Advances in Microbiology
Vol.3 No.4(2013), Article ID:35533,7 pages DOI:10.4236/aim.2013.34052
Effect of Antibiotic Consumption on Resistance of Pseudomonas aeruginosa Isolated from Lebanese Patients with Emphasis on MBL Production*
1Faculty of Health Sciences, University of Balamand, El-Koura, Lebanon
2MICROB Unit, Faculty of Medicine and Medical Sciences, University of Balamand, El-Koura, Lebanon
3Faculty of Medicine and Medical Sciences, University of Balamand, El-Koura, Lebanon
4Infectious Diseases Department, Centre Hospitalier du Nord, Zgharta, Lebanon
Email: #Ziad.daoud@balamand.edu.lb
Copyright © 2013 Rima Mouawad et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 15 May, 2013; revised 15 June, 2013; accepted 15 July, 2013
Keywords: Pseudomonas Aeruginosa; MBL; Antibiotic Consumption; Bacterial Resistance
ABSTRACT
The relationship between antibiotic consumption and resistance has been widely evaluated. Pseudomonas aeruginosa is one of the most important opportunistic pathogens in the nosocomial setting, and its resistance to antibiotics is increaseing. Production of metallo-β-lactamases (MBLs) is currently the most fearful resistance mechanism due to the potential of dissemination. This study aimed to evaluate the correlation between antibiotic consumption (expressed in DDD/100 bed days) and resistance (expressed in % of isolates and patients) in different time periods for P. aeruginosa between 2006 and 2009 at Saint George Hospital University Medical Center (SGH-UMC), Beirut. Pearson correlation coefficients (r) were calculated and linear regression was performed. Detection of MBL-producing Imipenem resistant P. aeruginosa (IRPA) isolates between 2008 and 2009 was performed using three MBL screening methods: MBL Etest®, Imipenem/EDTA combined disk test and EDTA disk potentiation with four cephalosporins. The modified Hodge test was also performed. From 2006 till 2009, there was a trend of increasing resistance of P. aeruginosa to all antibiotics, and the highest % of resistance was for Ofloxacin. Concerning resistance expressed by isolates, high correlation coefficients resulted among Imipenem, Ciprofloxacin and Tazobactam consumption and resistance to these agents in the same year correlation; Ceftazidime and Ofloxacin consumption and resistance in the next year correlation; Gentamicin and Ofloxacin consumption and the change in resistance (ΔR). Concerning resistance expressed by patients, results were similar except for Ceftazidime and Ofloxacin correlation in the next year correlation. In MBL screening, three isolates gave accordance among 4 methods which showed a positive result. The correlation between antibiotic consumption and resistance is highly dependent on the kind of antibiotic, the organism and the time of correlation. Various MBL screening phenotypic methods on one isolate can increase accuracy and eliminate false positive and negative results.
1. Introduction
Treatment failure resulting from antimicrobial resistance is a rising problem facing clinicians worldwide [1]. Antibiotic overuse and abuse is partially responsible for the increasing incidence of resistant microorganisms [2]. Some studies show a significant correlation between antibiotic use and resistance among clinically significant Gram-negative bacteria, such as E. coli, P. aeruginosa and Klebsiella spp. [3,4]. P. aeruginosa is an opportunistic pathogen that is responsible for a large number of nosocomial infections [5]. It is innately resistant to a wide range of antimicrobial agents due to the low permeability of its cell membrane [6]. This bacterium may also acquire resistance to antimicrobial agents through several mechanisms including production of β-lactamases, upregulation of efflux pumps, and alteration or downregulation of outer membrane porins [7].
