Open Journal of Medical Microbiology
Vol.3 No.2(2013), Article ID:33307,7 pages DOI:10.4236/ojmm.2013.32022
Surveillance of Fluoroquinolone-Resistant Clinical Isolates of Pseudomonas aeruginosa
1Pharmaceutical Unit, Ohashi Medical Center, Toho University, Tokyo, Japan
2School of Pharmaceutical Sciences, Toho University, Funabashi, Chiba, Japan
Email: *fkato@phar.toho-u.ac.jp
Copyright © 2013 Hideki Kobayashi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 13, 2013; revised May 14, 2013; accepted May 22, 2013
Keywords: Pseudomonas aeruginosa; Fluoroquinolone Resistant; Type II Topoisomerases; Efflux Pump
ABSTRACT
Ciprofloxacin (CPFX) and pazufloxacin (PZFX) have strong antibacterial activity against Pseudomonas aeruginosa. We investigated the sensitivity of P. aeruginosa to CPFX and PZFX in 373 strains isolated from inpatients (321 strains) and outpatients (52 strains) during September 2010 to September 2011 at Toho University Ohashi Medical Center. The percentage of CPFX-non-susceptible (≥3.91 µg/mL) among inpatients was 22.4%, but that among outpatients was 1.9%. As the major resistance mechanism to fluoroquinolones in P. aeruginosa involves modification of type II topoisomerases (DNA gyrase and topoisomerase IV), we examined mutations in the quinolone-resistance-determining regions (QRDRs) of gyrA and parC of P. aeruginosa isolates. Among the 373 isolates, 73 isolates had reduced CPFX-susceptibility and 88 had reduced PZFX-susceptibility. Sequencing of gyrA and parC revealed base substitutions that resulted in amino acid replacements in QRDR of GyrA in 70 P. aeruginosa isolates, while Thr83Ile (in GyrA) and Ser87Leu (in ParC) substitutions were found in 12 strains. These replacements were clearly associated with reduced susceptibility to CPFX and PZFX. However, we also found strains with high MICs to quinolones without mutations in either gyrA or parC. We then investigated the effect of efflux pumps in CPFX-resistance in these isolates. In the presence of an efflux pump inhibitor, MIC values in 12 of 66 strains decreased to 1/23. We also sequenced genes related to overexpression of efflux pumps, viz., mexZ, mexR, and nfxB. Eight of the strains without mutations in QRDRs had a mutation in mexZ, 7 strains had a mutation in mexR, but no mutation was identified in nfxB.
1. Introduction
Pseudomonas aeruginosa, a ubiquitous Gram-negative soil organism, is widely distributed in nature. P. aeruginosa is noted for its metabolic versatility and its exceptional ability to colonize a wide variety of environments, and also for its intrinsic resistance to a wide variety of antimicrobial agents. Such characteristics result in a higher prevalence of this organism in hospital environments, where it is a clinically significant pathogen causing opportunistic infections and nosocomial outbreaks [1, 2].
P. aeruginosa typically infects injured, burned, immune deficient, or immunocompromised patients and causes a variety of infections. The bacterium has various virulence determinants, such as adhesins [3], toxins [4], capsules [5], and biofilms [6], and is also frequently resistant to many commonly used antibiotics [7]. Though many strains of P. aeruginosa are susceptible to cephalosporins, carbenicillin, gentamicin, tobramycin, colistin, polymyxin, and fluoroquinolones, strains resistant to those antibiotics are increasing in medical care facilities and isolation of the multidrug-resistant strain have been reported [8-12].
One of the challenges in managing P. aeruginosa infections is an inherent resistance mechanism, referred to as intrinsic resistance. Because of its virulence and the limited choices of effective antimicrobial agents, treatment of P. aeruginosa infection is often complicated. The present study aimed to study the antimicrobial susceptibility patterns of P. aeruginosa isolated from outpatients and inpatients, using the agar dilution method, and by defining mutations in gyrA, parC, mexZ, mexR and nfxB, which contribute to fluoroquinolon resistance, in these isolates.
