Advances in Microbiology
Vol. 3 No. 2 (2013) , Article ID: 32423 , 9 pages DOI:10.4236/aim.2013.32029
Comparative Analysis of Pathogenicity Assays and PCR for Listeria monocytogenes Isolated from Animal, Raw Food and Environmental Sources in Korea
1Seoul Metropolitan Government Research Institute of Public Health and Environment, Yangjae-Dong, Seocho-Gu, Seoul, Korea
2Animal Disease Diagonsis Division, National Veterinary Research and Quarantine Service, Anyang 6-Dong, Anyang City, Korea
3Veterinary Pharmacology Division, National Veterinary Research and Quarantine Service, Anyang 6-Dong, Anyang City, Korea
4College of Veterinary Medcine, Chungnam National University, Daejeon, Korea
Email: *sanghun93@seoul.go.kr
Copyright © 2013 Sanghun Park et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received February 13, 2013; revised March 11, 2013; accepted April 11, 2013
Keywords: PFGE; Vero Cell; Listeria monocytogenes; in Vitro Assay
ABSTRACT
Results of PCR with oligonucleotide primers were designed from the assembled panel of four potential virulence genes (two of internalin gene and two of transcriptional regulator gene). Most of the isolates including reference strains were reactive by PCR, whereas the other strains (No.80, 81, and 83) isolated from pork, were non-reactive by PCR. In particular, all pork isolates were PCR-negative for two primers (lmo2672 and 2821) sets tested. However, No.82 was positive for lmo1134 primer, and No.84 was positive for lmo2470 of pork isolates. It was observed that all Listeria monocytogenes (L. monocytogenes) penetrate Vero cells, although the invasion efficiency of each strain varied (between 0.5 and 18.9%). When compared in cell assay with PFGE, the results were shown that the mean invasion efficiency for lineage II isolate (2.6%) was significantly lower (ANOVA-test, p < 0.05) than that for lineage I (12.9%) and III isolates (10.3%).
1. Introduction
Listeria monocytogenes (L. monocytogenes) are characterized as gram-positive and rod-shaped bacteria. Listeriosis caused by L. monocytogenes is a life-threatening disease in fetuses, newborns, immunocompromised people and the elderly [1,2]. L. monocytogenes continues to be a major food safety concern, a recent report from the Centers for Disease Control and Prevention (CDC) estimates 1600 cases and 255 deaths per year in the United States [3]. The most common route of infection by L. monocytogenes is via the gastrointestinal tract [4]. As evidenced by several outbreaks of listeriosis caused by the ingestion of contaminated food materials [5]. Numerous reports have been published dealing with the prevalence of L. monocytogenes in food products [6].
Despite frequent exposure of the population to this bacterium, the probability of contracting listeriosis is low, since the incidence of the disease ranges from 1.6 to 6 cases per million populations [7,8]. It has long been assumed that all L. monocytogenes strains are virulent. However, many strains in the environment may lack certain virulence properties [9]. Within the L. monocytogenes species there is great variability between the serotypes found in the environment or food. For example, 64% of the clinical isolates occurring in the UK (United Kingdom) are serotype 4b whereas 4% - 6% of this serotype is found in the environment [10]. The virulence of L. monocytogenes strains also varies within the same serotype [11].
Methods that have been developed for L. monocytogenes virulence assessment in vivo models frequently used to determine L. monocytogenes virulence include determination of 50% lethal dose in mice and recovery of the livers and spleens of immunocompetent or immunocompromised mice following intravenous or oral inoculation [9,12]. However, mice and rats are not natural hosts for L. monocytogenes [13]. Futhermore, mouse and rat E-cadherin differ from human E-cadherin at an important amino acid residue that renders cells resistant to InlA-mediated invasion [14]. Therefore, the use of laboratory animals is impractical for a particular study, or the best available animal models have significant limitations. Since evaluation of the role of strain differences in human must be important meaning for the protection of listeriosis and presumption of public health measures, it is advisable to develop and easy, fast laboratory test that can reliable verify the virulence of a given strain. In Vitro virulence test using cell culture models that have been used to characterize L. monocytogenes virulence include the plaque-forming assay, cytotoxicity-based assays, and invasion and multiplication assay [15]. Also, in vitro assays that characterize virulence are often relatively fast and simple to perform.
No reliable molecular method for prediction of L. monocytogenes virulence has been developed. But, PCR detection of known L. monocytogenes genes, such as internalin and transcriptional regulator have the potential to provide an alternative method for distinguishing virulent from avirulent isolates [16].