Imipenem is one of the most effective antimicrobial agents used for treatment of infections with P. aeruginosa. However, increasing rates of Imipenem resistance among P. aeruginosa isolates are reported [8]. Although the downregulation of the porin OprD (with or without additional mechanisms) is the most common cause of carbapenem resistance in P. aeruginosa [9], the most threatening mechanism is the production of MBL. This is mainly due to this enzyme’s ability to hydrolyze virtually all β-lactams and its presence on mobile genetic elements such as plasmids and transposons [10]. Rapid detection of MBL producing P. aeruginosa strains is crucial for the prevention of its transmission and the implication of effective infection control measures [10]. The genotypic detection of MBL producing strains using PCR is expensive, requires specialized instruments, and is limited to detecting already described MBL-encoding genes. The availability of rapid and easy-to-perform phenotypic tests for the detection of MBL producing organisms is crucial for the prevention of outbreaks [11]. Such tests exist and include the Double Disk Synergy Test (DDST) and the MBL E-test with the use of EDTA as a metal chelating agent, the Modified Hodge Test (MHT), EDTA disk potentiation, and the Combined Disk Test (CDT) [12]. There is currently no recommendation by the CLSI for the use of a particular test in the screening for MBL [13]. However, the CDT and the DDST were found to be of high sensitivity for the detection of MBL [10].
A study done in a 600 bed hospital over a period of three years showed a significant correlation between Imipenem consumption and Imipenem resistance among P. aeruginosa isolates [14]. Another nationwide study conducted in Germany also showed a positive correlation between Imipenem usage and resistance to this antimicrobial agent in P. aeruginosa isolates [15]. Data concerning the correlation between antibiotic consumption and resistance among P. aeruginosa isolates in Lebanon, as well as the occurrence of MBL among these bacteria, is scarce. In this study, our aim was to evaluate the relationship between antibiotic consumption and resistance in P. aeruginosa from the Saint Georges Hospital University Medical Center (SGH-UMC) between 2006 and 2009 and to evaluate the occurrence of MBL production among these isolates using phenotypic screening methods.
2. Materials & Methods
2.1. Antibiotic Consumption
The antibiotic consumption data from 2006 to 2008 and the percentage of Imipenem Resistant P. aeruginosa (IRPA) strains from 2006 to 2009 were collected at the Saint George Hospital—University Medical Center (SGH-UMC), a 300-bed hospital localized in Beirut and considered as one of the busiest hospitals of the country, receiving referrals from all over Lebanon. The correlation between these two parameters was determined by taking the percentage of Imipenem Resistant Pseudomonas aeruginosa (IRPA) as the dependent variable and the antibiotic consumption (measured in DDD/100 bed days) as the independent variable. The antibiotics that were included in this analysis were: Amikacin, Aztreonam, Cefepime, Ceftazidime, Ciprofloxacin, Gentamicin, Imipenem, Ofloxacin and Piperacillin/Tazobactam. Three Pearson correlation coefficients (r) were calculated for each antibiotic: (r)1 described the correlation between consumption and resistance from 2006 till 2008 (same year correlation); (r)2 described the correlation from 2007 until 2009 (next year correlation); and (r)3 described the correlation between consumption (2006 till 2008) and the change in the percentage of resistance (ΔR) in two consecutive years (2007-2006, 2008-2007, and 2009-2008). The correlation coefficient was calculated twice for each antibiotic using resistance by isolate for the first set and resistance by patient for the second set. Linear regression analysis was then performed in order to further emphasize the obtained results. The statistical analysis was done using SPSS version 17.0.
2.2. Phenotypic Detection of Resistance
Phenotypic detection of MBL in 20 isolates of Imipenem resistance Pseudomonas aeruginosa from 2008 to 2009 was done using all the following tests.
1) The Imipenem-EDTA CDT was performed by inoculating 10 [8] CFU/mL of the tested strain on Mueller Hinton Agar (MHA) and dispensing two 10 µg Imipenem disks 3 mm apart. 0.1 M EDTA was added to one of the disks. An increase in the inhibition zone of >4 mm upon EDTA induction after an overnight incubation was indicative of MBL production [10].
2) The EDTA Disk potentiation was performed by dispensing Ceftazidime, Ceftizoxime, Cefepime, and Cefotaxime (30 µg each) on a lawn of the tested strain on MHA 25 mm apart from a central disk containing only 10 µL of 0.1 M EDTA. The observation of synergy or enhancement of the inhibition zone of any of the above disks after an overnight incubation was considered positive for MBL production [12].
3) The MBL E-test was done by dispensing the MBL E-test Strip® (Biomerieux) on a lawn of the strain to be tested on MHA. The plates were incubated overnight and an MIC ratio of Imipenem/Imipenem + EDTA of ≥8 mm was considered positive for MBL production [16].
