Advances in Microbiology
Vol.4 No.1(2014), Article ID:42267,8 pages DOI:10.4236/aim.2014.41009
Cloning of a New Truncated cry1Ac Gene from an Indian Isolate of Bacillus thuringiensis
1Department of Agricultural Microbiology, Tamil Nadu Agricultural University, Coimbatore, India
2Department of Plant Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India
Email: *udayvar@yahoo.com
Copyright © 2014 A. Ramalakshmi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property A. Ramalakshmi et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received November 10, 2013; revised December 10, 2013; accepted December 16, 2013
KEYWORDS
Bacillus thuringiensis; cry1Ac; Cloning
ABSTRACT
Transgenic Bt crops producing insecticidal crystal proteins from Bacillus thuringiensis (Bt), so-called Cry toxins, have proved useful in controlling insect pests. Among the cry toxins, Cry1A toxins are important because of high toxicity to lepidopteran pests and their widespread distribution among Bt strains. In Cry1A proteins, toxin fragment is comprised of about 620 amino acids of N-terminal region and C-terminal half is not required for toxicity. Four indigenous isolates of Bt viz., T15, T16, T20 and T31 were screened by PCR-RFLP for 3’-truncated cry1A gene(s) corresponding to toxin fragment. RFLP analysis of cry1A amplicons obtained from the four isolates of Bt showed presence of cry1Ac-type gene alone in three isolates. One of the cry1Ac-postive isolates, T15 which showed 100 percent mortality in Helicoverpa armigera, was selected for cloning of DNA fragment of about 2.1 kb containing 3’-truncated cry1Ac gene. Nucleotide sequence data generated for 3’-truncated cry1Ac gene of T15 showed 98 to 99 percent homology with 1958 bp of already reported sequences of all cry1Ac genes (cry1Ac1 to cry1Ac24). Deduced amino acid sequence of cry1Ac of Bt strain, T15 showed one to four percent variation in comparison to all reported Cry1Ac holotypes (Cry1Ac1 to Cry1Ac24) by differing at 5 to 19 positions. This suggests that the cry1Ac toxin of Bt isolate, T15 is a new kind of its group.
1. Introduction
The Bt has been used as a successful biological insecticide for more than 40 years and is a uniquely specific, safe, and effective tool for the control of a wide variety of insect pests [1]. The advancement in molecular biology led to the cloning of Bt crystal protein (cry) gene for the first time in 1981 [2]. Till date more than 412 cry genes have been successfully cloned and characterized for their insecticidal potential. Till August 2009, the Cry proteins are classified into 59 families based on their amino acid sequence similarity (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/). Among the Cry toxins, Cry1A toxins are important because of high toxicity to lepidopteran pests and their widespread distribution among Bt strains [3,4].
The cry1Ac gene is used into first version of insect resistant transgenic Bt-cotton. However, continuous exposure of a single Bt protein in Bt cotton can lead to resistance development in lepidopteran insects [5,6]. Genes developed in one country may not be much more effective against insect pests present in other countries. Cry1Ac protein is about 30 fold less toxic to H. armigera than to Heliothis virescens, the original target pest of transgenic cotton in USA [7]. Therefore a large number of Bt strains have been isolated and many types of insecticidal crystal proteins genes have been cloned. The diversity of Bt strains facilitates isolation of new types of cry genes. New variants of the already known cry gene subgroups could encode crystal proteins with significant difference in the level and spectrum of toxicity due to variation in their sequences [8]. Variation of even a single amino acid residue at certain positions of Cry proteins can remarkably influence the level of toxicity [9]. Apart from the fulllength genes, truncated genes which produce insecticidally active protein have been expressed in different crops like potato [2] and rice [10]. The present study describes screening of indigenous isolates of Bt for cry1A genes by PCR-RFLP and cloning of a new truncated cry1Ac gene from an Indian isolate of Bt.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids
Four indigenous Bt strains (T15, T16, T20 and T31) and reference strain, Bt subsp kurstaki (HD1) are from the collection of Bt strains maintained by the corresponding author in the Department of Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India. The reference strain, HD1 were originally obtained from Bacillus Genetic Stock Centre, Ohio state university, Columbus, Ohio, USA. The T/A cloning vector, pGEM-T Easy used in the present study was purchased from Promega BioScience, INC.