2. Materials and Methods
2.1. Antibacterial Agents and Bacterial Strains
Ciprofloxacin, meropenem, and amikacin were purchased from Wako Pure Chemical Industries, Ltd., Japan, and pazufloxacin was purchased from Toyama Chemical Co., Ltd., Japan.
In this study, 373 clinical isolates of P. aeruginosa were collected during September 2010 to September 2011 at Toho University Ohashi Medical Center. Of these, 321 isolates of P. aeruginosa were obtained from inpatients with respiratory tract infections, urinary tract infections, surgical site infection, gastrointestinal tract infections, or bacteremia, whereas the remaining 52 isolates were obtained from outpatients with tympanitis, bacterial skin infections, respiratory tract infections, or urinary tract infections. All the P. aeruginosa strains isolated were maintained in MicrobankTM (Pro-Lab, Toronto, Canada) and kept at –80˚C.
2.2. Determination of MIC
MICs were determined by the standard agar dilution assay, according to guidelines of the National Committee for Clinical Laboratory Standards (NCCLS), using Mueller-Hinton (MH) agar (Becton, Dickinson and Company, USA). Strains preserved on MicrobankTM were inoculated into 2 mL of MH broth and incubated at 37˚C for 18 - 20 h. After dilution with MH broth, 2 μL of the bacterial suspension containing 104 CFU was spotted onto drug-containing agar plates. The plates were incubated at 37˚C for 24 h. The MIC was defined as the lowest drug concentration that prevented visible growth of bacteria.
2.3. Statistical Analysis
Mann-Whitney’s U-test was used to evaluate the relationship between the distribution of MIC of CPFX, MEPM, PZFX and AMK in the strains of P. aeruginosa isolated from outpatients and inpatients.
2.4. PCR Amplification and DNA Sequence Determination and Analysis
The strains were incubated in MH broth at 37˚C overnight, and 1 mL of the culture was centrifuged (15,000 rpm, 10 min, at 4˚C). A sterilized toothpick was used to lightly touch the precipitate, and cells on the toothpick were suspended in 25 µL of PCR mixture: KOD FX 0.5 U (TOYOBO Co., Ltd., Japan), 0.3 µM of each primer, 0.4 mM of each dNTP. Primers used for PCR are listed in Table 1. The amplification procedure comprised denaturation at 95˚C for 2 min, followed by 40 cycles, each consisting of denaturation for 10 s at 98˚C, annealing for 30 s at 55˚C, and polymerization for 30 s at 74˚C. Amplified DNA was directly sequenced by the dideoxy chain termination method, employing a Thermo Sequenase II dye terminator cycle sequencing kit (Amersham Pharmacia Biotech Inc., NJ, USA), BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Inc., CA, USA) according to the manufacturer’s protocol. The products were automatically analyzed in a model ABI 3500 DNA autosequencer (Applied Biosystems, Inc.). DNA sequences have been deposited in DDBJ (Accession numbers: AB753299-AB753440).
2.5. Impact of Efflux Pump Inhibitor (EPI) on the MICs of Ciprofloxacin
Strains of P. aeruginosa of which the ciprofloxacin MIC exceeded 3.91 μg/mL were inoculated on agar plates containing various concentrations of ciprofloxacin, with or without 20 μg/mL of the efflux pump inhibitor, phenylalanine arginyl-β-naphtylamide (Sigma-Aldrich, MO, USA). MICs of ciprofloxacin in the presence of EPI were
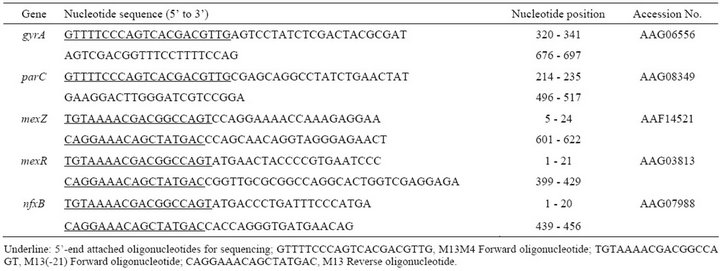
Table 1. Primers for amplification and sequencing of gyrA, parC, mexZ, mexR and nfxB genes.
determined after overnight incubation of isolates at 37˚C.