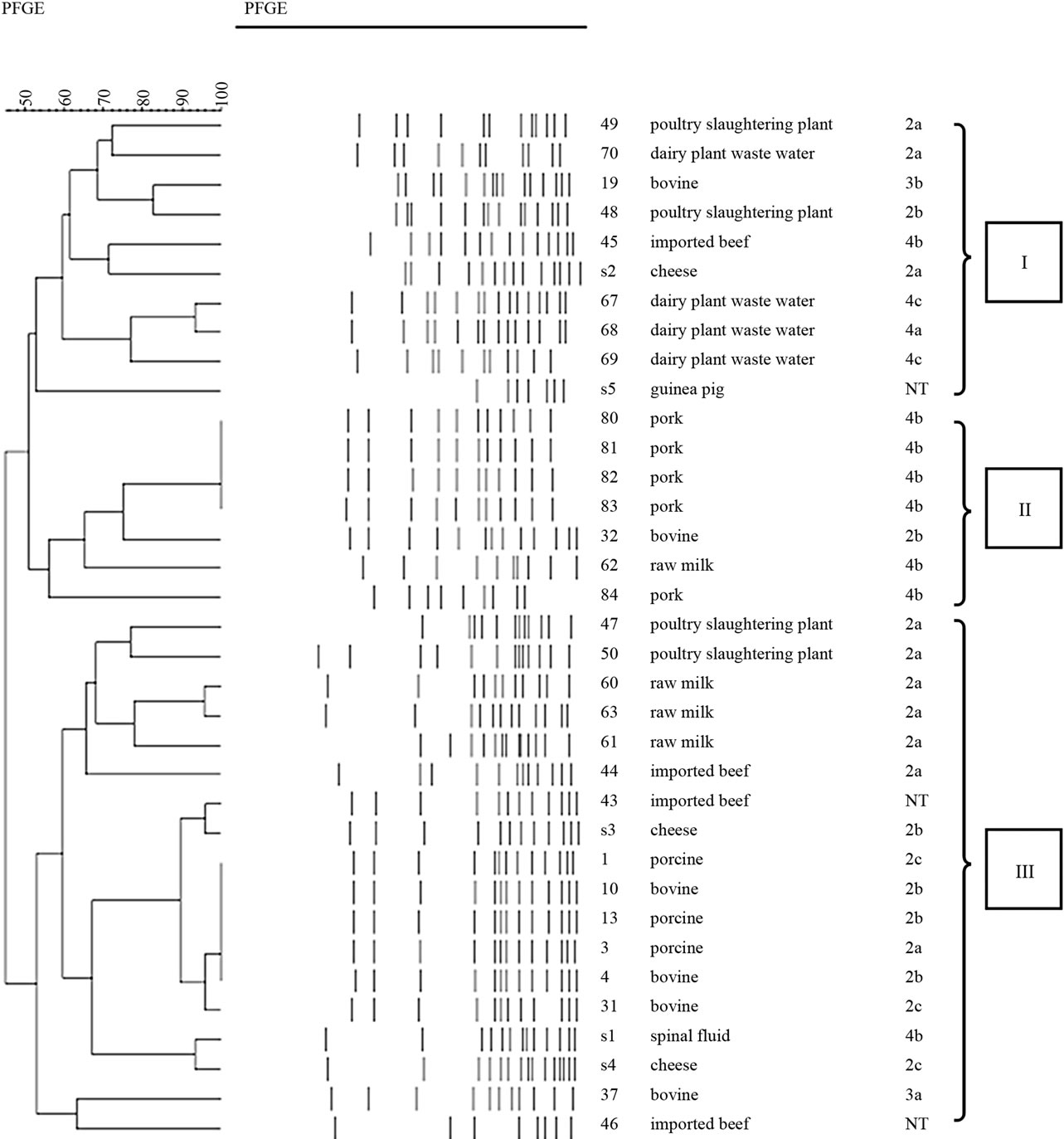
PFGE is a method with high discriminatory power and it has shown to be very accurate and reproducible for fine structure comparison and molecular typing of L. monocytogenes [17]. PFGE type and invasion into cell culture assay have considerable correlation [18].
In this study, we tested the virulence of L. monocytogenes strains found in Korea by using PCR primers that were derived from four L. monocytogenes and to compare invasion and multiplication in the Vero cell to PFGE types on 35 strains from food, animal, and environmental isolates.
2. Materials and Methods
2.1. Bacterial Strains
A total of 35 L. monocytogenes isolates were used for experiment. 16 L. monocytogenes isolates were obtained from Dr. Byeong Yeal Jung (National Veterinary Research & Quarantine Service, Anyang). In particular, L. monocytogenes obtained from different animal species (porcine and bovine), environment (poultry slaughtering plant and dairy plant waste water) and meat (imported beef and pork) and from different foods (milk and cheese) from 1997 to 2007, were used. And five additional strains were purchased from the ATCC. The Listeria strains and their virulence, serotype and origin are in presented in Table 1. Bacteria for the experiments were pre-cultivated in brain heart infusion (BHI; Difco) agar or BHI broth at 37˚C for 18 h. Cell lines: Vero cells (African green monkey kidney) were cultured in Eagle’s Minimum Essential Medium (MEM, Gibco BRL) supplemented with 5% fetal bovine serum (Intergen) at 37˚C in a 5% CO2 atmosphere.
2.2. Cell Invasion Assay
To assess the abilities of the invasion and muliplication in cell, we used an intracellular growth assay described [15,19,20]. Briefly, cell monolayers of each cell line grown in 24-well tissue culture plates for 72 h were washed twice with Phospahte Buffered Salines (PBS; JBI, Korea) before inoculation of the bacteria. Simultaneously, after trypsin treatment, cells collected from a separate part of the culture plate were used for determination of the cell number using a haemocytometer. For invasion and intracellular growth assays, 18 h pre-cultivated bacteria were inoculated into brain heart infusion (BHI) broth (Merk, USA) incubated at 37˚C for 13 h, washed twice with PBS, diluted with Minimum Essential Media Eagle (MEM), and then the appropriate bacterial count (5 × 105 - 5 × 107/ml) was inoculated onto the cell monolayer. The inoculated bacterial number was obtained from the colony count (colony forming units, CFU) after incubation on trypticase soy agar (TSA; Oxoid, Korea), containing 0.6% (w/v) yeast extract, for 48 h at 37˚C. Bacterial suspensions (0.3 ml per well) were added to the prepared cell monolayer, incubated at 37˚C for 2 h, washed twice with PBS and then incubated in 1 ml of MEM supplemented with 40 μg/ml of gentamicin at 37˚C for 2 h to kill extracellular bacteria. Monolayers were washed twice with PBS and cells lysed in 1 ml of distilled water containing 0.5% Triton X-100 on ice for 10 min. To measure invasion, the number of bacteria released from the cells was determined by plating serial dilutions onto TSA plates. To measure intracellular growth (Multiplication), MEM supplemented with 40 μg/ml of gentamicin was used as the growth medium after infection and infected cells were incubated fro 18 h after removal of extracellular bacteria. The number of intracellular bacteria was then enumerated at 18 h post-infection as described above and intracellular doubling times between 3.5 and 21.5 h post-infection were calculated. The experiment was repeated two times.