2.2. Amplification of Bt DNA by PCR
Total DNA from Bt strains, T15, T16, T20 and T31 was extracted as described earlier [11] and used as a template for the Polymerase Chain Reaction (PCR) amplification. Based on the published sequence of cry1A gene subtypes, cry1Aa1, cry1Ab1 and cry1Ac1 (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/), cloning primers (1AF and 1AR) specific for 3’-truncated cry1A gene(s) were designed and listed in Table 1. These primers are specific to upstream and internal region of the following genes: cry1Aa1, cry1Ab1 and cry1Ac1. The PCR was accomplished using an Eppendorf thermal cycler in 25 µl reaction volume containing 50 ng of total genomic DNA of Bt, 2.5 µl of 10X PCR buffer (10 mMTris-HCl; pH: 9.0, 50 mM KCl, 1.5 mM MgCl2), 75 μM each of dNTPs, 50 ng each of forward and reverse primers and 1.5 Units of Taq DNA polymerase. The PCR was performed for 30 cycles as follows: 94˚C for 1 min, 58˚C for 45 s, 72˚C for 45 s and the final extension was performed for 7 min at 72˚C.
2.3. RFLP for 3’-Truncated Cry1A Gene(s) by EcoRI
The amplicon (3’-truncated cry1A gene(s)) obtained by PCR was verified on 1.2 percent agarose gel. The PCR product was column purified with PCR clean up kit as per the manufacturer’s instruction provided by Sigma. The final concentration of the purified product was
Table 1. Primers used for amplification of 3’-truncated cry1A gene(s).
checked by resolving in 1.2 per cent agarose gel. The column purified product of 3’-truncated cry1A gene(s) was digested by EcoRI as per the manufacturer’s instruction. Restriction digestion was set up for 20 µl as follows: DNA: 500 ng, Buffer (10X): 2.0 µl, BSA (10X): 2.0 µl, EcoRI enzyme (10U): 0.5 µl , sterile distilled water : to 20 µl. The restriction digestion was carried out at 37˚C for 1 h and 30 min. The digested product was analyzed by agarose 1.2 per cent gel electrophoresis.
2.4. Cloning of 3’-Truncated cry1Ac Gene from Bt Isolate, T15
The column purified PCR product of 3’-truncated cry1Ac gene (~2.1 kb) from Bt isolate, T15 was ligated into T/A vector (pGEM-T Easy, Promega) as per the manufacturer’s instruction. The ligated mixture was transformed into E. coli as per the standard procedure [12]. The transformed colonies of E. coli were screened by colony PCR with M13F and M13R primers for checking the presence of insert (3’-truncated cry1A gene of Bt isolate, T15).
2.5. Nucleotide Sequencing of Recombinant Plasmids
The plasmid DNA was isolated from the E. coli transformants containing truncated cry1Ac gene of Bt isolate, T15 and Nucleotide sequence of recombinant plasmids was carried out by automated sequencing (Ist Base, Singapore and Chromous Biotech Pvt. Ltd., Bangalore, India). The sequence data generated for upstream region of about 147 bp and toxin fragment of 1958 bp were subjected to homology search through Basic Local Alignment Search Tool (BLAST) of National Centre for Biotechnological Information (NCBI) (www.ncbi.nlm.nih.gov/Blast). The deduced amino acid sequence was generated by BioEdit [13].
3. Result
3.1. PCR-RFLP for 3’-Truncated Cry1A Gene(s)
The four indigenous isolates of Bt viz., T15, T16, T20 and T31 which showed 90 to 100 per cent mortality in H. armigera (data not shown) were positive for amplification by cry1 genes specific primers described by [14]. Hence, these four isolates were selected for amplification of truncated cry1A gene(s) corresponding to toxin fragment and PCR-RFLP to know the novelty in their sequences. Total genomic DNA isolated from four indigenous isolates of Bt was used as a template for amplification of DNA fragment containing 3’-truncated cry1A gene(s) sequence. An intact band of DNA fragment of ~2.1 kb corresponding to 3’-truncated cry1A gene(s) was amplified from four Bt isolates, with 1AF and 1AR primers by PCR, without any nonspecific amplification (Figure 1).