3. Results and Discussion
3.1. Antibiogram Pattern of Pseudomonas aeruginosa
The results of MH agar-based antibiogram pattern study of the strains of P. aeruginosa isolated from inpatients (n = 321) and outpatients (n = 52) is shown in Figure 1. The MIC range of CPFX in the strains isolated from outpatients, except for 1 strain, was between <0.06 and 1.95 μg/mL, but that of strains isolated from inpatients was between <0.06 and >62.5 μg/mL (Figure 1(a)). Thus, strains from outpatients were sensitive to CPFX, except for 1 strain that was isolated from an outpatient who had received ofloxacin treatment for chronic otitis media. The percentage of CPFX-non-susceptible strains (≥3.91 μg/mL) among inpatients was 22.4%, but that among outpatients was 1.9%.
The MIC range of the strains from outpatients for PZFX was between 0.06 and 7.81 μg/mL, except for 1 strain, and that from inpatients was between <0.06 and 62.5 μg/mL (Figure 1(c)).
The MIC ranges of the strains from outpatients and inpatients for MEPM were between <0.49 and 7.81 μg/mL, and between <0.49 and >125 μg/mL, respectively. The percentage of MEPM-non-susceptible strains (≥15.6 μg/mL) among inpatients was 30.5%, but there were no MEPM-non-susceptible strains among outpatients (Figure 1(b)).
The MIC ranges of the strains from outpatients for AMK was between <0.49 and 15.62 μg/mL, and that from inpatients was between <0.49 and 62.5 μg/mL. The percentage of AMK-non-susceptible strains (≥31.25 μg/ mL) from inpatients was 5.6%, but no AMK-non-susceptible strains were found among outpatients (Figure 1(d)).
Our results indicated that most of the strains isolated from outpatients were susceptible to CPFX and PZFX, but 22.4% of the strains isolated from inpatients were resistant to CPFX; strains isolated from outpatient were susceptible to MEPM, but 30.5% of the strains isolated from inpatients were resistant to MEPM (Figure 1(b)). Similarly, strains isolated from outpatients were susceptible to AMK, but 5.6 % of the strains isolated from inpatients were resistant to this antimicrobial agent (Figure 1(d)).
Statistical analysis of the distribution of MIC of CPFX, MEPM, PZFX and AMK in the strains of P. aeruginosa isolated from outpatients and inpatients showed significant differences (P < 0.05) in each antibiotics. Surveys of antimicrobial susceptibilities in clinical isolates of P. aeruginosa in 2004 and 2008 in Japan showed that 22% of the 90 isolates collected from 13 medical facilities
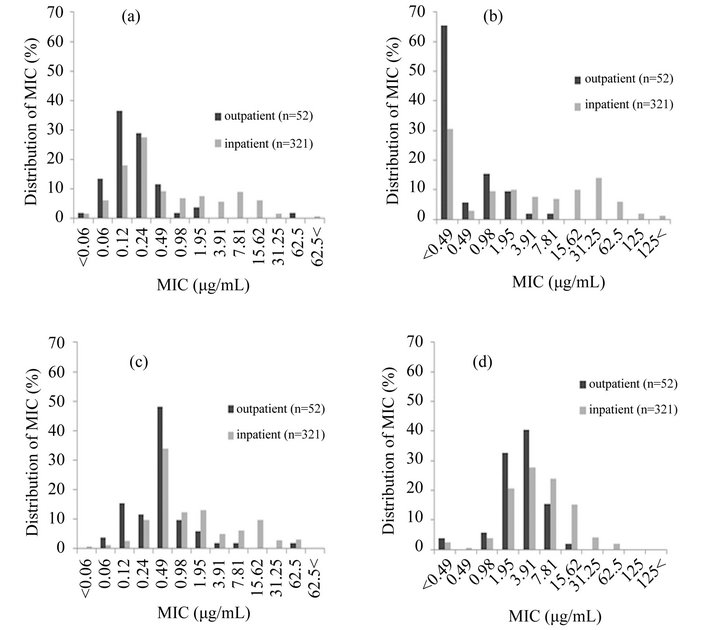
Figure 1. Distribution of MIC of CPFX (a); MEPM (b); PZFX (c); and AMK (d) in outpatients and inpatients.
