The membrane was blocked with 2% BSA/TBS-T (w/v) for 2 hours at room temperature. Phosphorylation of PI3K, p38, JNK and ERK mitogen-activated protein kinases and nuclear factor kappa B inhibitor alpha (IkBα) degradation were evaluated using Phospho-PI3K p85 (Tyr458)/p55 (Tyr199) antibody (p-PI3K, #4228); PI3K kinase p85 antibody (PI3K, #4292); Phospho-p38 MAPK (Thr180/Tyr182) antibody (p-p38, #9211); p38 MAPK antibody (p38, #9212); Phospho-SAPK/JNK (Thr183/Tyr185) antibody (p-JNK, #9251); SAPK/JNK antibody (JNK, #9252); Phospho-p44/42 MAP kinase (Thr202/Thy204) antibody (p-ERK, #9101); p44/42 MAP (Erk 1/2) antibody (ERK, #9102); I kappaB-alpha antibody (IkBa, #9242) from Cell Signaling Technology (Beverly, MA, USA) at 1000 times dilution of their original antibodies overnight at room temperature. After washing with TBS-T, the membrane was incubated alkaline phosphatase conjugated anti-rabbit IgG (#A3937, Sigma) at 2000 times dilution for 1 hour at room temperature. After washing with TBS-T, signals were generated by overlaying the membrane with ECFTM substrate (GE Healthcare, Piscataway, NJ, USA) for 5 min at room temperature under dark conditions. The Attophos (Ex; 440 nm, Em; 560 nm) was detected by Molecular Imager Fx (Bio-Rad, Hercules, CA, USA). The densitometry of western blots was carried out by using Image J software. The relative density was expressed as the area of the peak in the densitogram, in which expressed as 1.0 at 0 min, and compared with that of ETEC control at each time point.
2.6. Statistical Analysis
Normalized relative expression of each control data (showed as the ratio to β-actin mRNA expression) was transferred to a normal distribution with a mean of 1.0. In order to normalize the control data, they were fitted by using the function: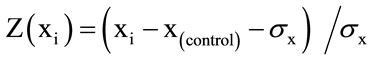 , in which Z(xi): all adjusted data; xi: ith experimental datax(control): a mean of repeated control data; and σx was a standard deviation of repeated control or trial data. Similarly, normalized relative expression for heat-killed ETEC and bifidobacteria data was fitted to this function to show them as a fold value compared to the control data. Each of data number repeated in a same condition was from 8 to 10. Statistical analysis was performed by using SAS computer program, GLM procedure. The multiple comparisons among means of fold expression were carried out by Fisher’s least significance differential test, LSD method. Differences were significant at 5% level and were showed in graphs with superscripts letters.
, in which Z(xi): all adjusted data; xi: ith experimental datax(control): a mean of repeated control data; and σx was a standard deviation of repeated control or trial data. Similarly, normalized relative expression for heat-killed ETEC and bifidobacteria data was fitted to this function to show them as a fold value compared to the control data. Each of data number repeated in a same condition was from 8 to 10. Statistical analysis was performed by using SAS computer program, GLM procedure. The multiple comparisons among means of fold expression were carried out by Fisher’s least significance differential test, LSD method. Differences were significant at 5% level and were showed in graphs with superscripts letters.
3. Results
3.1. Effect of Bifidobacteria on Heat-Killed ETEC-Induced Inflammatory Response in BIE Cells
Our studies showed that TLR4 is expressed at comparably higher level in BIE cells (Takanashi et al., 2013. BMC Microbiol, In press), and that challenge of BIE cells with heat-killed ETEC significantly increased the expression of pro-inflammatory cytokines MCP-1 and IL-6 (Figure 1). Then, we first aimed to evaluate if the bifidobacteria strain were able to modulate the expression of these cytokines in heat-killed ETEC-challenged BIE cells. For this reason, BIE cells were stimulated for 48 hours with each bifidobacteria strain or the synthetic TLR2 agonist Pam3CSK4 and then challenged with heatkilled ETEC. Twelve hours after stimulation levels of
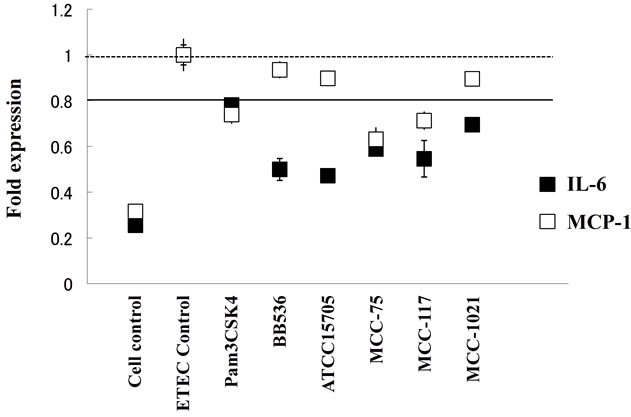
Figure 1. Expression of cytokines in BIE cells after stimulation with heat-killed Enterotoxigenic Escherichia coli (ETEC). BIE cells were pre-treated with bifidobacteria for 48 h and stimulated with heat-killed ETEC. The expression of MCP- 1 and IL-6 mRNAs were studied at 12 h post-stimulation. The results represent four independent experiments. Significant differences are showed under the solid line (P < 0.05).
MCP-1 and IL-6 were evaluated (Figure 1). Stimulation of BIE cells with all bifidobacteria strains significantly decreased the production of IL-6 in response to heatkilled ETEC challenge. However, levels of MCP-1 were significantly lower than controls only in PIE cells treated with B. adolescentis MCC-75 and B. breve MCC-117 (Figure 1). In addition, we observed that Pam3CSK4 was able to reduce levels of IL-6 and MCP-1 in PIE cells, although the effect was significantly lower when compared with B. adolescentis MCC-75 and B. breve MCC- 117 (Figure 1).
3.2. Effect of Bifidobacteria on MAPK, PI3K and NF-κB Pathways in BIE Cells
We next evaluated whether bifidobacteria were able to attenuate heat-killed ETEC-mediated pro-inflammatory responses by modulating the NF-κB pathway. Challenge of BIE cells with heat-killed ETEC significantly reduced the levels of the counter-regulatory factor IκBa (Figure 2A). BIE cells previously stimulated with B. longum BB536 or B. infantis MCC-1021 did not show a significant degradation of IκBa early from minute 5 after challenge, indicating an inhibitory effect in NF-κB pathway (Figure 2A). B. adolescentis ATCC15705 and B. adolescentis MCC-75 were also able to reduce NF-κB pathway activation 20 minutes after the challenge, while BIE cells treated with B. breve MCC-117 did not show differences when compared with control BIE cells (Figure 2A). Challenge of BIE cells with heat-killed ETEC significantly increased phosphorylation of PI3K (Figure 2B). BIE cells previously stimulated with MCC-117 and MCC-75 showed significantly lower levels of phosphorylated PI3K on minutes 20 and 40 when compared with control BIE cells. On the contrary, BB536, ATCC15705 and MCC-1021 strains significantly increased levels of phosphorylated PI3K, being the effect of ATCC15705 strain higher than the other strains (Figure 2B).