The expected restriction fragment sizes of the known cry1A truncated genes with EcoRI were listed in Table 2. Restriction analysis of the reference strain of Bt, HD1 was performed for comparison of indigenous isolates of Bt. The column purified PCR products of truncated cry1A gene(s) from four Bt isolates along with reference strain, HD1 were digested with restriction enzyme, EcoRI. Data from agarose gel electrophoresis of cry1A amplicons digested by EcoRI showed that three of four indigenous Bt isolates viz., T15, T16 and T20 had a cry1Ac-type RFLP pattern. Another isolate (T31) showed fragments corresponding to cry1Aa and/or cry1Ab along with cry1Ac genes as in the case of reference strain, HD1 (Figure 2 and Table 3).
3.2. Cloning and Sequence Analysis of 3’-Truncated cry1Ac Gene from Bt Isolate, T15
Based on PCR-RFLP data, the Bt isolate, T15 which showed 100 per cent mortality in H. armigera and having cry1Ac gene alone was selected for cloning of DNA fragments of ~2.1 kb corresponding to 3’-truncated cry1A gene. The column purified PCR product (~2.1 kb) of cry1Ac truncated gene from Bt isolate, T15 was cloned into pGEM-T easy vector (T/A vector). The recombinant clones (white colonies) were selected on LB agar containing X-gal, IPTG and ampicillin. Presence of insert was confirmed in recombinant E. coli colonies, by colony PCR with M13 forward and M13 reverse primers. Agarose gel electrophoresis showed amplification of ex-

Table 2. RFLP for EcoRI enzyme in 3’-truncated cry1A genes of ~2.1 kb.

Table 3. Restriction analysis of 3’-truncated cry1A gene fragments of indigenous isolates of B. thuringiensis by EcoRI enzyme.
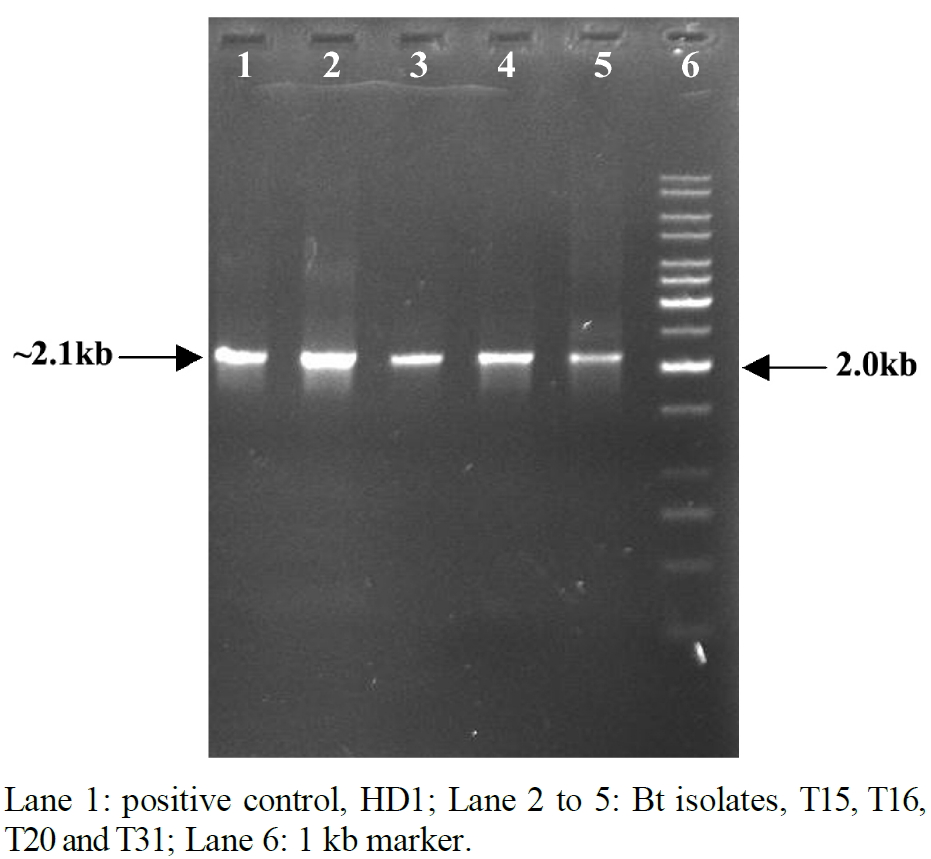
Figure 1. Amplification of 3’-truncated cry1A gene(s) from indigenous isolates of Bt.