2.3. PFGE
PFGE was performed according to the PulseNet standardized protocol [21]. Bacteria were grown on BHI agar plates at 37˚C for 16 - 18 h. Bacterial culture were embedded in 1.2% SeaKem Gold agarose (Cambrex Bio Science, USA), 1% sodium dodecyl sulfate (Sigma, USA), 0.2 mg/ml proteinase K (Roche Molecular Biochemicals,
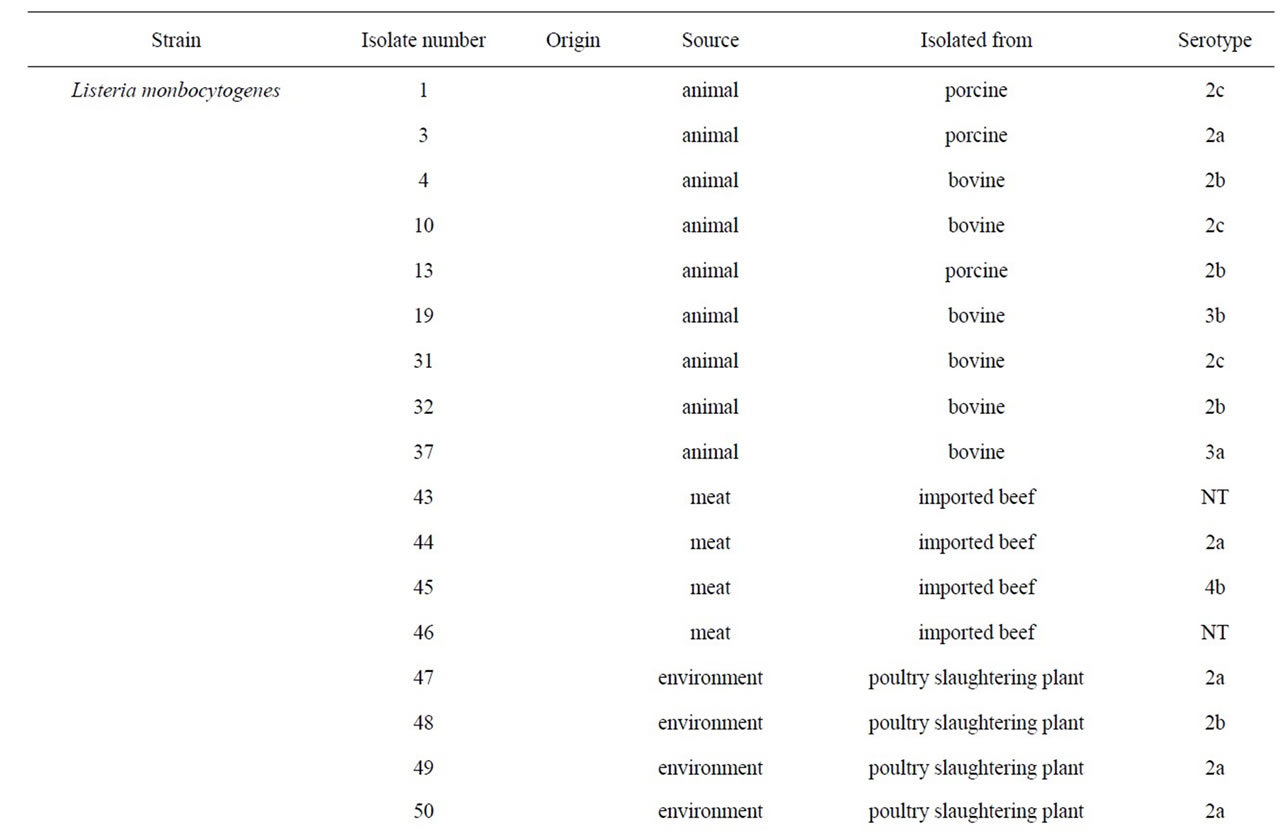
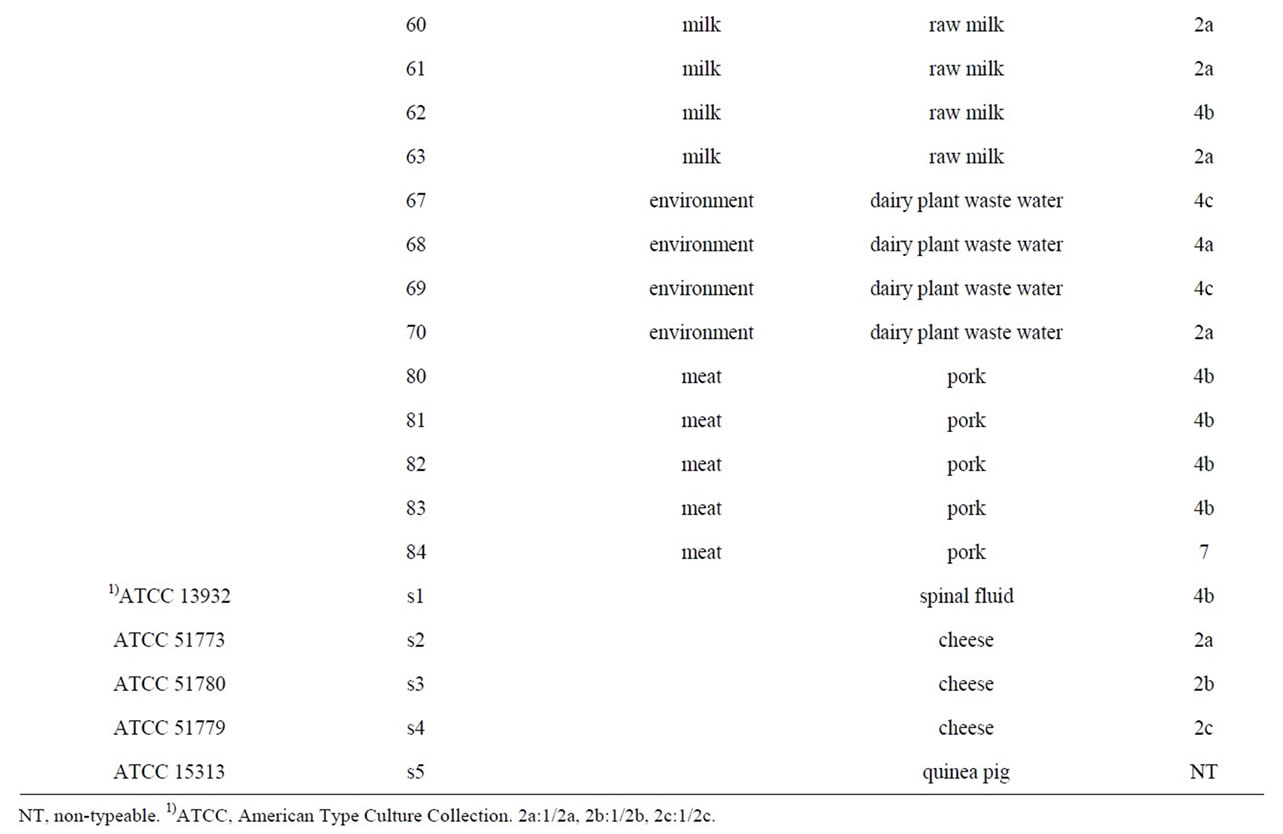
Table 1. List of the strains used in this study.
Germany) (SSP), lysed, washed, and digested with the restriction enzymes ApaI for 5 h at 3.0˚C. The DNA restriction fragments in plugs were separated by electrophoresis through 1% SeaKem Gold agarose gel in 0.5 × solution of Tris-borate-EDTA (prepared by diluting 10 × TBE) buffer (Bioneer, Korea) at 14˚C in a CHEF-Mapper PFGE apparatus (Bio-Rad, USA). The electrophoretic parameters used were as follows; initial switch time, 4.0 s; final switch time, 40.0 s. PFGE profiles were separated in to three main lineages showing about 50% genetic similarity.
2.4. PCR
Extraction, purification and quantification of DNA from each strain were performed as previously described. All of the primers used for specific PCR amplifications of virulent genes are reported in Table 2 [16]. PCR was performed in a PCR 9600 thermal cycler (Perkin-Elmer Corporation). A 50-ul aliquot contained buffer (10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2 [pH 8.3]), the dNTP mixture (TaKaRa, Japan) 2.5 mM each, 10 pM primer, 25 ng of DNA, and 0.8 U of Taq DNA polymerase (TaKaRa, Japan). The cycling conditions were the template DNA was denatured at 94˚C for 2 min followed by 25 cycles of amplification (each cycle consisted of denaturation at 94˚C for 20 s, annealing at 60˚C for 20 s and elongation at 72˚C for 45 s) and finally 72˚C for 2 min. 5 microlitre of the amplified products was separated by electrophoresis in 1.5% agarose gel containing ethidium bromide, and visualized under UV.
2.5. Statistical Analysis
Differences among results from PFGE lineage and cell culture assay were evaluated with one-way ANOVA test in SPSS ver10.0. Differences were considered significant at p < 0.05.
3. Results