4) The Modified Hodge Test (MHT) was done by preparing a lawn of E. coli (ATCC 25922) on MHA and dispensing a 10 µg Imipenem disk in the center of the plate. The tested strain was then streaked from the edge of the disk to the periphery of the plate. The plate was incubated overnight and a distortion in the inhibition zone of the Imipenem disk was indicative of carbapenemase production [17].
3. Results
3.1. Resistance of P. aeruginosa
Resistance to Ofloxacin was the highest in all four years (Table 1). The percent of resistance by isolates to all antibiotics except Aztreonam increased from 2006 to 2008, and then decreased in 2009 with the exception of Ceftazidime, which remained nearly constant in the last two years. By patients, resistance also increased from 2006 to 2008 in all antibiotics except Aztreonam. However, it remained approximately the same from 2008 to 2009 except for Ceftazidime and gentamycin (Table 1).
3.2. Consumption and Resistance
Data obtained from the correlation of the consumption and the percentage of resistance by isolates showed that the highest positive correlation coefficient for the same year correlation was for Piperacillin/Tazobactam (r1 = 0.999). Ciprofloxacin and Imipenem also showed high correlation coefficients (0.964 and 0.871, respectively), while negative correlation coefficients were observed for Cefepime, Gentamycin, and Ofloxacin. In the next year correlation, Ceftazidime showed the highest correlation coefficient (r2 = 0.986). Ofloxacin also showed a high correlation coefficient (r2 = 0.856) while Amikacin, Aztreonam, and Cefepime showed a negative one. Using ΔR, the highest coefficient was found for Gentamycin (r3 = 0.964). Ofloxacin also had a high coefficient (r3 = 0.813) while Amikacin, Aztreonam, Ciprofloxacin, Imipenem, and Piperacillin/Tazobactam showed negative correlation coefficients. The data is presented in Figure 1.
Data obtained from the correlation of the consumption and the percentage of resistance by patient showed that the highest positive correlation coefficient for the same year correlation was for Imipenem (r1 = 0.987). Piperacillin/tazobactam and Ceftazidime also showed high correlation coefficients (0.928 and 0.894 respectively) while negative correlation coefficients were observed for Cefepime, Gentamycin, and Ofloxacin. In the next year correlation, Aztreonam showed the highest correlation coefficient (r2 = 1.000) while Cefepime and Ciprofloxacin showed negative ones. Using ΔR, the highest correlation coefficient was for Gentamycin (r3 = 0.979). Amikacin, Aztreonam, Ceftazidime, Ciprofloxacin, Imipenem, and Piperacillin/Tazobactam showed negative correlation coefficients. The data is presented in Figure 2.
3.3. Detection of Metallo-Beta-Lactamases
MBL detection varied among the 20 IRPA strains depending on the test being used. Table 2 shows the detection of MBL in each of the phenotypic tests that were done.
Among the 20 tested IRPA isolates, 14 showed a positive result by the MBL E-test, 18 were positive in the CDT test, and 9 were positive in the disk potentiation test. Among the 9 Ceftazidime susceptible isolates, 6 showed an MBL positive E-test, all 9 were positive on the CDT test, and 3 were positive in the disk potentiation test. Only 8 isolates showed an MBL positive result in all
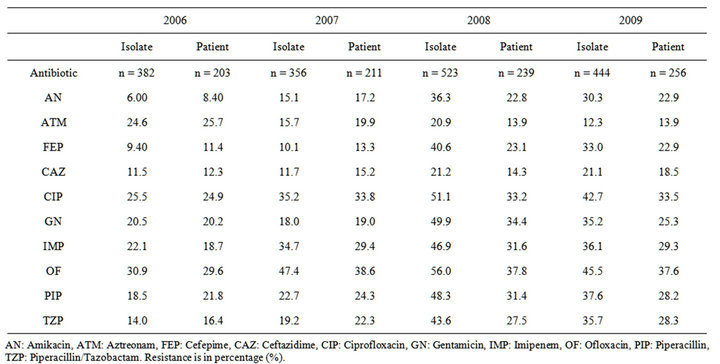
Table 1. % of Resistance of P. aeruginosa from 2006 till 2009 by isolates and by patients.