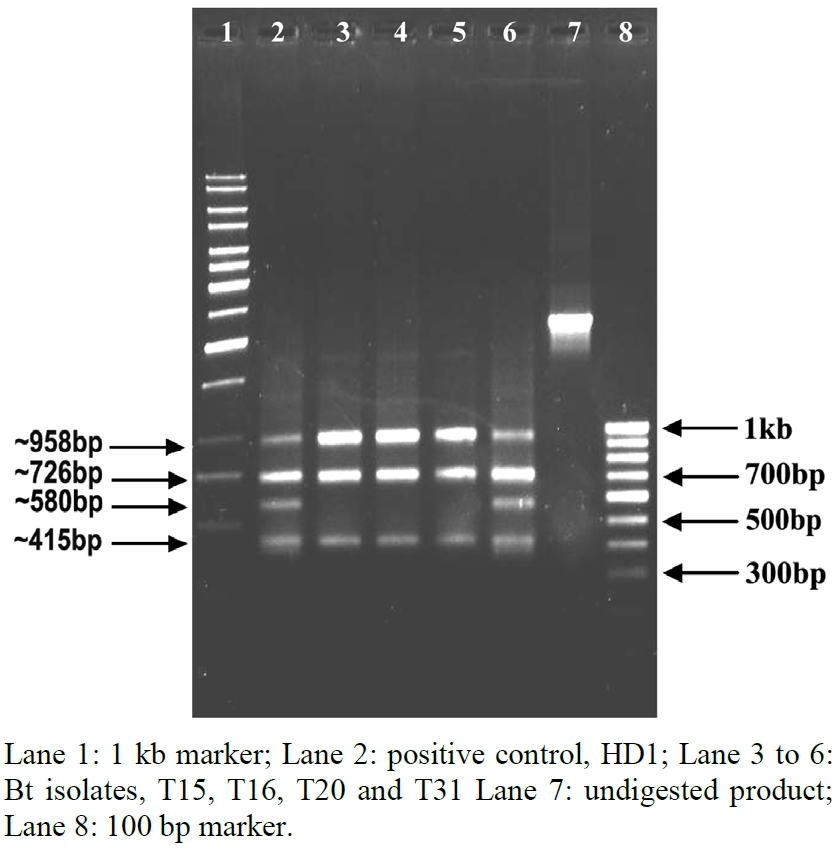
Figure 2. RFLP analysis of 3’-truncated cry1A gene(s) from indigenous isolates of Bt.
pected size of ~2.3 kb corresponding to the sum of insert DNA of 2.1 kb and vector sequence of about 200 bp.
Recombinant plasmid isolated from three of the E. coli clones were used to determine the nucleotide sequence of the 3’-truncated cry1Ac gene of Bt strain, T15 by automated DNA sequencing. The nucleotide sequence revealed the presence of upstream (147 bp) and internal region (1958 bp) corresponding to cry1Ac gene. The internal region of 1958 bp starting from ATG encodes 652- amino acids (Figure 3). The computer-based homology

Figure 3. Nucleotide sequence and deduced amino acid sequence of truncated cry1Ac gene of Bt strain, T15.