Results of PCR with oligonucleotide primers were designed from the assembled panel of four potential virulence genes (Table 3). Most of the isolates including reference strains were reactive by PCR, whereas the other strains (No.80, 81, and 83) isolated from pork, were non-reactive by PCR. In particular, all pork isolates were PCR-negative for two primers (lmo2672 and 2821) sets tested. However, No.82 was positive for lmo1134 primer, and No.84 was positive for lmo2740 of pork isolates.
The ability of L. monocytogenes isolates to invade and multiply was evaluated by using and in vitro model with Vero cells (Figures 1 and 2). We previously studied that three types of PFGE pattern showed at 50% relative genetic similarity in L. monocytogenes isolates (Figure 3). As the number of deposited bacteria varied with the strain, a percentage of entry and an index of multiplication were determined. Under these conditions, it was observed that all L. monocytogenes penetrate Vero cells, although the invasion efficiency of each strain varied (between 0.5% and 18.9%). Besides, these strains multiplied broadly the number of bacteria was enhanced 22 to 2548 times 3.5 and 21 h of infection. Entry and intracellular multiplication proficiency varied greatly among L. monocytogenes isolates.
The mean invasion efficiency for lineage II isolate (2.6%) was significantly lower (ANOVA-test, p < 0.05) than that for lineage I (12.9%) and III isolates (10.3%). And the mean multiplication rate for lineage II isolate (124) was significantly lower (ANOVA-test, p < 0.05) than that for lineage I (801) and III isolates (1188).
4. Discussion
It has long been presumed that all L. monocytogenes strains are pathogens. However, much evidence to suggest that, under natural conditions, some L. monocytogenes strains may lack virulence properties [22]. In this study, the virulence of various isolates of the L. monocytogenes was compared to that of the well-known reference strains to know their capacity to invade and multiplicate in Vero cell and using PCR of putative virulence gene. We use Vero cell to determine for virulence of L.

Table 2. List of L. monocytogenes and primers used in the PCR assay.