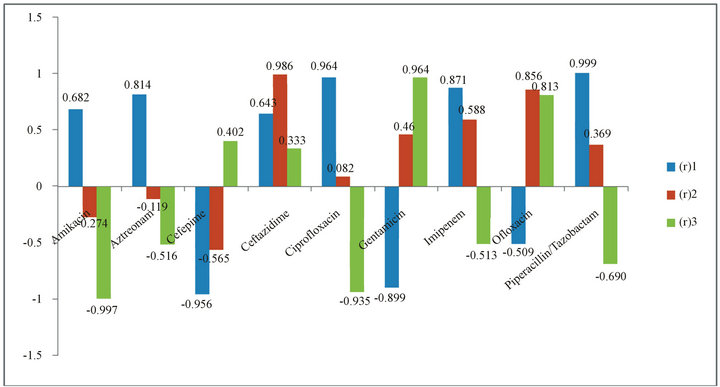
Figure 1. Correlation Coefficients Variation depending on the Years of Resistance by Isolate and the Change of Resistance Rates (∆R). (r)1: correlation coefficient between antibiotic consumption in 2006-2008 and resistance in 2006-2008; (r)2: correlation coefficient between antibiotic consumption in 2006-2008 and resistance in 2007-2009; (r)3: correlation coefficient between antibiotic consumption in 2006-2008 and change in resistance (∆R).
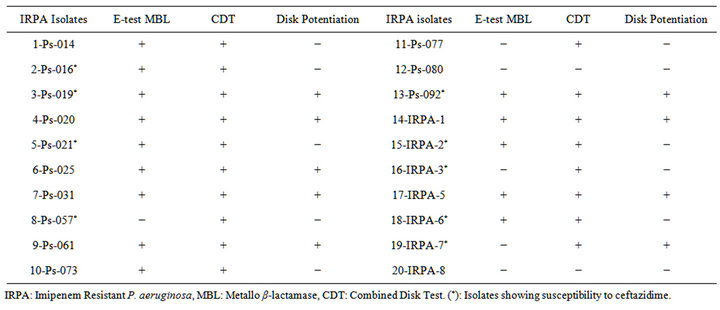
Table 2. MBL Detection for IRPA Isolates.
three phenotypic tests.
The results of the MHT showed that only 3 carbapenem producing strains were detected by this test (Figure 3).
4. Discussion
The increasing rates of resistance encountered among P. aeruginosa isolates worldwide makes the surveillance of resistance trends highly important in the choice of the best antimicrobial agent for therapy [18]. Our current study showed that the trends of resistance varied from one antimicrobial agent to another. However, the percentage of resistant organisms in 2009 was higher than that of 2006, with the exception of Aztreonam. There was a drop in the percentage of resistance by isolates in 2009 as compared to 2008; however it remained almost the same by patients. This difference could be attributed to the lower number of patients in 2008 (n = 239) as compared to 2009 (n = 256) which could be reflected by a difference in percentages. This data indicates a trend towards increasing percentages of antimicrobial resistance towards the tested antibiotics and poses an escalat-
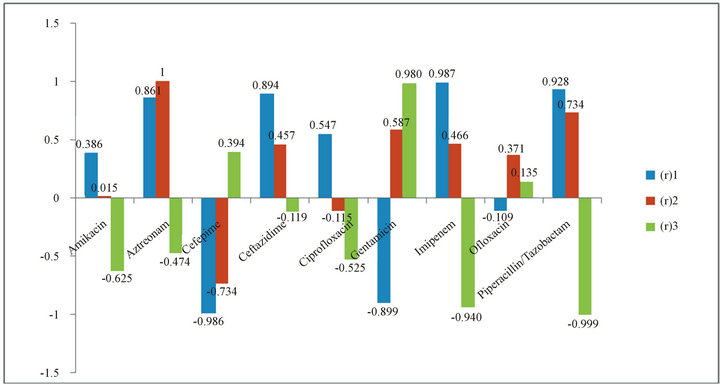
Figure 2. Correlation Coefficients Variation depending on the Years of Resistance by Patient and the Change of Resistance Rates (∆R). (r)1: correlation coefficient between antibiotic consumption in 2006-2008 and resistance in 2006-2008; (r)2: correlation coefficient between antibiotic consumption in 2006-2008 and resistance in 2007-2009; (r)3: correlation coefficient between antibiotic consumption in 2006-2008 and change in resistance (∆R).