search program of the National Centre for Biotechnology Information revealed that it is a new cry1Ac gene. It has 98 to 99 per cent homology with 1958 bp of already reported sequences of all cry1Ac genes (cry1Ac1 to cry1Ac24 till Aug, 2009). Deduced amino acid sequence of truncated cry1Ac of Bt isolate, T15 showed one to four per cent variation from all the other Cry1Ac sequences (Cry1Ac1 to Cry1Ac24, till Aug, 2009) by differing at 5 to 19 positions. Minimum variation at five positions (44, 61, 62, 276 and 293) was observed with six reported Cry1Ac sequences (Cry1Ac1, Cry1Ac7, Cry1Ac8, Cry1Ac9, Cry1Ac10 and Cry1Ac16). Maximum variation at 19 positions was observed in truncated Cry1Ac sequence Bt strain, T15 when compared to Cry1Ac17 (Tables 4 and 5).
4. Discussion
Restriction fragment length polymorphic (RFLP) analysis of cry genes amplified from novel Bt isolates could provide preliminary information about the diversity [15]. This is a two-step approach in which PCR amplification with specific primers is followed by restriction analysis of the PCR products. In the earlier studies, a novel cry1A-type gene and cry1Ie1 gene were detected in Bt isolates using RFLP of cry genes by [16,17], respec-

Table 4. Comparison of deduced amino acid sequence of cry1Ac gene of Bt isolate T15 with other sequences of Cry1Ac*.

Table 5. Comparison of deduced amino acid sequence of cry1Ac gene of Bt isolate T15 with other sequences of Cry1Ac*.
tively. In the present study, EcoRI digestion of 3’-truncated cry1A gene(s) fragments amplified from four indigenous isolates of Bt and the reference strain, HD1 showed the presence of cry1Ac gene alone in three of the four Bt isolates. Therefore, the banding patterns of the three indigenous isolates are different from that of the reference strain, HD1.
Lepidopteran active insecticidal crystal proteins are protoxins of molecular weight about 135 kDa. These protoxins are proteolyticallty cleaved into smaller active forms of molecular weight of 60 - 70 kDa derived from the N-terminal half of the protein [18]. The C-terminal half of 135-kDa Cry1 protoxins is not required for toxicity, if it could be eliminated and the cellular resources could be redirected to synthesize an equivalent additional amount of the N-terminal half, the specific toxicity—i.e., the toxicity per unit of mass of bacterial insecticides— might be improved [19]. This would in essence convert Cry1 proteins by truncation into toxins like Cry2A or Cry3A.
In the present study, the 3’-truncated derivative of cry1Ac of a Bt strain, T15, was amplified and cloned into a T/A vector. Nucleotide sequencing data of the newly cloned (3’-truncated) cry1Ac gene showed 98 to 99 percent similarity with sequences of already reported cry1Ac genes. Deduced amino acid sequence (652 residues) of cry1Ac of Bt isolate, T15 showed variation from already reported Cry1Ac sequences at 5 to 19 positions. Even slight variation in amino acid sequence within a Cry protein class can dramatically impact insecticidal activity [20]. Lee et al. [21] reported that Cry1Ac proteins differing at only two amino acid positions exhibited a 10-fold difference in toxicity towards the gypsy moth, Lymantria dispar.
Tertiary structure of Cry1A toxins are formed of three domains. The domain Ι is extending from residue 33 to residue 253, containing eight helices. Residues from 265 - 461 and 463 to 609 are domain II and domain ΙΙΙ, respectively [22]. Different mutational studies have demonstrated that domain Ι of Cry proteins involved in pore formation and toxicity [23]. In the present study, the amino acid variations were observed in domain Ι of Cry1Ac protein too (3 to 8 variations). Because of the variations in the Cry1Ac amino acid sequence of Bt isolate, T15 may influence the level of toxicity. Further studies on expression of the newly cloned truncated cry1Ac gene in recombinant bacteria will reveal its insecticidal property prior to its use in the development of indigenous Bt crops.
Acknowledgements
Authors are thankful to Dr. P. Balasubramanian, Prof. TNAU, India and Dr. P.U. Krishnaraj (Project Coordinator), UAS, Dharwad, India for their encouragement during the course of study. This study was supported by research grant from the Department of Biotechnology, Government of India, New Delhi (BT/PR5349/AGR/02/ 274/2004).