were resistant to CPFX, while 22% were resistant to MEPM [13]; furthermore, 42.9% of 207 isolates from 1 medical facility were resistant to CPFX, while 12.6% were resistant to MEPM [14]. In another study, among 188 strains isolated from 139 cases in a number of medical facilities, 24% were resistant to CPFX, whereas 18% were resistant to MEPM [15]. The differences in resistance rates among the present study and the surveys mentioned above could reflect differences in clinical usage of antimicrobial agents and periods of administration, etc.
3.2. Relationship between MIC and Mutations in gyrA and parC
As mentioned above, 22.4% of the P. aeruginosa strains isolated from inpatients were not susceptible to CPFX; we then set out to identify mutations in gyrA and parC that may underlie this CPFX-resistance. Strains of P. aeruginosa repeatedly isolated from the same patient, or strains with MICs exceeding 3.91 μg/mL for CPFX or PZFX were selected. Mutations in gyrA and parC of 131 strains were analyzed by PCR and DNA sequencing.
As shown in Figure 2(a), we found no mutations in the PCR-amplified gyrA and parC fragments from 52 of 60 strains with MICs less than 3.91 μg/mL for CPFX. The remaining 8 strains had a mutation only in gyrA; these mutations resulted in a Thr83Ile substitution in GyrA in 4 strains, an Asp87Gly substitution in 3 strains, and an Asp87Asn substitution in 1 strain. Nine of 46 strains with a MIC range from ≥3.91 μg/mL to <15.6 μg/mL for CPFX had no mutation in gyrA and parC, whereas 35 of these 46 strains had a mutation in only gyrA. Two strains had mutations in both gyrA and parC. Two of 25 strains with MICs for CPFX of ≥15.6 μg/mL had no mutation, 15 of these 25 strains had a mutation only in gyrA, and 8 strains had mutations in both gyrA and parC.
A similar analysis was performed in strains with MICs for PZFX of less than 3.91 μg/mL, ≥3.91 μg/mL to <15.6 μg/mL, and ≥15.6 μg/mL (Figure 2(b)). No strains with MICs of less than 3.91 μg/mL for PZFX had mutations in gyrA and parC. In the 27 strains with a MIC range of ≥3.91 μg/mL to <15.6 μg/mL for PZFX, 7 had no mutations in gyrA or parC, 16 strains had a mutation only in gyrA, and 4 strains had mutations in both gyrA and parC. Two of 52 strains with MICs for PZFX of 15.6 μg/mL or more had no mutations, 42 of the 52 strains had a mutation only in gyrA, and 8 strains had mutations in both gyrA and parC.