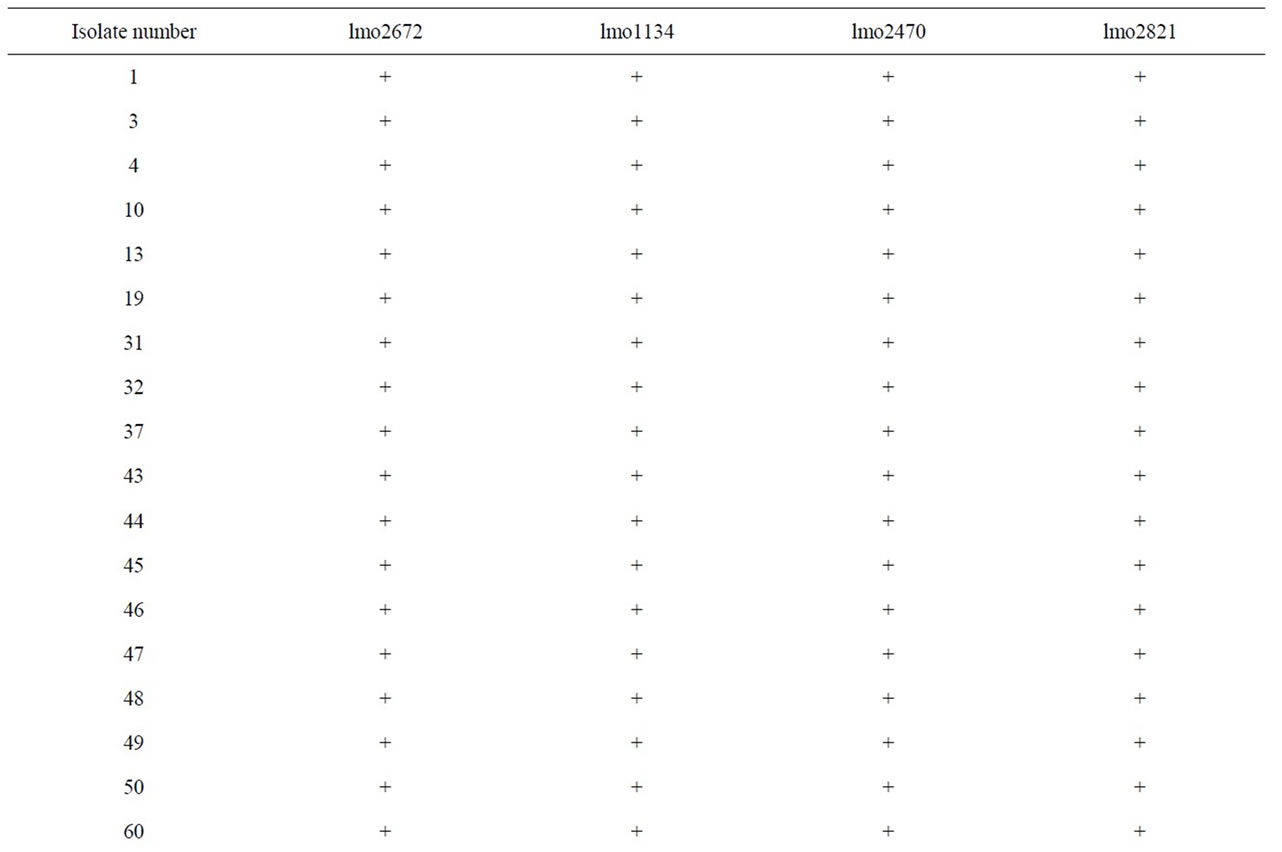

Table 3. PCR results using primers derived from four putative L. monocytogenes transcriptional regular and internalin genes.
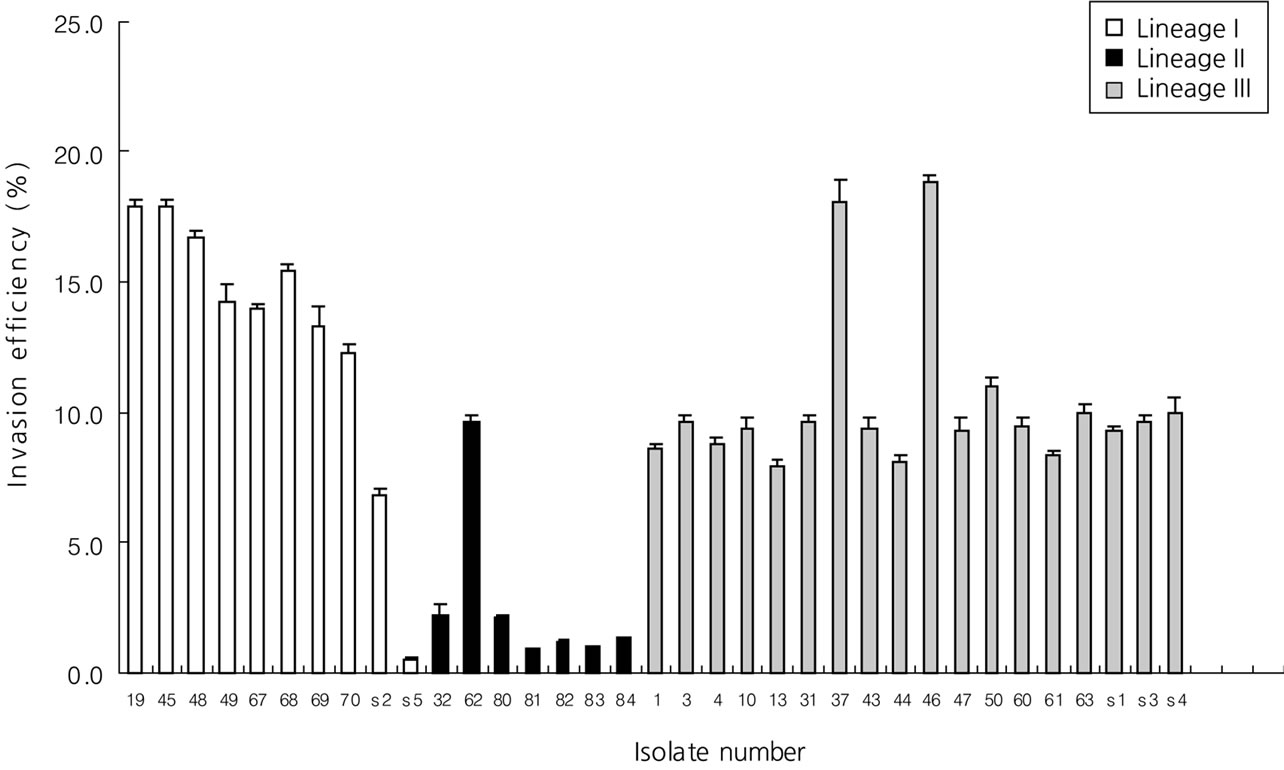
Figure 1. Distribution of invasion efficiency of selected lineage I, II, and III isolates in Vero cells.