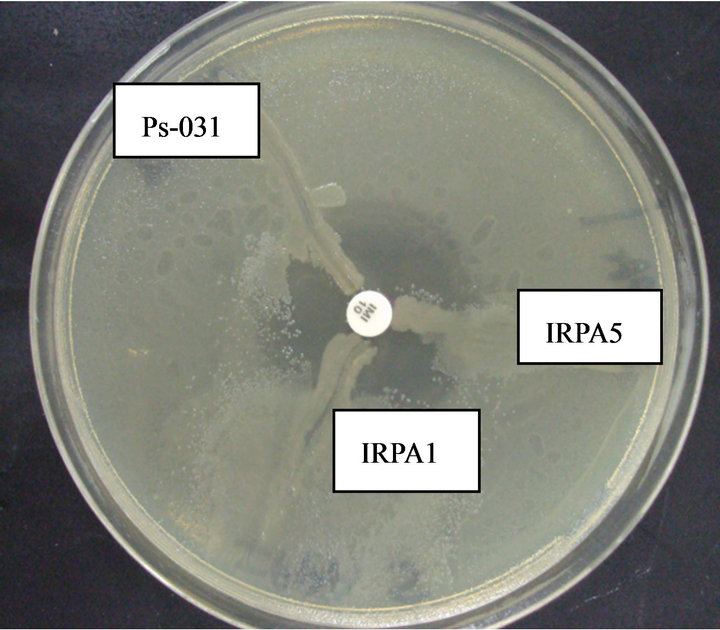
Figure 3. Postitive Hodge Test for Three Isolates IRPA 1, IRPA 5, Ps-031 showing distortion in the zone of inhibition and yielding a positive Hodge test.
ing threat on the success of treatment.
The view of bacterial resistance in comparison with antibiotic consumption has been a very useful tool for the evaluation of the selective pressure exerted by the different antibiotics [19]. The high correlation coefficients for the same year analysis by isolate for Ciprofloxacin, Imipenem, and Piperacillin/Tazobactam observed in this study are in line with data collected worldwide [14,15, 20]. This indicates that consumption of these antibiotics is having an effect on the emergence of bacterial resistance against these antimicrobial agents during the same year. Nevertheless, the antimicrobial agents had low correlation coefficients for the next year correlation, indicating that the effect on resistance for these antibiotics is limited to the year itself. However, data reported elsewhere showed that Imipenem had a high correlation coefficient for the next year analysis [14,20]. This discrepancy may be due to the relatively small population investtigated in this study. Evaluation of the ΔR showed that consumption of Gentamycin and Ofloxacin was correlated to the change in resistance in two consecutive years. All the other antibiotics investigated failed to show this effect. This goes in line with a study by Mutnick, et al. that investigated antibiotic consumption and its effect on the change in resistance [4].
When the same correlation coefficients were calculated by patient, the data obtained was slightly different. In the same year correlation, Imipenem and Piperacillin/ Tazobactam maintained a high correlation coefficient (r1 = 0.987 and r1 = 0.928, respectively). However, that of Ciprofloxacin was low (r1 = 0.547). This indicates that the observed effect on resistance on the isolate level is not necessarily seen on the patient level for Ciprofloxacin. Aztreonam had a correlation coefficient of 1 in the next year analysis indicating a strong effect of the consumption of this antibiotic on the emergence of resistance in subsequent years. Gentamycin also had a high correlation coefficient when ΔR was used suggesting that the effect of the consumption of this antibiotic is observed on both the patient and the isolate levels.
MBL producing P. aeruginosa strains have been well known to cause nosocomial outbreaks [21]. The rapid detection of this enzyme is crucial for the proper infection control and for the success of treatment [11]. There are currently no specific guidelines by the CLSI that indicate which is the best test for the detection of MBL. In this study, the MHT was used for the detection of carbapenemases in general and the MBL E-test, the Imipenem-EDTA combined disk test, and the EDTA disk potentiation using Ceftazidime, Ceftizoxime, Cefepime and Cefotaxime tests were used in order to screen for MBL production.