We found no CPFXor PZFX-resistant strain with a mutation only in parC. In Gram-positive cocci, mutations in parC are seen commonly in fluoroquinolone-resistant strains, and it is well-known that these mutations precede
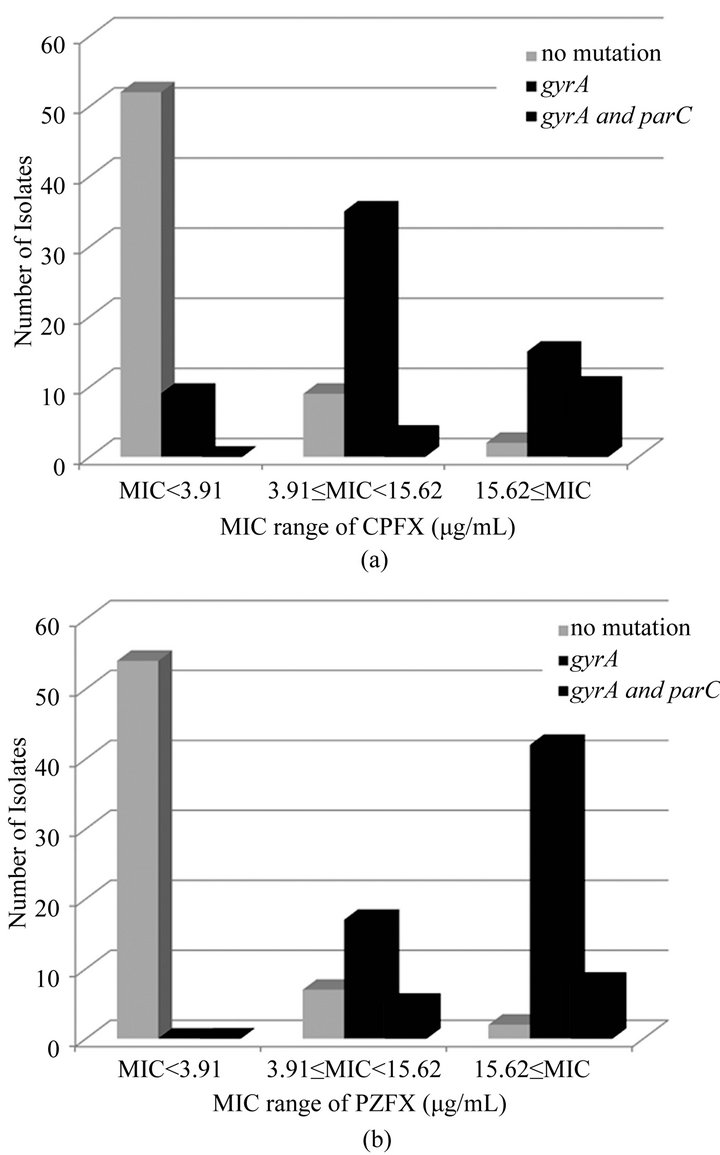
Figure 2. The relation with MICs of CPFX (a) or PZFX (b) and mutations in gyrA and parC genes.
mutations in gyrA [16,17]. The inverse, i.e., that mutations in gyrA precede mutations in parC, is typically seen in Gram-negative bacteria. In this study, CPFX-resistant strains had mutations in gyrA, while some strains had mutations in both gyrA and parC, similar to the finding in fluoroquinolone-resistant Gram-negative bacteria. Moreover, the strains with mutations in both gyrA and parC had higher MICs for CPFX [18,19].
3.3. Impact of Efflux Pump Inhibition on MICs of Ciprofloxacin
Interestingly, we found strains with high MICs for fluoroquinolones without mutations in either gyrA or parC. These higher CPFX-resistant strains were assumed to have other fluoroquinolone-resistance mechanisms, such as an overproduction of efflux pumps. To investigate this possibility, we used phenylalanine arginyl β-naphtylamide (PAβN), which inhibits efflux pumps in P. aeruginosa. [20]. Table 2 shows the effect of PAβN on the MICs of CPFX-resistant strains. Twelve of 66 CPFX-
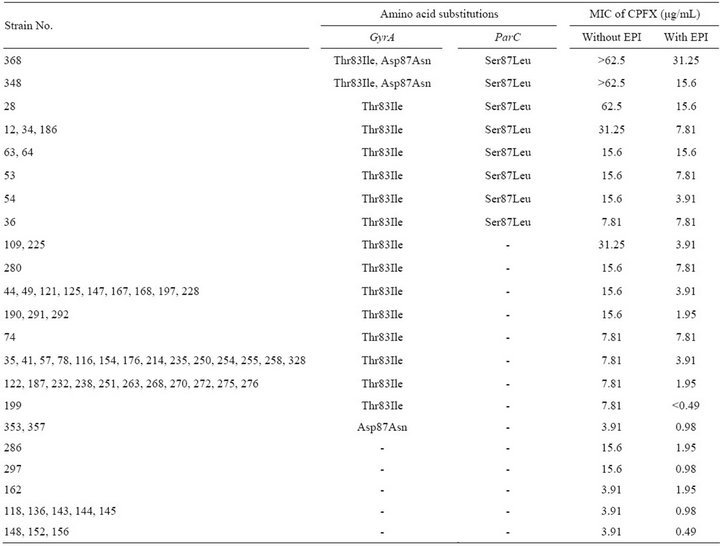
Table 2. Impact of efflux pump inhibitor on the MICs of ciprofloxacin.