We also examined the relationship between MAPK activation and regulation of pro-inflammatory cytokines in BIE cells by bifidobacteria. BIE cells were stimulated with each bifidobacteria strain or control medium and; the activation profiles of p38, ERK and JNK were compared. As shown in Figure 3 heat-killed ETEC induced the phosphorylation of p38 and ERK, which reached a maximum between 5 and 10 minutes. Pretreatment of BIE cells with B. longum BB536 significantly increased the levels of phosphorylated p38, JNK and ERK on minutes 5 and 40 (Figure 3). We also observed significantly higher levels of phosphorylated p38 in B. adolescentis MCC-75-treated BIE cells and higher levels of phosphorylated ERK in BIE cells treated with both MCC-75 and MCC-117 strains. In addition, B. adolescentis ATCC15705 reduced the activation of p38 and JNK pathways while B. infantis MCC-1021-tretaetd BIE cells showed decreased levels of phosphorylated p38 and and ERK (Figure 3).
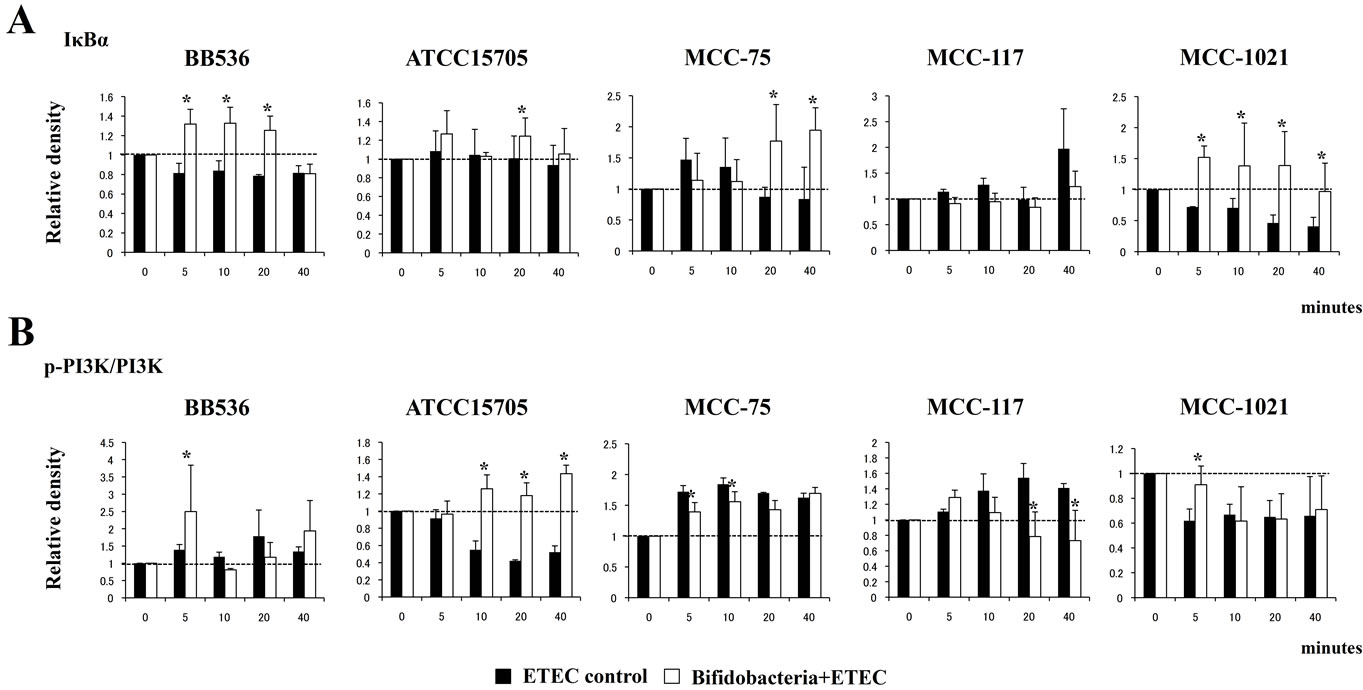
Figure 2. Western blot analysis of NF-κB (A) and PI3K (B) pathways activation on BIE cells after challenge with heat-killed ETEC. BIE cells were pre-treated with different bifidobacteria strains for 48 h and stimulated with heat-killed ETEC. Levels of the counter-regulatory factor IκBα and phosphorylated PI3K were studied at the indicated times post-stimulation. *Indicates significant differences against ETEC control at each time point (P < 0.05).
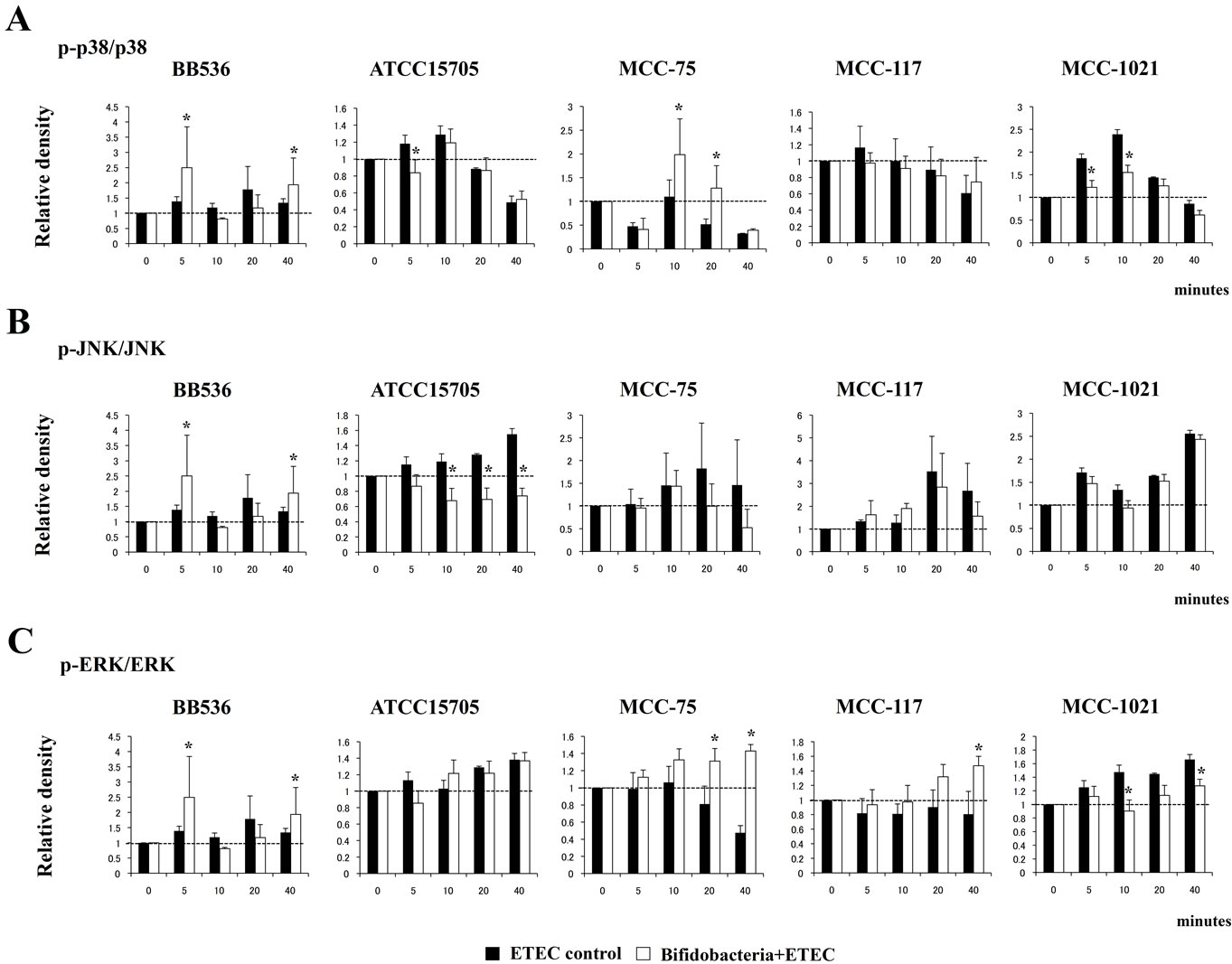
Figure 3. Western blot analysis of activation of p38 (A), JNK (B) and ERK (C) mitogen-activated protein kinases in BIE cells after challenge with heat-killed ETEC. BIE cells were pre-treated with different bifidobacteria strains for 48 h and then stimulated with heat-killed ETEC. Phosphorylation of p38, JNK, and ERK was studied at the indicated times post-stimulation. *Indicates significant differences against ETEC control at each time point (P < 0.05).