REFERENCES
- E. W. Nester, L. S. Thomashow, M. Metz and M. Gordon, “100 Years of Bacillus thuringiensis Bacillus thuringiensis: A Critical Scientific Assessment. American Society for Microbiology,” Washington DC, 2002. http://www.asmusa.org
- E. Schnepf and H. R. Whiteley, “Cloning and Expression of the B. thuringiensis Protein Gene in E. coli,” Proceedings of the National Academy of Sciences, Vol. 78, No. 5, 1981, pp. 2893-2897. http://dx.doi.org/10.1073/pnas.78.5.2893
- A. Bravo, S. Sarabia, L. Lopez, H. Ontiveros, C. Abarca, A. Ortiz, M. Ortiz, L. Lina, F. J. Villalobos, G. Pena, M. E. Nunez-Valdez, M. Soberon and R. Quintero, “Characterization of Cry Genes in a Mexican B. thuringiensis Strain Collection,” Applied and Environmental Microbiology, Vol. 64, 1998, pp. 4965-4972.
- D. Uribe, W. Masrtinez and J. Ceron, “Distribution and Diversity of Cry Genes in Native Strains of B. thuringiensis Obtained from Different Ecosystems from Colombia,” Journal of Invertebrate Pathology, Vol. 82, No. 2, 2003, pp. 119-127. http://dx.doi.org/10.1016/S0022-2011(02)00195-7
- R. J. Akhurst, W. James, L. Bird and C. Beard, “Resistance to the Cry1Ac-Endotoxin of B. thuringiensis in the Cotton Bollworm, H. armigera (Lepidoptera: Noctuidae),” Journal of Economic Entomology, Vol. 96, No. 4, 2003, pp. 1290-1299. http://dx.doi.org/10.1603/0022-0493-96.4.1290
- B. E. Tabashnik, “Evolution of Resistance to B. thuringiensis,” Annual Review of Entomology, Vol. 39, 1994, pp. 47-79. http://dx.doi.org/10.1146/annurev.en.39.010194.000403
- C. Liao, D. G. Heckel and R. Akhurst, “Toxicity of B. thuringiensis Insecticidal Protein for Helicoverpa armigera and H. punctigera (Lepidoptera: Noctuidae), Major Pests of Cotton,” Journal of Invertebrate Pathology, Vol. 80, No. 1, 2002, pp. 55-63. http://dx.doi.org/10.1016/S0022-2011(02)00035-6
- J. Xue, G. M. Liang, N. Crickmore, H. Li, K. He, F. Song, X. Feng, D. Huang and J. Zhang, “Cloning and Characterization of a Novel Cry1A Toxin from Bacillus thuringiensis with High Toxicity to the Asian Corn Borer and Other Lepidopteran Insects,” Microbiology Letters, Vol. 280, No. 1, 2008, pp. 95-101. http://dx.doi.org/10.1111/j.1574-6968.2007.01053.x
- V. Udayasuriyan, A. Nakamura, A. Mori, H. Masaki and T. Uozumi, “Cloning of a New crylA(a), Gene from B. thuringiensis Strain FU-2-7 and Analysis of Chimeric cry1A(a) Proteins of Toxicity,” Bioscience, Biotechnology, and Biochemistry, Vol. 58, No. 5, 1994, pp. 830-835. http://dx.doi.org/10.1271/bbb.58.830
- H. Fujimoto, K. Itoh, M. Yamamoto, J. Kyozuka and K. Shimamoto, “Insect Resistant Rice Generated by Introduction of a Modified δ-Endotoxin Gene of Bacillus thuringiensis,” Biotechnology, Vol. 11, 1993, pp. 1151-1155. http://dx.doi.org/10.1038/nbt1093-1151
- S. Kalman, K. L. Kiehne, N. Cooper, M. S. Reynoso and T. Yamamoto, “Enhanced Production of Insecticidal Proteins in Bacillus thuringiensis Strains Carrying an Additional Crystal Protein Gene in the Chromosomes,” Applied and Environmental Microbiology, Vol. 61, 1995, pp. 3063-3068.