resistant strains showed an 8-fold decrease in CPFX-MIC in the presence of PAbN. Strains 109 and 225 had an alteration only in GyrA (Thr83Ile), and the MICs for CPFX of these strains (31.25 µg/mL) were higher than those of strains with the same GyrA alteration. However, MICs of these strains were decreased to 3.91 µg/mL in the presence of PAbN. Similar MIC decreases were observed in strains with no alterations in GyrA and ParC. Therefore, overexpression of efflux pumps appears to contribute to the higher CPFX-resistance in these 12 strains.
3.4. Mutations in gyrA, parC, mexZ, mexR and nfxB in Clinical Isolates from Inpatients
As mentioned above, not only mutations in DNA topoisomerase II, but also those in genes regulating efflux pumps could contribute to CPFX-resistance in P. aeruginosa. We sequenced gyrA, parC, mexZ, mexR, and nfxB in 120 P. aeruginosa strains isolated from inpatients. The sequences were compared with the corresponding sequences of P. aeruginosa PAO1 (Pseudomonas Genome Database, http://www.pseudomonas.com/). The isolates were grouped into 12 groups on the basis of the amino acid substitutions found in GyrA, ParC, MexZ, MexR, and NfxB as shown in Table 3. No mutation were found in these genes in 52 strains (Group I), but MICs of CPFX in 11 of these strains were ≥3.91 µg/mL. Strains with mutations in mexZ, or mexZ and mexR, were sensitive to 3.91 µg/mL CPFX. The mutation most frequently found in mexZ was a single base substitution that resulted in a missense mutation (C490T; 15 of 27 strains). Other mutations identified in mexZ were single base deletions leading to frame-shifts after Glu114 and Glu146, and 5-bp deletions leading to frame-shifts following Ala21 or Gly148. However, when comparing the MIC ranges of Groups V and VII, the contribution of these mexZ mutations to CPFX-resistance was not significant.
Amino acid substitutions in MexR were found in 31 of 120 strains; the Val126Glu substitution was found in 30 of 31 strains, and 1 strain carried substitutions Val126- Glu and Ser92Asn. As for mexZ mutations, the mexR mutations found in this study did not have a significant
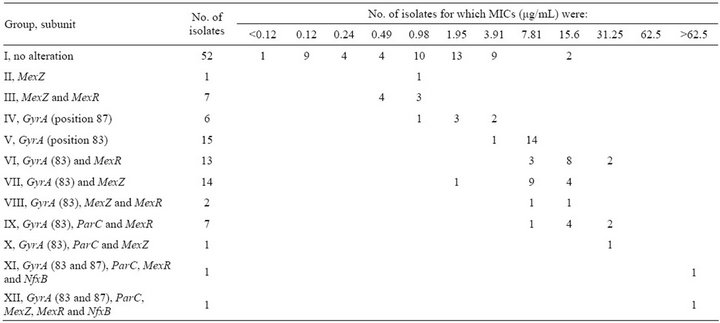
Table 3. Alterations in GyrA, ParC, MexZ, MexR and NfxB subunits and correlation of MICs of ciprofloxacin.
effect on CPFX-resistance. Two strains had an 11-bp deletion in nfxB leading to a frame-shift following Val35; these strains also had other alterations affecting GyrA, ParC, MexZ, and MexR.
Amino acid substitutions in quinolone resistance-determining region (QRDR) of GyrA (Thr83Ile or Asp- 87Asn) were found in 60 of 120 isolates; moreover, the Thr83Ile substitution was the mutation most frequently reported previously in CPFX-resistant clinical isolates of P. aeruginosa [21,22]. Fifteen strains in Group V with an amino acid substitution in QRDR of GyrA (Thr83Ile) were resistant to 3.91 μg/mL CPFX, but 2 of 6 strains in Group IV with another amino acid substitution in QRDR of GyrA (Asp87Asn) were resistant to 3.91 μg/mL CPFX.