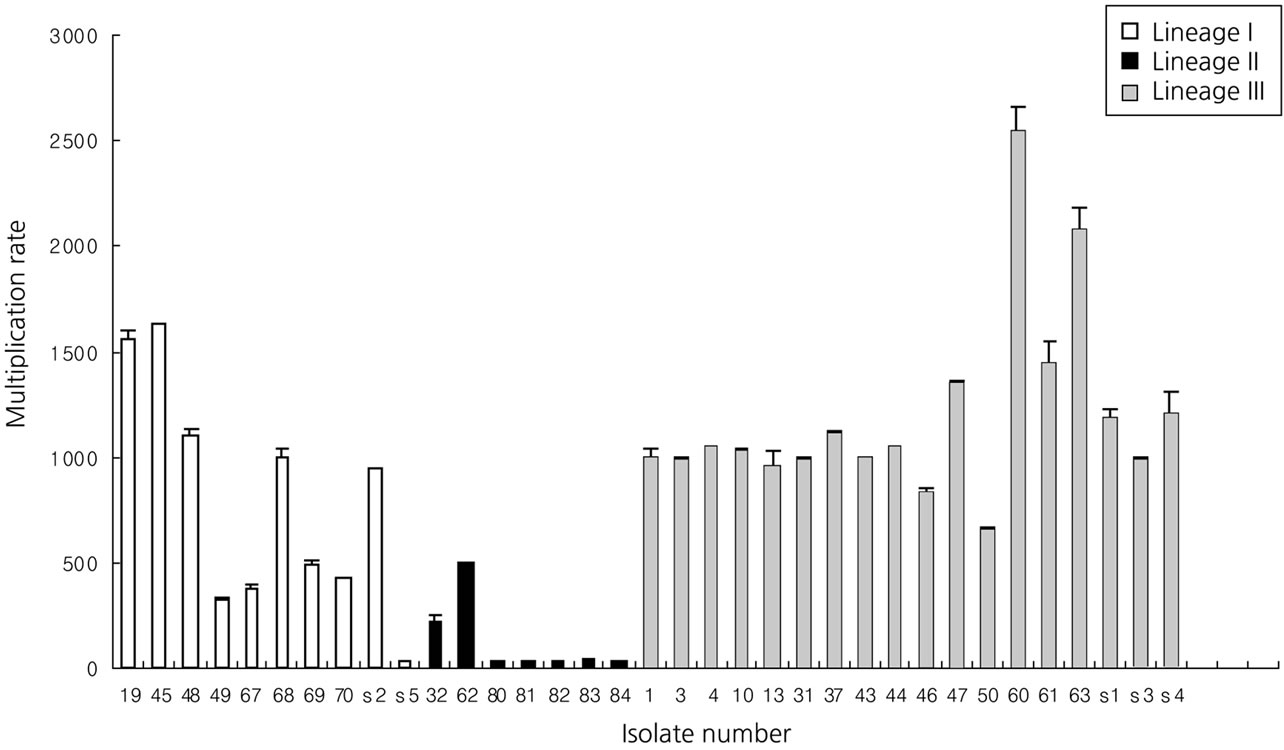
Figure 2. Distribution of multiplication rate of selected lineage I, II and III isolates in Vero cells.
monocytogenes isolated from various source. Our results indicated that most of isolates had a significantly higher (p < 0.05) invasion efficiency and significantly higher (p < 0.05) multiplication rate than some pork isolates (No.
80, 81, 82, 83 and 84) in cell culture assay.
A number of studies conducted on food and clinical isolates using in vitro tests produced a pattern of results similar to those of the in vivo studies [23]. Another study demonstrated that two human feces carriage isolates of L. monocytogenes were compared to reference strains with regard to their invasiveness towards Caco-2 cells and strains virulent entered Caco-2 cells with great penetration efficiency, whereas strains with attenuated virulence showed low entry levels [24]. Host variation is an important element in deciding virulence of a pathogen. A specific strain of L. monocytogenes may have low or no virulence in most individuals, but be highly virulent in a susceptible host. In fact, most invasive listeriosis occurs in persons with a predisposing condition, primarily related to a defect in immune function [25]. For in vivo studies, these models may be used such as knockout mouse, in which genes coding for immune function proteins such as cytokines are disrupted [26]. However, because of the high cost associated with knockout mice, these models were difficult to conduct.