3.3. Role of TLR2 in the Anti-Inflamamtory Activity of Bifidobacteria
In order to study the role of TLR2 in the immunomodulatory effect of bifidobacteria, we next performed comparative studies with anti-TLR2 blocking antibodies (Figure 4). The use of anti-TLR2 antibodies abolished the reduction of IL-6 mRNA levels induced by BB536, ATCC15705, MCC-75 and MCC-117 strains in heatkilled ETEC-challenged BIE cells. In addition, no effect was observed in the levels of MCP-1 (Figure 4).
3.4. Effect of Bifidobacteria on Negative Regulators of the TLRs Signaling Pathway in BIE Cells
We studied the negative regulators that are known to mediate the TLR signaling pathway. First, we evaluated the changes in TLRs negative regulators that regulate the activation of several pathways involved in TLR4 signaling at the same time. We consider the intracellular TLR inhibitors Toll interacting protein (Tollip), A20-binding inhibitor of nuclear factor kappa B activation 3 (ABIN-3), and interleukin-1 receptor-associated kinase M (IRAK-M) as well as the TLR membrane-bound suppressor single immunoglobulin IL-1-related receptor (SIGIRR) (Figure 5). We observed a significant reduction of SIGIRR in hour 3 after the challenge with ETEC in all groups. BIE cells treated with MCC-117 and MCC-1021 restored the normal levels of SIGIRR on hours 6 and 12 respectively. Differently, BIE cells treated with BB536, ATCC15705 or MCC-75 showed significantly higher levels of SIGIRR after hour 3 post-challenge (Figure 5). All the bifidobacteria strains upregulated the expression of Tollip and ABIN-3, however the effect of B. adolescentis MCC-75 was significantly higher than the other strains (Figure 5). In addition, BB536, ATCC15705 and MCC-75 upregu-
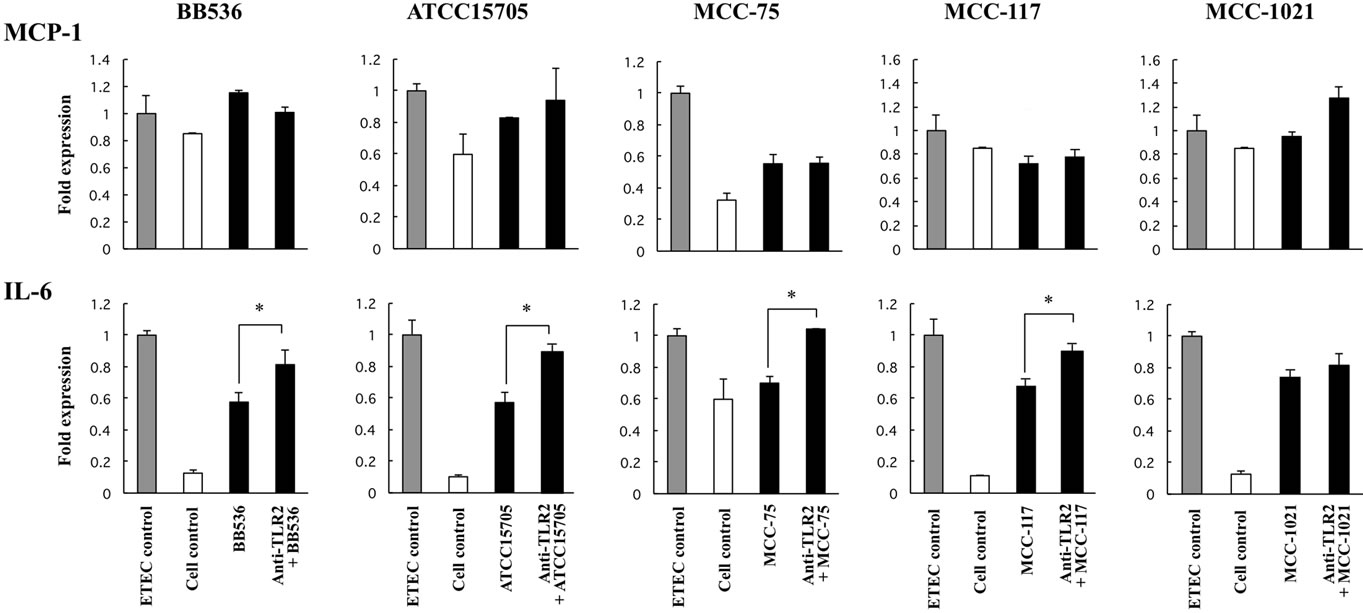
Figure 4. Role of TLR2 in the immunomodulatory effect of bifidobacteria in BIE cells. BIE cells were pre-treated with bifidobacteria strains in the presence or absence of anti-TLR2 antibodies. Untreated BIE cells were used as controls. BIE cells were stimulated with heat-killed ETEC, and then the expression of MCP-1 and IL-6 was studied at 12 h post-stimulation. The results represent four independent experiments. *Indicates significant differences between presence or absence of anti-TLR2 antibodies (P < 0.05).