There is conflicting data present regarding the sensitivity, specificity, and positive predictive value of the MBL E-test. One study by Samuelsen, et al. showed a sensitivity of the E-test of 100%, a specificity of 86% and a positive predictive value of 20% [22]. Another study by Bergès, et al. showed a sensitivity of 100%, a specificity of 86.4% and a positive predictive value of 86.9% [23]. In contrast, Qu, et al. showed an 85.7% sensitivity, 100% specificity, and 100% positive predictive value [24]. A study by Lee, et al. concluded that the MBL E-test is highly sensitive and specific for detecting IMP-1- and VIM-2 types of MBL [25]. In our study, 14 MBL positive strains were detected by the MBL E-test. Nevertheless, 6 of these isolates were susceptible to Ceftazidime and therefore could not be MBL producers. The MBL positive results for these strains may have been false positive results produced by this test.
In the Imipenem-EDTA CDT, 18 isolates were MBL positive. The use of EDTA has been known to produce false positive results due to this chemical’s ability to degrade porins on the bacterial cell membrane [23]. Moreover, several studies have shown that the ImipenemEDTA CDT could produce false positive results [12,23]. Therefore, the higher amount of MBL producers detected by this test as compared to the MBL E-test may have been false positive detection.
The use of the EDTA disk potentiation using Ceftazidime, Ceftizoxime, Cefepime and Cefotaxime test showed only 9 MBL producing strains. 3 of these strains were susceptible to Ceftazidime. By comparing these values to the values obtained from the other tests, we may assume that the use of this method is not highly specific for the detection of MBLs.
The sensitivity and specificity of the MHT was found to be 100% and 88% respectively [17]. However, a study by Galani, et al. showed that the MHT produced many false positive and false negative results [13]. Only 3 strains were found to produce carbapenems using this test.
Overall, 8 isolates were MBL positive on the three MBL specific phenotypic tests and only 3 isolates were MBL positive on the four phenotypic tests that were used (the MHT included). Therefore, we can assume that at least 3 out of 20 isolates are MBL producers with the possibility of adding up to 5 MBL producing isolates. Further molecular analysis could help determine the exact amount of MBL producing isolates. However, the high cost and the requirement of use of specialized machinery makes screening for MBL with molecular techniques an unfavorable process. As an alternative, it is recommended that different phenotypic techniques be implemented for the screening of MBL in order to obtain higher accuracy for the detection of this enzyme.
The percentage of resistance among P. aeruginosa isolates is increasing throughout the years in the SGHUMC. There was a correlation between the consumption of certain antibiotics and the emergence of resistance during the same year and the following years at that same hospital. Screening for MBL among IRPA is crucial for the rapid detection of this enzyme. There is no one phenotypic technique that is adequate for screening and the use of several phenotypic techniques in the screening process is recommended.
REFERENCES
- F. Baquero, G. Baquero-Artigao, R. Canton and C. Garcia-Rey, “Antibiotic Consumption and Resistance Selection in Streptococcus pneumoniae,” Journal of Antimicrobial Chemotherapy, Vol. 50, Suppl. 3, 2002, pp. 27- 38. doi:10.1093/jac/dkf504
- R. Sharma, C. L. Sharma and B. Kapoor, “Antibacterial Resistance: Current Problems and Possible Solutions,” Indian Journal of Medical Sciences, Vol. 59, No. 3, 2005, pp. 120-129.
- J. M. Loeffler, J. Garbino, D. Lew, S. Harbarth and P. Rohner, “Antibiotic Consumption, Bacterial Resistance and Their Correlation in a Swiss University Hospital and its Adult Intensive Care Units,” Scandinavian Journal of Infectious Diseases, Vol. 35, No. 11-12, 2003, pp. 843- 850. doi:10.1080/00365540310016646
- A. Mutnick, P. Rhomberg, H. Sader and R. Jones, “Antimicrobial Usage and Resistance Trend Relationships from the MYSTIC Programme in North America (1999- 2001),” Journal of Antimicrobial Chemotherapy, Vol. 53, No. 2, 2004, pp. 290-296. doi:10.1093/jac/dkh039
- T. Slama, “Gram-Negative Antibiotic Resistance: There Is a Price to Pay,” Journal of Critical Care, Vol. 12, Suppl. 4, 2008, p. S4.