The only amino acid substitution found in QRDR of the ParC subunit of topoisomerase IV, another target of CPFX, was Ser87Leu; this alteration was always found along with a GyrA alteration (Groups IX-XII). These results suggested that CPFX-resistance in P. aeruginosa is mainly mediated through alterations in GyrA, and that alterations in ParC are secondary, as reported previously [23]. Alterations in ParC, MexZ, MexR, or NfxB in addition to mutation in GyrA (Thr83Ile) increased the MICs for CPFX, although the number of strains with multiple alterations was limited.
Our results revealed that multiple alterations in GyrA, ParC, MexZ, MexR, and NfxB were associated with a higher-level resistance to CPFX than that caused by a single alteration in GyrA. Although 2 strains in Group I (Table 3), in which we could not identify mutatition in QRDRs of gyrA, parC, or the efflux pump regulator genes mexZ, mexR, and nfxB, were resistant to >7.81 µg/mL CPFX, the MICs of CPFX in strains 286 and 297 decreased from 15.6 µg/mL to 1.95 or 0.98 µg/mL in the presence of PAbN, respectively (Table 2). A similar effect of PAbN on the MICs of Group III strains, which had a GyrA (Thr83Ile) alteration, but no alterations in efflux pump regulators, was observed in 9 of 14 strains. These results suggested that over expression of other active efflux systems or decreased permeability could contribute to CPFX-resistance in these strains [24].
4. Acknowledgements
We thank Mieko Yoshida, Yukitaka Itou, and Masanobu Ohtsuka at the Clinical Microbiology Laboratories at Toho University Ohashi Medical Center.
REFERENCES
- M. D. Obritsch, D. N. Fish, R. MacLaren and R. Jung, “Nosocomial Infections Due to Multidrug-Resistant Pseudomonas aeruginosa: Epidemiology and Treatment Options,” Pharmacotherapy, Vol. 25, No. 10, 2005, pp. 1353-1364. doi:10.1592/phco.2005.25.10.1353
- T. Strateva and D. Yordanov, “Pseudomonas aeruginosa— A Phenomenon of Bacterial Resistance,” Journal of Medical Microbiology, Vol. 58, No. 9, 2009, pp. 1133-1148. doi:10.1099/jmm.0.009142-0
- H. P. Hahn, “The Type-4 Pilus Is the Major VirulenceAssociated Adhesin of Pseudomonas aeruginosa,” Gene, Vol. 192, No. 1, 1997, pp. 99-108. doi:10.1016/S0378-1119(97)00116-9
- L. L. Blackwood, R. M. Stone, B. H. Iglewski and J. E. Pennington, “Evaluation of Pseudomonas aeruginosa Exotoxina and Elastase as Virulence Factors in Acute Lung Infection,” Infection and Immunity, Vol. 39, No. 1, 1983, pp. 198-201.
- R. H. George, “Pseudomonas Infections in Cystic Fibrosis,” Archives of Disease in Childhood, Vol. 62, pp. 438- 439. doi:10.1136/adc.62.5.438
- R. Mittal, S. Sharma, S. Chhibber and K. Harjai, “A Time Course Study of Production of Virulence Factors by Biofilms of Pseudomonas aeruginosa,” American Journal of Biomedical Sciences, Vol. 1, No. 3, 2009, pp. 178-187. doi:10.5099/aj090300178
- R. E. W. Hancock and D. P. Speert, “Antibiotic Resistant in Pseudomonas aeruginosa: Mechanisms and Impact on Treatment,” Drug Resistance Updates, Vol. 3, No. 4, 2000, pp. 247-255. doi:10.1054/drup.2000.0152