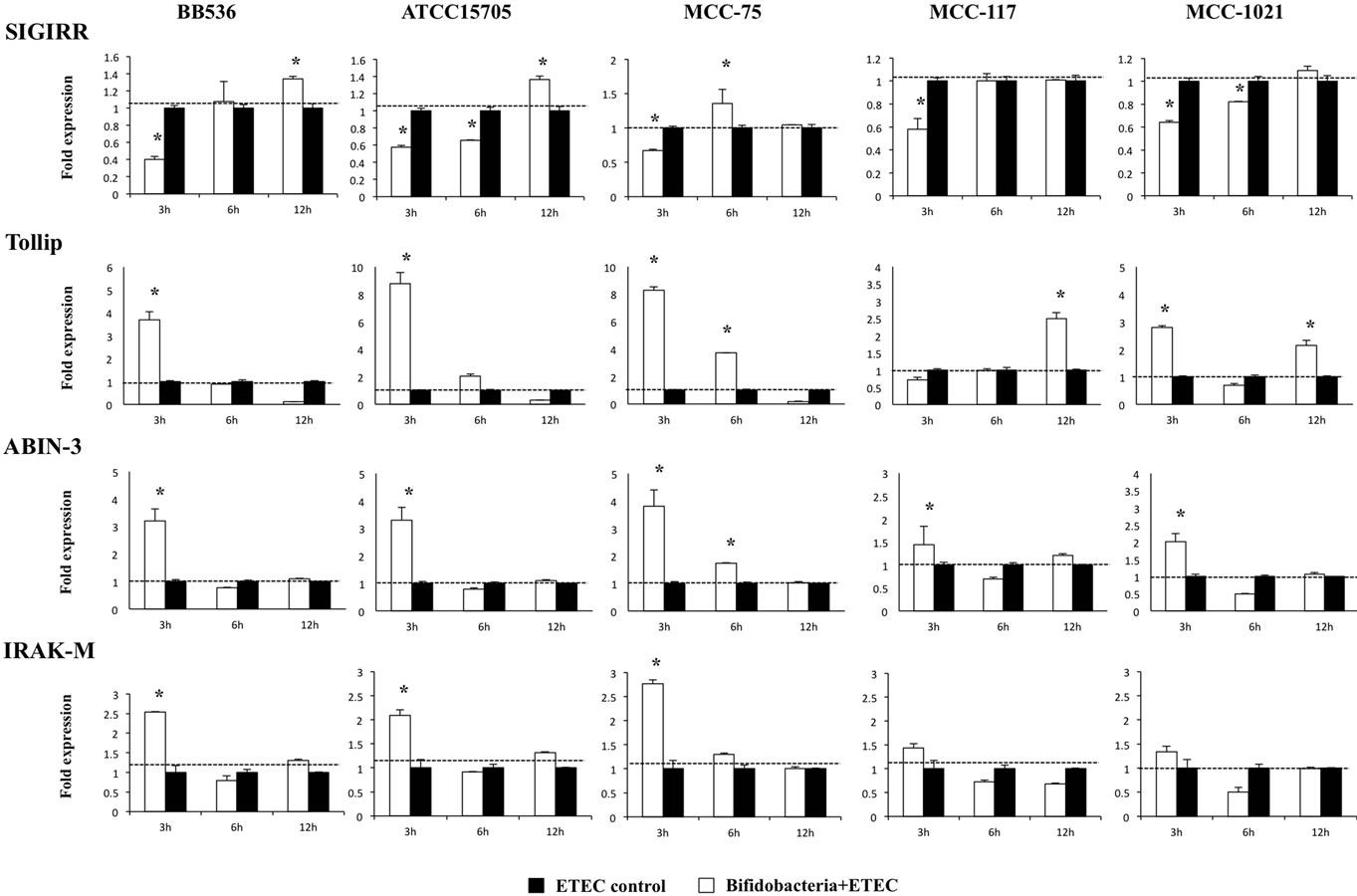
Figure 5. Expression of toll-like receptor negative regulators in BIE cells. BIE cells were pretreated with different bifidobacteria strains for 48 h, stimulated with heat-killed ETEC, and the expression of SIGIRR, Tollip, ABIN-3, and IRAK-M negative regulators was studied at the indicated time points. The results represent four independent experiments. *Indicates significant differences against ETEC control at each time point (P < 0.05).
lated the expression of IRAK-M at hour 3 post-ETEC challenge, while strains MCC-117 and MCC-1021 did not induce changes in the mRNA levels of this TLR negative regulator (Figure 5).
We also studied the effect of bifidobacteria on the expression of the NF-κB pathway inhibitor B-cell lymphoma 3-encoded protein (Bcl-3) and the MAPK pathway inhibitor mitogen-activated protein kinase 1 (MKP-1) (Figure 6). BB536, ATCC15705 and MCC-75 strain upregulated the expression of Bcl-3 in ETEC challengedBIE cells, being the effect of B. adolescentis MCC-75 significantly higher than the other strains (Figure 6). In addition, we observed a significant increase of MKP-1 in hour 3 after the challenge with heat-killed ETEC in all groups. However, the levels of MKP-1 in BB536- and ATCC15705-treated BIE cells were significantly higher that those observed in the other groups (Figure 6).
4. Discussion
The use of probiotics in animal feeding could be enhanced by preliminary in vitro screening such us the potential to adhere to IECs, production of inhibitory substances, survival in the gastrointestinal tract, antibiotic susceptibility [15] or immunoregulatory activities in immune and IECs [12] that can be analyzed to evaluate functionality and safety [7]. In this sense, we developed a method for evaluating the immune responses to immunobiotic bifidobacteria by constructing a porcine TLR2 (pTLR2)-expressing transfectant (HEKpTLR2 cells) [16,17]. We used the HEKpTLR2 immunoassay system to evaluate various bifidobacteria, belonging to different species, on the activation pattern of pTLR2-overexpressing cells [12]. As it has been observed in studies in which bifidobacteria were evaluated using immune cells [18], we observed a strain-specific capacity of bifidobacteria to stimulate HEKpTLR2 cells. Of the strains tested, B. breve MCC-117 was the one that showed the highest NF-κB activity in the HEKpTLR2 immunoassay system, while other strains of the same species were less effective or did not modify this parameter [12]. We selected B. breve MCC-117 and evaluated its anti-inflammatory effect using co-cultures of porcine intestinal epithelial cell line (PIE cells) and immune cells from porcine PPs and as well as heat-killed ETEC as inflammatory challenge [12]. We demonstrated that B. breve MCC-117 treatment was able to significantly reduce the expression of inflammatory cytokines in PIE cells in response to heat-killed ETEC. In addition, B. breve MCC-117 treatment was able to significantly reduce the levels of IFN-γ in both CD4+ and CD8+ lymphocytes and improved IL-10 levels in CD4+CD25high Foxp3+ lymphocytes [12].
These previous works demonstrated that it would be possible to modulate the intestinal immune system of animals by using appropriate bifidobacteria strains. So far, the majority of the studies evaluating the effect of probiotics in live stock animals were performed in swine and only few in the cattle [10,15]. Therefore, we were interesting in selecting immunoregulatory bifidobacteria for their use in cattle. In this work we used a previously established a bovine intestinal epithelial cell line (BIE cells) and TLR4-mediated inflammatory response to select bifidobacteria strains with immunomodulatory capacity. TLR4 is conserved among different species and its expression appears to be a characteristic feature of IECs [19], therefore, the presence of TLR4 in BIE cells resembles IECs of other species. In addition, the ETEC 987P strain used in this study does not express flagellin and we have demonstrated that the main molecule responsible for the inflammatory response triggered by this bacterium is the LPS present on its surface [13,20]. Then,
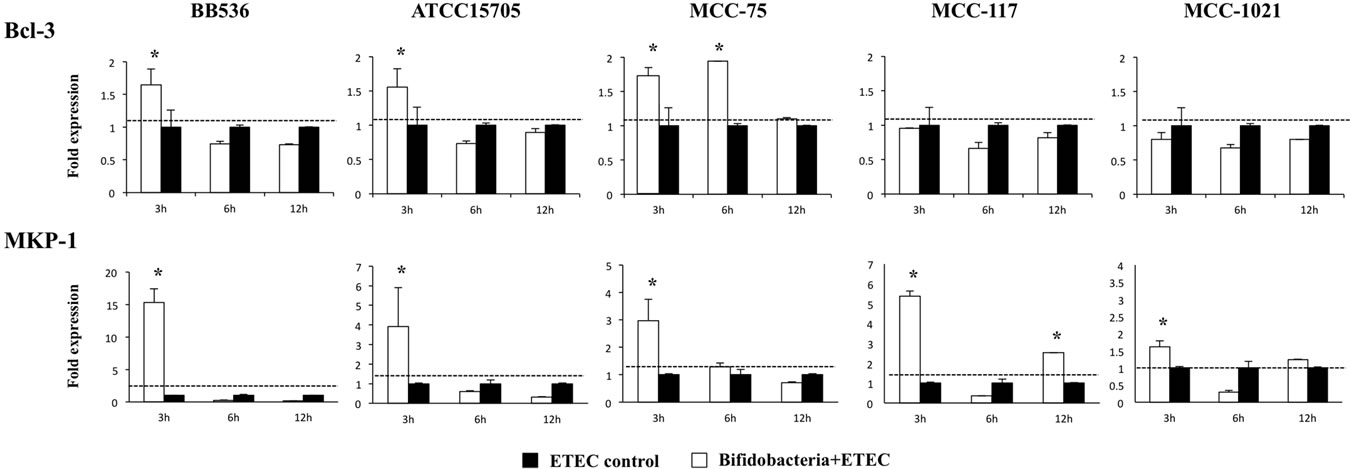
Figure 6. Expression of toll-like receptor negative regulators in BIE cells. BIE cells were pre-treated with different bifidobacteria strains for 48 h, stimulated with heat-killed ETEC, and the expression of MKP-1 and Bcl-3 negative regulators was studied at the indicated time points. The results represent four independent experiments. *Indicates significant differences against ETEC control at each time point (P < 0.05).