- S. Pournaras, M. Maniati, N. Spanakis, A. Ikonomidis, P. T. Tassios, A. Tsakris, N. J. Legakis and N. J. Maniatis, “Spread of Efflux Pump-Overexpressing, Non-Metallo-β- Lactamase Producing, Meropenem-Resistant But Ceftazidime-Susceptible Pseudomonas aeruginosa in a Region with BlaVIM Endemicity,” Journal of Antimicrobial Chemotherapy, Vol. 56, No. 4, 2005, pp. 761-764. doi:10.1093/jac/dki296
- C. Lagatolla, E. Edalucci, L. Dolzani, M. L. Riccio, F. Luca, E. Medessi, G. M. Rossolini and E. A. Tonin, “Molecular Evolution of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa in a Nosocomial Setting of HighLevel Endemicity,” Journal of Clinical Microbiology, Vol. 44, No. 7, 2006, pp. 2348-2353. doi:10.1128/JCM.00258-06
- P. Onguru, A. Erbay, H. Bodur, G. Baran, E. Akinci, N. Balaban and M. Cevik, “Imipenem-Resistant Pseudomonas aeruginosa: Risk Factors for Nosocomial Infections,” Journal of Korean Medical Sciences, Vol. 23, No. 6, 2008, pp. 982-987. doi:10.3346/jkms.2008.23.6.982
- O. Gutiérrez, C. Juan, E. Cercenado, F. Navarro, E. Bouza and P. Coll, “Molecular Epidemiology and Mechanisms of Carbapenem Resistance in Pseudomonas aeruginosa Isolates from Spanish Hospitals,” Antimicrobial Agents and Chemotherapy, Vol. 51, No. 12, 2007, pp. 4329-4335. doi:10.1128/AAC.00810-07
- C. Franklin, L. Liolios and A. Y. Peleg, “Phenotypic Detection of Carbapenem-Susceptible Metallo-β-LactamaseProducing Gram-Negative Bacilli in the Clinical Laboratory,” Journal of Clinical Microbiology, Vol. 44, No. 9, 2006, pp. 3139-3144. doi:10.1128/JCM.00879-06
- R. C. Picão, S. S. Andrade, A. G. Nicoletti, E. H. Campana, G. C. Moraes, R. E. Mendes and A. C. Gales, “Metallo-β-Lactamase Detection: Comparative Evaluation of Double-Disk Synergy versus Combined Disk Tests for IMP-, GIM-, SIM-, SPM-, or VIM-Producing Isolates,” Journal of Clinical Microbiology, Vol. 46, No. 6, 2008, pp. 2028-2037. doi:10.1128/JCM.00818-07
- B. Behera, P. Mathur, A. Das, A. Kapil and V. Sharma, “An Evaluation of Four Different Phenotypic Techniques for Detection of Metallo-β-Lactamase Producing Pseudomonas aeruginosa,” Indian Journal of Medical Microbiology, Vol. 26, No. 3, 200, 233-237. doi:10.4103/0255-0857.39587
- I. Galani, P. Rekatsina, D. Hatzaki, D. Plachouras, M. Souli and H. Giamarellou, “Evaluation of Different Laboratory Tests for the Detection of Metallo-β-Lactamase Production in Enterobacteriaceae,” Journal of Antimicrobial Chemotherapy, Vol. 61, No. 3, 2008, pp. 548- 553. doi:10.1093/jac/dkm535
- P. M. Lepper, E. Grusa, H. Reichl, J. Hogel and M. Trautmann, “Consumption of Imipenem Correlates with β-Lactam Resistance in Pseudomonas aeruginosa,” Antimicrobial Agents and Chemotherapy, Vol. 46, No. 9, 2002, pp. 2920-2925. doi:10.1128/AAC.46.9.2920-2925.2002
- M. S. Lapatschek and A. Hartinger, “Antibiotic Usage and Resistance Development in an Intensive Care Unit,” International Journal of Antimicrobial Agents, Vol. 29, 2007, p. S633.