- J. Sambrook, E. F. Fritsch and T. M. Maniatis, “Molecular Cloning: A Laboratory Manual,” 2nd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1989.
- T. A. Hall, “BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis,” 1999. http://www.mbio.ncsu.edu/BioEdit/bioedit.html
- E. Ben-Dov, A. Zaritsky, E. Dahan, Z. Barak, R. Sinai, R. Manasherob, A. Khamraev, E. Troitskaya, A. Dubitsky, N. Berezina and Y. Margalith, “Extended Screening by PCR for Seven cry Group Genes from Field-Collected Strains of B. thuringiensis,” Applied and Environmental Microbiology, Vol. 63, 1997, pp. 4883-4890.
- W. S. Kuo and K. F. Chak, “Identification of Novel cryType Genes from B. thuringiensis Strains on the Basis of Restriction Fragment Length Polymorphism of the PCRAmplified DNA,” Applied and Environmental Microbiology, Vol. 62, 1996, pp. 1367-1377.
- J. Wang, A. Boets, J. van Rie and G. Ren, “Characterization of cry1, cry2 and cry9 Genes in B. thuringiensis Isolates from China,” Journal of Invertebrate Pathology, Vol. 82, No. 1, 2003, pp. 63-71. http://dx.doi.org/10.1016/S0022-2011(02)00202-1
- F. Song, J. Zhange, A. Gu, Y. We, L. Han, K. He, Z. Chan, J. Yao, Y. Hu, G. Li and D. Huang, “Identification of cry1I-Type Genes from B. thuringiensis Strains and Characterization of a nOvel cry1I Type Gene,” Applied and Environmental Microbiology, Vol. 69, No. 9, 2003, pp. 5207-5211. http://dx.doi.org/10.1128/AEM.69.9.5207-5211.2003
- H. Hofte and H. R. Whiteley, “Insecticidal Crystal Proteins of B. thuringiensis,” Microbiology Reviews, Vol. 53, 1989, pp. 242-255.
- H. W. Park, D. K. Bideshi and B. A. Federici, “Molecular Genetic Manipulation of Truncated Cry1C Protein Synthesis in Bacillus thuringiensis to Improve Stability and Yield,” Applied and Environmental Microbiology, Vol. 66, No. 10, 2000, pp. 4449-4455. http://dx.doi.org/10.1128/AEM.66.10.4449-4455.2000
- E. Schnepf, N. Crickmore, J. Van Rie, D. Lerecurs, J. Baum, J. Feitelson, J. D. R. Zeigler and D. H. Dean, “B. thuringiensis and Its Pesticidal Crystal Proteins,” Microbiology and Molecular Biology Reviews, Vol. 62, 1998, pp. 775-806.
- M. K. Lee, T. H. You, A. Curtiss and D. H. Dean, “Involvement of Two Amino Acid Residues in the Loop Region of Bacillus thuringiensis Cry1Ab Toxin in Toxicity and Binding to Lymantria dispar,” Biochemical and Biophysical Research Communications, Vol. 229, No. 1, 1996, pp. 139-146. http://dx.doi.org/10.1006/bbrc.1996.1770
- P. Grochulski, L. Masson, S. Borisova, M. Pusztai-Carey, J. L. Schwartz, R. Brousseau and M. Cygler, “B. thuringiensis CryIA(a) Insecticidal Toxin: Crystal Structure and Channel Formation,” Journal of Molecular Biology, Vol. 254, No. 3, 1995, pp. 447-464. http://dx.doi.org/10.1006/jmbi.1995.0630
- E. Gazit, P. L. Rocca, M. S. P. Sansom and Y. Shai, “The Structure and Organization within the Membrane of the Helices Composing the Pore-Forming Domain of Bacillus thuringiensis Delta-Endotoxin Are Consistent with an ‘Umbrella-Like’ Structure of the Pore,” Proceedings of the National Academy of Sciences, Vol. 95, No. 21, 1998, pp. 12289-12294. http://dx.doi.org/10.1073/pnas.95.21.12289
NOTES
*Corresponding author.