we showed that heat-killed ETEC significantly enhanced the production of IL-6 and MCP-1 in BIE cells by activating NF-κB, PI3K and MAPK pathways. Moreover, in the present study, we demonstrated for the first time that bifidobacteria can modulate the response of bovine intestinal epithelial cells to TLR4 activation via modulation of different signaling pathways.
Five bifidobacteria strains were evaluated in this study and according to their capacity to modulate the production of IL-6 and MCP-1 in ETEC challenged-BIE cells, they can be divided in the following two groups: 1) strains able to reduce both IL-6 and MCP-1 (B. adolescentis MCC-75 and B. breve MCC-117) and 2) strains able to reduce only IL-6 (B. longum BB536, B. adolescentis ATCC15705 and B. infantis MCC-1021). When we analyzed the activation of MAPK, NF-κB and PI3K pathways in BIE cells we found that each strain induces unique changes in TLR4 signaling. Another interesting feature of our study relates to the modulation of TLR negative regulators by bifidobacteria. When we evaluated the effect of the bifidobacteria strains on TLRs negative regulators expression in BIE cells we also found straindependent changes. Despite the unique effect of each strain, four general conclusions can be made when comparing the effect of the two different immunoregulatory groups: high anti-inflammatory activity (B. adolescentis MCC-75) and moderate anti-inflammatory activity (B. adolescentis ATCC15705) (Figure 7).
1) The upregulation of TLR negative regulators and the intensity of that upregulation would be related to the different immunomodulatory capacity of each bifidobacteria strain. It has been reported that several TLRs negative regulators have important roles in the regulation of inflammatory responses in IECs. A20 deficiency in enterocytes renders mice sensitive to TNF-α-induced lethal inflammation, leading to disruption of the epithelial barrier and infiltration of commensal bacteria that initiate a systemic inflammatory response [21], suggesting that
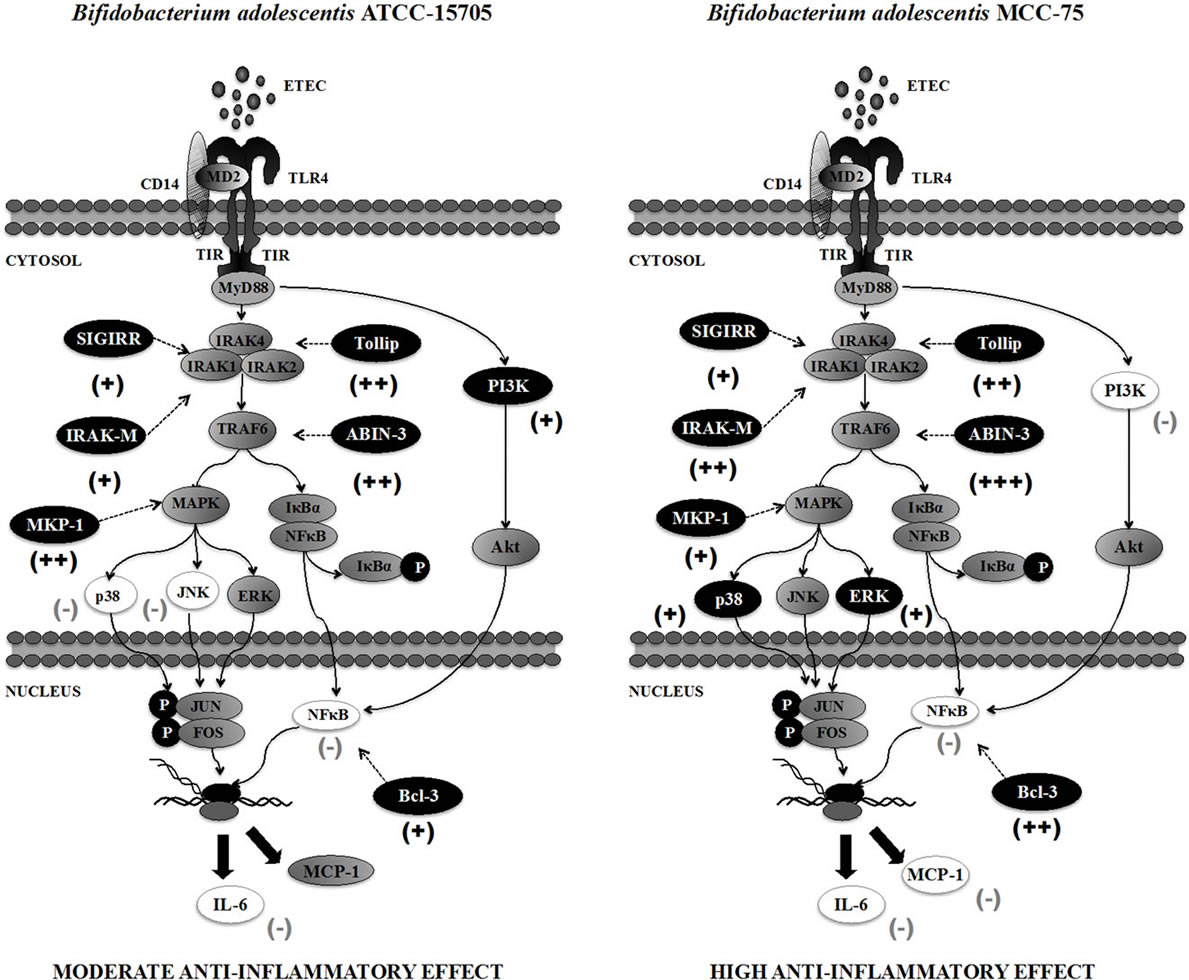
Figure 7. Proposed mechanism for the immunomodulatory effect of Bifidobacterium adolescentis ATCC-15705 and Bifidobacterium adolescentis MCC-75 in bovine intestinal epithelial cells. (−) inhibition; (+) stimulation.