More recently, test methods based on detection of specific virulence genes may offer alternative approaches for identifying of L. monocytogenes virulence [15]. We selected two genes (lmo2672 and lmo1134) encoding transcriptional regulator gene which had the lowest nucleotide identity with any sequences from the L. innocua genome (non-virulent strain) [16]. Results of PCR using lmo2672 were correlated with invasion assay in Vero cell but not using lmo1134. Especially, lmo2672 allow pork isolates to be identified negative results of PCR. PCR results also correlated with ATCC reference, which isolated from an outbreak of listeriosis. However, our result showed that ATCC 15313, which was avirulent isolate, was positive for lmo2672 and lmo1134. That was consistent with D. Liu et al. [16]. D. Liu et al., characterize to virulent and avirulent L. monocytogenes strains by PCR amplification of four putative transcriptional regulator genes. And it demonstrated that ATCC 15313 has not maintained its ability to cause cytopathogenic effects in Caco-2 monolayer but positive for all tested primers. ATCC 15313 was originally isolated from an infected rabbit during an outbreak of listeriosis, but it later became avirulent after successive laboratory subculturing. It has been demonstrated that ATCC 15313 does not express listeriolysin, a well-known virulence factor this occurred spontaneously after its original isolation [27]. So, it was assumed that ATCC 15313, which is derived from a virulent L. monocytogenes isolates, would retain many of its virulence properties [16].
We also selected two genes (lmo2470 and lmo2821) encoding putative internalin genes. Results of PCR using lmo2821 were positive for most isolates but not for pork samples. Lmo2470 gene was detected in most L. monocytogenes but not pork isolates (No.80, 81, 82, and 83). These results were correlated with invasion assay in Vero cell but not using lmo2470. PCR results also correlated with ATCC reference, which isolated from an outbreak of listeriosis. However, our result showed that ATCC 15313, which was avirulent isolate, was positive for lmo2470 and lmo2821. The presence of virulence genes in L. monocytogenes isolates appears to be so common that such tests are relatively poor discriminators of relative disease-causing potential [15].
We also compare levels of virulence of three different PFGE lineages of L. monocytogenes isolates using cell culture assay. PFGE lineage II expressed a significantly higher level of invasion rate in cell culture assays than the others (p < 0.05). The same results showed in multiplication assay. These noted variations in invasive and multiplication assay might account for why certain PFGE types were more virulent than others. In other previous studies [18], PFGE type and invasion into cell culture assay have considerable correlation. Thus, combining assay using PFGE and invasion assay in L. monocytogenes would seem to be of crucial value for the interpretation of the virulence.
The four primer sets and Invasion assay using Vero cell were evaluated by PCR and cell culture system against Listeria isolated various source. Most of the isolates were positive for at least one PCR primer. However, some pork isolates (No.80, 81, and 83) were not positive any of the primer. Therefore, most of isolates are considered to be more invasive than some pork isolates and could be potential food-borne pathogens. It was insisted that a single molecular, cell culture or animal assay cannot detect all virulence-attenuated isolates. Rather, it is a combination of cell culture and molecular screening assays that would offer the best chance of most accurately and reliably identifying naturally virulence-attenuated L. monocytogenes isolates [28].
Therefore, combining studies of differences in the expression of virulence genes with the present study could give more information on the complexity of virulence of L. monocytogenes. Invasion assay using Vero cells and PCR-based assay for putative virulence genes could be used as an alternative to test for determent the pathogenicity of Listeria and reduce expensive and time-consuming animal testing.
REFERENCES
- A. Schuchat, B. Swaminathan and C. V. Broome, “Epidemiology of Human Listeriosis,” Clinical Microbiology Reviews, Vol. 99, No. 4, 1991, pp. 169-183.
- H. Hof, “History and Epidemiology of Listeriosis,” FEMS Immunology and Medical Microbiology, Vol. 35, No. 3, 2003, pp. 199-202. doi:10.1016/S0928-8244(02)00471-6
- A. R. Datta, P. Laksanalamai and M. Solomotis, “Recent Developments in Molecular Sub-Typing of Listeria monocytogenes,” Food Additives and Contaminants Part A, 2012, pp. 1-9.
- W. F. Schlech, “New Perspectives on the Gastrointestinal Mode of Transmission in Invasive Listeria monocytogenes Infection,” Clinical and Investigative Medicine, Vol. 7, No. 4, 1984, pp. 321-324.
- W. F. Schlech, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls and C. V. Broome, “Epidemic Listeriosis-Evidence for Transmission by Food,” New England Journal of Medicine, Vol. 308, No. 4, 1983, pp. 203-206. doi:10.1056/NEJM198301273080407
- B. Swaminathan and P. Gerner-Smidt, “The Epidemiology of Human Listeriosis,” Microbes and Infection, Vol. 9, No. 10, 2007, pp. 1236-1243. doi:10.1016/j.micinf.2007.05.011
- S. Notermans, J. Dufrenne, P. Teunis and T. Chackraborty, “Studies on the Risk Assessment of Listeria monocytogenes,” Journal of Food Protection, Vol. 61, No. 2, 1998, pp. 244-248.
- J. Rocourt, C. Jacquet and A. Reilly, “Epidemiology of Human Listeriosis and Seafoods,” International Journal of Food Microbiology, Vol. 62, No. 3, 2000, pp. 197-209. doi:10.1016/S0168-1605(00)00336-6
- H. Hof, “Virulence of Different Strains of Listeria monocytogenes Serovar 1/2a,” Medical Microbiology and Immunology, Vol. 173, No. 4, 1984, pp. 207-218. doi:10.1007/BF02122112
- K. G. Kerr, P. Kite, J. Heritage and P. M. Hawkey, “Typing of Epidemiologically Associated Environmental and Clinical Strains of Listeria monocytogenes by Random Amplification of Polymorphic DNA,” Journal of food protection, Vol. 58, No. 6, 1995, pp. 609-613.