- H. Sader, M. Castanheira, R. Mendes, M. Toleman, T. Walsh and R. Jones, “Dissemination and Diversity of Metallo-β-Lactamases in Latin America: Report from the SENTRY Antimicrobial Surveillance Program,” International Journal of Antimicrobial Agents, Vol. 25, No. 1, 2005, pp. 57-61. doi:10.1016/j.ijantimicag.2004.08.013
- K. Lee, Y. Chong, H. Shin, Y. Kim, D. Yong and J. Yum, “Modified Hodge and EDTA-Disk Synergy Tests to Screen Metallo-β-Lactamase-Producing Strains of Pseudomonas and Acinetobacter Species,” Clinical Microbiology and Infection, Vol. 7, No. 2, 2001, pp. 88-102. doi:10.1046/j.1469-0691.2001.00204.x
- T. Kirikae, Y. Mizuguchi and Y. Arakawa, “Investigation of Isolation Rates of Pseudomonas aeruginosa with and without Multidrug Resistance in Medical Facilities and Clinical Laboratories in Japan,” Journal of Antimicrobial Chemotherapy, Vol. 61, No. 3, 2008, pp. 612-615. doi:10.1093/jac/dkm537
- M. A. Borg, P. Zarb, E. A. Scicluna, O. Rasslan, D. Gur, S. B. Redjeb, Z. Elnasser and Z. Daoud, “Antibiotic Consumption as a Driver for Resistance in Staphylococcus aureus and Escherichia coli within a Developing Region,” American Journal of Infection Control, Vol. 38, No. 3, 2010, pp. 212-216. doi:10.1016/j.ajic.2009.07.010
- H. Kallel, F. Mahjoubi, H. Dammak, M. Bahloul, C. Hamida, H. Chelly, N. Rekik, A. Hammami and M. Bouaziz, “Correlation between Antibiotic Use and Changes in Susceptibility Patterns of Pseudomonoas aeruginosa in a Medical-Surgical Intensive Care Unit,” Indian Journal of Critical Care Medicine, Vol. 12, No. 1, 2008, pp. 18-23. doi:10.4103/0972-5229.40945
- A. Varaiya, N. Kulkarni, M. Kulkarni, P. Bhalekar and J. Dogra, “Incidence of Metallo Beta Lactamase Producing Pseudomonas aeruginosa in ICU Patients,” Indian Journal of Medical Research, Vol. 127, No. 4, 2008, pp. 398- 402.
- Ø. Samuelsen, L. Buarø, C. Giske, G. Simonsen, B. Aasnæs and A. Sundsfjord, “Evaluation of Phenotypic Tests for the Detection of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa in a Low Prevalence Country,” Journal of Antimicrobial Chemotherapy, Vol. 61, No. 4, 2008, pp. 827-830. doi:10.1093/jac/dkn016
- L. Bergès, H. Rodriguez-Villalobos, A. Deplano and M. J. Struelens, “Prospective Evaluation of Imipenem/EDTA Combined Disc and Etest for Detection of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa,” Journal of Antimicrobial Chemotherapy, Vol. 56, No. 4, 2007, pp. 812-813. doi:10.1093/jac/dkm001
- T. Qu, J. Zhang, J. Wang, J. Tao, Y. Yu, Y. Chen, C. Zhou and L. Li , “Evaluation of Phenotypic Tests for Detection of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa Strains in China,” Journal of Clinical Microbiology, Vol. 47, No. 4, 2009, pp. 1136-1142. doi:10.1128/JCM.01592-08
- K. Lee, D. Yong, J. H. Yum, Y. S. Lim, A. Bolmström, A. Qwärnström, A. Karlsson and Y. Chong, “Evaluation of Etest MBL for Detection of blaIMP-1 and blaVIM-2 Allele-Positive Clinical Isolates of Pseudomonas spp. and Acinetobacter spp,” Journal of Clinical Microbiology, Vol. 43, No. 2, 2005, pp. 942-944. doi:10.1128/JCM.43.2.942-944.2005
NOTES
*Acknowledgement: This work was supported by a grant from the Lebanese Council for Scientific Research (CNRS).
#Corresponding author.