A20 is important for the inhibition of innate immune responses in the gut [22]. ABINs have been described as three different proteins (ABIN-1, ABIN-2, ABIN-3) that bind A20. Overexpression of ABINs inhibits NF-κB activation by TNF-α and several other stimuli. Similar to A20, ABIN-3 expression is NF-κB dependent, implicating a potential role for the A20/ABIN complex in the negative feedback regulation of NF-κB activation [23]. On the other hand, Bcl-3 protein functions as an inhibitor of NF-κB activity by stabilizing repressive NF-κB homodimers in a DNA-bound state and preventing binding of dimers that activate transcription. In fact, stabilization of repressive complexes through the induction of Bcl-3 expression is proposed to function in the process of LPS tolerance [24]. Treatment of macrophages with IL- 10 induces the expression of Bcl-3, and Bcl-3 expression leads to inhibition of LPS-induced TNF-α production [25]. In addition, IECs deficient in SIGIRR are more susceptible to commensal-dependent intestinal inflamemation indicating the intrinsic expression of SIGIRR by IECs regulates the communication between commensal bacteria and host immune system [26]. Tollip and IRAKM are also known to be expressed at high levels in IECs, and to thereby contribute to the hyporesponsiveness of IECs to commensals [27,28]. Therefore, induction of these five negative regulators by bifidobacteria in BIE cells may be important for establishing tolerance against ETEC challenge (Figure 7). Moreover, the fold expression increase should be also important since the levels of ABIN-3, IRAK-M and Bcl-3 were significantly higher in B. adolescentis MCC-75-treated BIE cells when compared with BIE cells treated with the moderate anti-inflammatory strain B. adolescentis ATCC15705 (Figure 7).
2) The balance between MAPK activation and MKP-1 upregulation would be related to the anti-inflammatory effect of bifidobacteria in BIE cells. The MAPK pathway is involved in the upregulation of several inflammatory genes, such those encoding inducible nitric oxide synthase, IL-8, and IL-6. In addition, MKP-1 plays a role in the inhibition of proinflammatory mRNA expression, because it can inactivate MAPK pathway. An MKP-1 deficiency leads to hyperresponsiveness to LPS stimulation in innate immune cells [29]. Moreover, MKP-1 desensitizes enterocytes to TLR ligands by inactivating the p38 signaling pathway [30]. Therefore, we expected that bifidobacteria with high anti-inflammatory activity significantly upregulate MKP-1 expression and reduce MAPK activation. However, when we analyzed ERK, p38 and JNK MAPK activation and MKP-1 expression in BIE cells treated with bifidobacteria, we found that bifidobacteria with high anti-inflammatory potential activated ERK MAPK pathway and only moderately upregulated MKP-1 (Figure 7). On the contrary, bifidobacteria strains with moderate anti-inflammatory activity such as B. adolescentis ATCC15705 strongly increased expression of MKP-1 and inhibit p38 and JNK pathways (Figure 7). The ERK pathway is invoked in various cell types by diverse extracellular stimuli. Stimulation of this pathway activates a series of protein kinases and finally activates ERK1/2, which in turn phosphorylates downstream targets involved in cellular events such as cell proliferation, differentiation, survival, and motility [31]. It has been demonstrated that TGF-β rapidly induces ERK1 activity in IECs and that this TGF-β and ERK interaction regulates gene expression that is crucial for cell growth and survival of IECs as well as IECs migration [32,33]. Moreover, TGF-β increases protein levels, collagen I, TGF-β, of type-1 inhibitor of plasminogen activator and the T TGF-β-converting enzyme furin in various IEC lines via ERK [34] indicating an important immunoregulatory role of the ERK pathway in maintaining homeostasis in IECs. Therefore, the activation of the ERK pathway by B. adolescentis MCC-75 and B. breve MCC- 117 during ETEC-mediated inflammation could have an important protective role against inflammatory damage.
3) The inhibition of PI3K pathway would be related to the anti-inflammatory effect of bifidobacteria in BIE cells. We showed that B. adolescentis MCC-75 and B. breve MCC-117, the strains able to significantly reduced expression of IL-6 and MCP-1 in BIE cells, were also able to inhibit PI3K pathway. The family of PI3K enzymes is largely responsible for the phosphorylation of phosphatidylinositol lipids in response to cellular stimuli. PI3K regulates TLR signaling in both positive and negative ways. It was showed that site-directed mutagenesis of specific tyrosine residues within the cytosolic domain of TLR2 resulted in a loss in the ability of p85 to associate with TLR2 and abrogated the ability of the TLR2 to induce NF-κB transcriptional activity. Furthermore, inhibition of PI3K during TLR2 stimulation, through use of either Ly294002 or dominant-negative Akt, has been shown to greatly reduce NF-κB activation [35]. Studies on PI3K knock out mice; however, support the idea that PI3K negatively regulates TLR activation as signaling by TLR2, TLR4, TLR5, and TLR9 is elevated in p85a deficient mice [36,37]. Then, it appears that PI3K is an important TLR activation component that affects signaling in different ways depending on cell type and readout. In this work we demonstrated that stimulation of BIE cells with heat-killed ETEC activated PI3K pathway, indicating that PI3K is positively involved in TLR4 signaling in BIE cells. Moreover, we demonstrated that bifidobacteria able to reduce activation PI3K pathway were the strains with the highest anti-inflammatory activity (Figure 7).
4) The anti-inflammatory effect of bifidobacteria in BIE cells is partially dependent on TLR2. In previous works evaluating the immunoregulatory activity of immunobiotics in porcine immune cells and IECs we demonstrated that the upregulation of some regulatory cytokines and downregulation of inflammatory mediators was dependent of TLR2 activation [13,20]. In this work we evaluated this possibility by using anti-TLR2 blocking antibodies. We further demonstrated that the reduction of IL-6 induced by bifidobacteria in BIE cells was abolished when anti-TLR2 antibodies were used. This is in line with other reports demonstrating an important regulatory role of TLR2 in the recognition of bifidobacteria which possess immunoinhibitory effect [6,38,39]. It is known that TLR2 can induce tolerance against LPS that is referred to as heterotolerance or crosstolerance. Therefore, it is possible that bifidobacteria could induce crosstolerance in BIE cells through their interaction with TLR2. In addition, we showed that the reduction of MCP-1 levels after challenge of BIE cells was not abolished when anti-TLR2 antibodies were used. This finding indicates that a PRR(s), other than TLR2, may mediate the anti-inflammatory effects of bifidobacteria in BIE cells. Identification of the unknown receptor or receptors is an interesting topic for future investigations.
5. Conclusion
We firstly reported in this study that BIE cells can be used for the selection of immunoregulatory bifidobacteria and for studying the mechanisms involved in the protective activity of immunobiotics against TLR4-induced inflammatory damage. The screening and precise selection of bifidobacteria strains that are able to beneficially modulate the immune system in the bovine host can provide useful information that may be used for the development of new immunologically functional feeds. In addition, we have demonstrated in this work that the antiinflammatory effect of bifidobacteria was achieved by a complex interaction of multiple TLRs negative regulators as well as the inhibition/activation of multiple signaling pathways (Figure 7).
6. Acknowledgements
This study was supported by a Grant-in-Aid for Scientific Research (B) (2) (No. 21380164, 24380146) and Challenging Exploratory Research (No. 23658216) from the Japan Society for the Promotion of Science (JSPS), Japan Racing Association and the Japan Dairy Association (J-milk) to Dr. H. Kitazawa. Dr. Julio Villena was supported by JSPS (Postdoctoral Fellowship for Foreign Researchers, Program No. 21-09335). We thank Leonardo Albarracin for his help with the design and development of figures.
REFERENCES
- M. Roselli, A. Finamore, M. S. Britti, S. R. Konstantinov, H. Smidt, W. M. de Vos and E. Mengheri, “The Novel Porcine Lactobacillus Sobrius Strain Protects Intestinal Cells from Enterotoxigenic Escherichia coli K88 Infection and Prevents Membrane Barrier Damage,” Journal of Nutrition, Vol. 137, No. 12, 2007, pp. 2709-2716.