- H. Barbour, A. Rampling and C. E. Hormaeche, “Comparison of the Infectivity of Isolates of Listeria monocytogenes Following Intragastric and Intravenous Inoculation in Mice,” Microbial pathogenesis, Vol. 20, No. 4, 1996, pp. 247-253. doi:10.1006/mpat.1996.0023
- H. Barbour, A. Rampling and C. E. Hormaeche, “Variation in the Infectivity of Listeria monocytogenes Isolates Following Intragastric Inoculation of Mice,” Infection and Immunity, Vol. 69, No. 7, 2001, pp. 4657-4660. doi:10.1128/IAI.69.7.4657-4660.2001
- M. Pentecost, G. Otto, J. A. Theriot and M. R. Amieva, “Listeria monocytogenes Invades the Epithelial Junctions at Sites of Cell Extrusion,” PLOS Pathogens, Vol. 2, No. 1, 2006, p. e3. doi:10.1371/journal.ppat.0020003
- M. Lecuit, S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner and P. Cossart, “A Single Amino Acid in ECadherin Responsible for Host Specificity towards the Human Pathogen Listeria monocytogenes,” The EMBO Journal, Vol. 18, No. 4, 1999, pp. 3956-3963. doi:10.1093/emboj/18.14.3956
- R. B. Raybourne, “Virulence Testing of Listeria monocytogenes,” Journal of AOAC International, Vol. 85, No. 2, 2002, pp. 516-523.
- D. Liu, A. J. Ainsworth, F. W. Austin and M. L. Lawrence, “Characterization of Virulent and Avirulent Listeria monocytogenes Strains by PCR Amplification of Putative Transcriptional Regulator and Internalin Genes,” Journal of Medical Microbiology, Vol. 52, No. 12, 2003, pp. 1065-1070. doi:10.1099/jmm.0.05358-0
- J. Bille and J. Rocourt, “WHO International Multicenter Listeria monocytogenes Subtyping Study-Rationale and Set-Up of the Study,” International Journal of Food Microbiology, Vol. 32, No. 3, 1996, pp. 251-262. doi:10.1016/S0168-1605(96)01140-3
- C. N. Larsen, B. Norrung, H. M. Sommer and M. Jakobsen, “In Vitro and in Vivo Invasiveness of Different Pulsed-Field Gel Electrophoresis Types of Listeria monocytogenes,” Applied and Environmental Microbiology, Vol. 68, No. 11, 2002, pp. 5698-5703. doi:10.1128/AEM.68.11.5698-5703.2002
- Z. Xiaohui, J. Xinan and M. Wiedmann, “Listeria monocytogenes in the Chinese Food System: Strain Characterization through Partial actA Sequencing and TissueCulture Pathogenicity Assays,” Journal of Medical Microbiology, Vol. 54, No. 3, 2005, pp. 217-224. doi:10.1099/jmm.0.45882-0
- F. Yamada, F. Ueda, Y. Ochiai, M. Mochizuki, H. Shoji, K. Ogawa-Goto, T. Sata, K. Ogasawara, A. Fujima and R. Hondo, “Invasion Assay of Listeria monocytogenes Using Vero and Caco-2 Cells,” Journal of Microbiological Methods, Vol. 66, No. 1, 2006, pp. 96-103. doi:10.1016/j.mimet.2005.10.017
- L. M. Graves and B. Swaminathan, “PulseNet Standardized Protocol for Subtyping Listeria monocytogenes by Macrorestriction and Pulsed-Field Gel Electrophoresis,” International Journal of Food Microbiology, Vol. 65, No. 1-2, 2001, pp. 55-62. doi:10.1016/S0168-1605(00)00501-8
- L. N. Van, E. Bottreau, S. Bailly, M. Tabouret, J. Marly, P. Pardon and P. Velge, “Tissue Culture Assays Using Caco-2 Cell Line Differentiate Virulent from NonVirulent Listeria monocytogenes Strains,” Journal of Applied Microbiology, Vol. 85, No. 2, 1998, pp. 337-346. doi:10.1046/j.1365-2672.1998.00515.x
- A. Gattuso, M. Gianfranceschi, R. Sessa, F. Taggi, M. Pourshaban and P. Aureli, “In Vivo and in Vitro Assessment of the Virulence of Listeria monocytogenes Strains,” New Microbiologica, Vol. 23, No. 3, 2000, pp. 289-295.
- M. Olier, F. Pierre, J. P. Lemaitre, C. Divies, A, Rousset and J. Guzzo, “Assessment of the Pathogenic Potential of Two Listeria monocytogenes Human Faecal Carriage Isolates,” Microbiology, Vol. 148, No. 6, 2002, pp. 1855- 1862.
- A. Schuchat, K. A. Deaver, J. D. Wenger, B. D. Plikaytis, L. Mascola, R. W. Pinner, A. L. Reingold and C. V. Broome, “Role of Foods in Sporadic Listeriosis. I. CaseControl Study of Dietary Risk Factors. The Listeria Study Group,” The journal of the American Medical Association, Vol. 267, No. 15, 1992, pp. 2041-2045. doi:10.1001/jama.1992.03480150047035
- R. U. Emil, “Why Listeriosis? A Perspective on Cellular Immunity to Infection,” Immunological Reviews, Vol. 158, No. 1, 1997, pp. 5-9. doi:10.1111/j.1600-065X.1997.tb00987.x
- S. Kathariou and L. Pine, “The Type Strain(s) of Listeria monocytogenes: A Source of Continuing Difficulties,” International Journal of Systematic and Evolutionary Microbiology, Vol. 41, No. 2, 1991, pp. 328-330.
- A, Roberts, Y. Chan and M. Wiedmann, “Definition of Genetically Distinct Attenuation Mechanisms in Naturally Virulence-Attenuated Listeria monocytogenes by Comparative Cell Culture and Molecular Characterization,” Applied and Environmental Microbiology, Vol. 71, No. 7, 2005, pp. 3900-3910. doi:10.1128/AEM.71.7.3900-3910.2005
NOTES
*Corresponding author.