- M., Roselli, A., Finamore, M. S. Britti and E. Mengheri, “Probiotic Bacteria Bifidobacterium Animalis MB5 and Lactobacillus rhamnosus GG Protect Intestinal Caco-2 Cells from the Inflammation-Associated Response Induced by Enterotoxigenic Escherichia coli K88,” British Journal of Nutrion, Vol. 95, 2006, pp. 1177-1184. doi:10.1079/BJN20051681
- G., Zanello, F. Meurens, M. Berri, C. Chevaleyre, S. Melo, E. Auclair and H. Salmon, “Saccharomyces Cerevisiae Decreases Inflammatory Responses Induced by F4+ Enterotoxigenic Escherichia coli in Porcine Intestinal Epithelial Cells,” Veterinary Immunology and Immunopathology, Vol. 141, No. 1-2, 2011, pp. 133-138. doi:10.1016/j.vetimm.2011.01.018
- G. Zanello, M. Berri, J. Dupont, P. Y. Sizaret, R. D’Inca, H. Salmon and F. Meurens, “Saccharomyces cerevisiae Modulates Immune Gene Expressions and Inhibits ETECMediated ERK1/2 and p38 Signaling Pathways in Intestinal Epithelial Cells,” PLoS One, Vol. 6, No. 4, 2011, Article ID: e18573. doi:10.1371/journal.pone.0018573
- D. O’Mahony, S. Murphy, T. Boileau, J. Park, F. O’Brien, G. Groeger, P. Konieczna, M. Ziegler, P. Scully, F. Shanahan, B. Kiely and L. O’Mahony, “Bifidobacterium animalis AHC7 Protects against Pathogen-Induced NF-κB Activation in vivo,” BMC Immunology, Vol. 11, 2010, p. 63. doi:10.1186/1471-2172-11-63
- C. Hoarau, L. Martin, D. Faugaret, C. Baron, A. Dauba, C. Aubert-Jacquin, F. Velge-Roussel and Y. Lebranchu, “Supernatant from Bifidobacterium Differentially Modulates Transduction Signaling Pathways for Biological Functions of Human Dendritic Cells,” PLoS One, Vol. 3, No. 7, 2008, Article ID: e2753. doi:10.1371/journal.pone.0002753
- F. Gaggìa, P. Mattarelli and B. Biavati, “Probiotics and Prebiotics in Animal Feeding for Safe Food Production,” International Journal of Food Microbiology, Vol. 141, 2010, pp. S15-S28. doi:10.1016/j.ijfoodmicro.2010.02.031
- J. Villena, H. Aso, S. Alvarez and H. Kitazawa, “Porcine Toll-Like Receptors and Their Crosstalk with Immunobiotics: Impact in the Regulation of Gut Inflammatory Immunity,” Probiotics: Sources, Types and Health Benefits, NOVA Science Publishers, Inc., New York, 2012, pp. 53- 84.
- K. Miyazawa, T. Hondo, T. Kanaya, S. Tanaka, I. Takakura, W. Itani, M. T. Rose, H. Kitazawa, T. Yamaguchi and H. Aso, “Characterization of Newly Established Bovine Intestinal Epithelial Cell Line,” Histochemical Cell Biology, Vol. 133, No. 1, 2010, pp. 125-134. doi:10.1007/s00418-009-0648-3
- E. Chiba, J. Villena, S. Hosoya, N. Takanashi, T. Shimazu, H. Aso, M. Tohno, Y. Suda, Y. Kawai, T. Saito, K. Miyazawa, F. He and H. Kitazawa, “A Newly Established Bovine Intestinal Epithelial Cell Line Is Effective for in Vitro Screening of Potential Antiviral Immunobiotic Microorganisms for Cattle,” Research in Veterinary Science, Vol. 93, No. 2, 2012, pp. 688-694. doi:10.1016/j.rvsc.2011.10.002
- J. Berkes, V. K. Viswanathan, S. D. Savkovic and G. Hecht, “Intestinal Epithelial Responses to Enteric Pathogens: Effects on the Tight Junction Barrier, Ion Transport, and Inflammation,” Gut, Vol. 52, 2003, pp. 439-451. doi:10.1136/gut.52.3.439
- H. Fujie, J. Villena, M. Tohno, K. Morie, T. Shimazu, H. Aso, Y. Suda, N. Iwabuchi, J. Xiao, K. Iwatsuki, Y. Kawai, T. Saito and H. Kitazawa, “Toll-Like Receptor-2 Activating Bifidobacteria Strains Differentially Regulate Inflammatory Cytokines in Porcine Intestinal Epithelial Cell Culture System: Finding New Anti-Inflammatory Immunobiotics,” FEMS Immunology and Medical Microbiology, Vol. 63, No. 1, 2011, pp. 129-139. doi:10.1111/j.1574-695X.2011.00837.x
- J. Villena, R. Suzuki, H. Fujie, E. Chiba, T. Takahashi, T. Shimazu, H. Aso, S. Ohwada, Y. Suda, S. Ikegami, H. Itoh, S. Alvarez, T. Saito and H. Kitazawa, “Immunobiotic Lactobacillus jensenii Modulates Toll-Like Receptor 4-Induced Inflammatory Response via Negative Regulation in Porcine Antigen Presenting Cells,” Clinical and Vaccine Immunology, Vol. 19, No. 7, 2012, pp. 1038- 1053. doi:10.1128/CVI.00199-12
- T. Yamamoto and M. Nakazawa, “Detection and Sequences of the Enteroaggregative Escherichia coli Heat-Killed Enterotoxin 1 Gene in Enterotoxigenic E. coli Strains Isolated from Piglets and Calves with Diarrhea,” Journal of Clinical Microbiology, Vol. 35, No. 1, 1997, pp. 223-227.
- N. C. Maldonado, C. Silva de Ruiz, M. C. Otero, F. Sesma and M. E. Nader-Macías, “Lactic Acid Bacteria Isolated from Young Calves—Characterization and Potential as Probiotics,” Research in Veterinary Science, Vol. 92, No. 2, 2012, pp. 342-349. doi:10.1016/j.rvsc.2011.03.017
- M. Tohno, T. Shimosato, Y. Kawai, H. Aso, S. Ikegami, N. Takemoto, T. Saito and H. Kitazawa, “Advanced Molecular Immunoassay System for Immunobiotic Lactic Acid Bacteria Using a Transfectant of Toll-Like Receptor 2,” Animal Science Journal, Vol. 78, No. 2, 2007, pp. 195-201. doi:10.1111/j.1740-0929.2007.00425.x
- H. Kitazawa, M. Tohno, T. Shimosato and T. Saito, “Development of Molecular Immunoassay System for Probiotics via Toll-Like Receptors Based on Food Immunology,” Animal Science Journal, Vol. 79, No. 1, 2008, pp. 11-21. doi:10.1111/j.1740-0929.2007.00491.x-i1
- P. López, M. Gueimonde, A. Margolles and A. Suárez, “Distinct Bifidobacterium Strains Drive Different Immune Responses in Vitro,” International Journal of Food Microbiology, Vol. 138, No. 1-2, 2010, pp. 157-165. doi:10.1016/j.ijfoodmicro.2009.12.023
- J. D. Lee, J. H. Mo, C. Shen, A. N. Rucker and E. Raz, “Toll-Like Receptor Signaling in Intestinal Epithelial Cells Contributes to Colonic Homoeostasis,” Current Opinion in Gastroenterology, Vol. 23, No. 1, 2007, pp. 27-33. doi:10.1097/MOG.0b013e3280118272
- T. Shimazu, J. Villena, M. Tohno, H. Fujie, S. Hosoya, T. Shimosato, H. Aso, Y. Suda, Y. Kawai, T. Saito, S. Makino, S. Ikegami, H. Itoh and H. Kitazawa, “Immunobiotic Lactobacillus jensenii Elicit Anti-Inflammatory Activity in Porcine Intestinal Epithelial Cells by Modulating Negative Regulators of the Toll-Like Receptor Signaling Pathway,” Infection and Immunity, Vol. 80, No. 1, 2012, pp. 276-288. doi:10.1128/IAI.05729-11
- L. Vereecke, M. Sze, C. Mc Guire, B. Rogiers, Y. Chu, M. Schmidt-Supprian, M. Pasparakis, R. Beyaert and G. van Loo, “Enterocyte-Specific A20 Deficiency Sensitizes to Tumor Necrosis Factor-Induced Toxicity and Experimental Colitis,” Journal of Experimental Medicine, Vol. 207, 2010, pp. 1513-1523. doi:10.1084/jem.20092474
- O. Shibolet and D. K. Podolsky, “TLRs in the Gut. IV. Negative Regulation of Toll-Like Receptors and Intestinal Homeostasis: Addition by Subtraction,” American Journal of Physiology Gastrointestestinal and Liver Physiology, Vol. 292, No. 6, 2007, pp. G1469-G1473. doi:10.1152/ajpgi.00531.2006
- L. Verstrepen, I. Carpentier, K. Verhelst and R. Beyaert, “ABINs: A20 Binding Inhibitors of NF-Kappa B and Apoptosis Signaling,” Biochemical Pharmacology, Vol. 78, No. 2, 2009, pp. 105-114. doi:10.1016/j.bcp.2009.02.009
- J. Wessells, M. Baer, H. A. Young, E. Claudio, K. Brown, U. Siebenlist and P. F. Johnson, “BCL-3 and NF-Kappa B p50 Attenuate Lipopolysaccharide-Induced Inflammatory Responses in Macrophages,” Journal of Biological Chemistry, Vol. 279, 2004, pp. 49995-50003. doi:10.1074/jbc.M404246200
- H. Kuwata, Y. Watanabe, H. Miyoshi, M. Yamamoto, T. Kaisho, K. Takeda and S. Akira, “IL-10-Inducible Bcl-3 Negatively Regulates LPS-Induced TNF-Alpha Production in Macrophages,” Blood, Vol. 102, No. 12, 2003, pp. 4123-4129. doi:10.1182/blood-2003-04-1228
- H. Xiao, M. F. Gulen, J. Qin, J. Yao, K. Bulek, D. Kish, C. Z. Altuntas, D. Wald, C. Ma, H. Zhou, V. K. Tuohy, R. L. Fairchild, C. de la Motte, D. Cua, B. A. Vallance and X. Li, “The Toll-Interleukin-1 Receptor Member SIGIRR Regulates Colonic Epithelial Homeostasis, Inflammation, and Tumorigenesis,” Immunity, Vol. 26, 2007, pp. 461- 475. doi:10.1016/j.immuni.2007.02.012
- N. Oshima, S. Ishihara, M. A. Rumi, M. M. Aziz, Y. Mishima, C. Kadota, I. Moriyama, N. Ishimura, Y. Amano and Y. Kinoshita, “A20 Is an Early Responding Negative Regulator of Toll-Like Receptor 5 Signalling in Intestinal Epithelial Cells during Inflammation,” Clinical and Experimental Immunology, Vol. 159, No. 2, 2010, pp. 185- 198. doi:10.1111/j.1365-2249.2009.04048.x
- Y. Sugi, K. Takahashi, K. Nakano, A. Hosono and S. Kaminogawa, “Transcription of the Tollip Gene Is Elevated in Intestinal Epithelial Cells through Impaired O-GlcNAcylation-Dependent Nuclear Translocation of the Negative Regulator Elf-1,” Biochemical and Biophysical Research Communications, Vol. 412, No. 4, 2011, pp. 704- 709. doi:10.1016/j.bbrc.2011.08.035
- Q. Zhao, X. Wang, L. D. Nelin, Y. Yao, R. Matta, M. E. Manson, R. S. Baliga, X. Meng, C. V. Smith and J. A. Bauer, “MAP Kinase Phosphatase 1 Controls Innate Immune Responses and Suppresses Endotoxic Shock,” Journal of Experimental Medicine, Vol. 203, No. 1, 2006, pp. 131-140. doi:10.1084/jem.20051794
- J. Wang, H. R., Ford and A. V. Grishin, “NF-Kappa BMediated Expression of MAPK Phosphatase-1 Is an Early Step in Desensitization to TLR Ligands in Enterocytes,” Mucosal Immunology, Vol. 3, 2010, pp. 523-534. doi:10.1038/mi.2010.35
- M. Kohno and J. Pouyssegur, “Targeting the ERK Signaling Pathway in Cancer Therapy,” Annals of Medicine, Vol. 38, No. 3, 2006, pp. 200-211. doi:10.1080/07853890600551037
- M. T. Hartsough and K. M. Mulder, “Transforming Growth Factor Beta Activation of p44MAPK in Proliferating Cultures of Epithelial Cells,” Journal of Biological Chemestry, Vol. 270, 1995, pp. 7117-7124. doi:10.1074/jbc.270.13.7117
- Y. Mamane, E. Petroulakis, O. LeBacquer and N. Sonenberg, “mTOR, Translation Initiation and Cancer,” Oncogene, Vol. 25, 2006, pp. 6416-6422. doi:10.1038/sj.onc.1209888
- F. Blanchette, N. Rivard, P. Rudd, F. Grondin, L. Attisano and C. M. Dubois, “Cross-Talk between the p42/p44 MAP Kinase and Smad Pathways in Transforming Growth Factor Beta 1-Induced Furin Gene Transactivation,” Journal of Biological Chemestry, Vol. 276, 2001, pp. 33986- 33994. doi:10.1074/jbc.M100093200
- L. Arbibe, J. P. Mira, N. Teusch, L. Kline, M. Guha and N. Mackman, “Toll-Like Receptor 2-Mediated NF-Kappa B Activation Requires a Rac1-Dependent Pathway,” Nature Immunology, Vol. 1, 2000, pp. 533-540. doi:10.1038/82797
- T. Fukao, M. Tanabe, Y. Terauchi, T. Ota, S. Matsuda and T. Asano, “PI3K-Mediated Negative Feedback Regulation of IL-12 Production in DCs,” Nature Immunology, Vol. 3, 2002, pp. 875-881. doi:10.1038/ni825
- Y. Yu, S. Nagai, H. Wu, A. S. Neish, S. Koyasu and A. T. Gewirtz, “TLR5-Mediated Phosphoinositide 3-Kinase Activation Negatively Regulates Flagellin-Induced Proinflammatory Gene Expression,” Journal of Immunology, Vol. 176, No. 10, 2006, pp. 6194-6201.
- J. M. Otte and D. K. Podolsky, “Functional Modulation of Enterocytes by Gram-Positive and Gram-Negative Microorganisms,” American Journal of Physiology Gastrointestinal and Liver Physiology, Vol. 286, No. 4, 2004, pp. G613-G626. doi:10.1152/ajpgi.00341.2003
- L. H. Zeuthen, L. N. Fink and H. Frøkiaer, “Toll-Like Receptor 2 and Nucleotide-Binding Oligomerization Domain-2 Play Divergent Roles in the Recognition of GutDerived Lactobacilli and Bifidobacteria in Dendritic Cells,” Immunology, Vol. 124, No. 4, 2008, pp. 489-495. doi:10.1111/j.1365-2567.2007.02800.x
NOTES
*Equal contribution.
#Corresponding author.