- E. Paramythiotou, J. C. Lucet, J. F. Timsit, D. Vanjak, C. Paugam-Burtz, J. L. Trouillet, S. Belloc, N. Kassis, A. Karabinis and A. Andremont, “Acquisition of MultidrugResistant Pseudomonas aeruginosa in Patients in Intensive Care Units: Role of Antibiotics with Antipseudomonal Activity,” Clinical Infectious Diseases, Vol. 38, No. 5, 2004, pp. 670-677. doi:10.1086/381550
- V. Aloush, S. Navon-Venezia, Y. Seigman-Igra, S. Cabili and Y. Carmeli, “Multidrug-Resistant Pseudomonas aeruginosa: Risk Factors and Clinical Impact,” Antimicrob Agents and Chemotherapy, Vol. 50, No. 1, 2006, pp. 43- 48. doi:10.1128/AAC.50.1.43-48.2006
- D. L. Paterson, “The Epidemiological Profile of Infections with Multidrug-Resistant Pseudomonas aeruginosa and Acinetobacter Species,” Clinical Infectious Diseases, Vol. 43, Suppl. 2, 2006, pp. S43-S48. doi:10.1086/504476
- S. Cabili and Y. Carmeli, V. Aloush, S. Navon-Venezia and Y. Seigman-Igra, “Multidrug-Resistant Pseudomonas aeruginosa: Risk Factors and Clinical Impact Antimicrob,” Antimicrobial Agents and Chemotherapy, Vol. 50, No. 1, 2006, pp. 43-48. doi:10.1128/AAC.50.1.43-48.2006
- E. Paramythiotou, J.-C. Lucet, J.-F. Timsit, D. Vanjak, C. Paugam-Burtz, J.-L. Trouillet, S. Belloc, N. Kassis, A. Karabinis and A. Andremont, “Acquisition of MultidrugResistant Pseudomonas aeruginosa in Patients in Intensive Care Units: Role of Antibiotics with Antipseudomonal Activity,” Clinical Infectious Diseases, Vol. 38, No. 5, 2004, pp. 670-677. doi:10.1086/381550
- I. Yoshida, T. Fujimura, Y. Itoh, M. Tachibana, M. Kaku, K. Kanemitsu, C. Takahashi, J. Shiotani, Y. Ono, H. Baba, S. Matsuo, S. Asari, K. Matsuoka, N. Kusano, M. Nose, T. Saikawa, K. Hiramatsu, S. Kohno, Y. Hirakata, N. Yamane, I. Nakasone and Y. Yamano, “Antimicrobial Susceptibility of Clinical Isolates of Aerobic Gram-Negative Bacteria in 2004,” Japanese Journal of Chemotherapy, Vol. 56, No. 5, 2008, pp. 562-579.
- N. Abe, M. Komatsu, M. Iwasaki, Y. Nagasaka, S. Fukuda and S. Matsuo, “In Vitro Activity of Antimicrobial Agents against Clinical Isolates of Pseudomonas aeruginosa,” The Japanese Journal of Antibiotics, Vol. 58, No. 5, 2005, pp. 445-451.
- A. Kanayama, M. Kida, T. Iyoda, K. Matsuzaki, S. Shibuya, M. Hasegawa, T. Saika, F. Ikeda and I. Kobayashi, “Annual Changes in Drug Susceptibility of Pseudomonas aeruginosa Isolated from Blood and Other Clinical Specimens,” Japanese Journal of Chemotherapy, Vol. 58, No. 1, 2010, pp. 7-13.
- D. C. Hooper, “Fluoroquinolone Resistance among GramPositive Cocci,” The Lancet Infectious Diseases, Vol. 2, No. 9, 2002, pp. 530-538. doi:10.1016/S1473-3099(02)00369-9
- X.-S. Pan, J. Ambler, S. Mehtar and L. M. Fisher, “Involvement of Topoisomerase IV and DNA Gyrase as Ciprofloxacin Targets in Streptococcus pneumonia,” Antimicrobial Agents and Chemotherapy, Vol. 40, No. 10, 1996, pp. 2321-2326.
- L. M. Weige, C. D. Steward and F. C. Tenover, “gyrA Mutations Associated with Fluoroquinolone Resistance in Eight Species of Enterobacteriaceae,” Antimicrobial Agents and Chemotherapy, Vol. 42, No. 10, 1998, pp. 2661- 2667.
- R. M. Salma, F. A. Dabboussi, I. O. Kassaa, R. H. Khudary and M. M. Hamze, “GyrA and ParC Mutations in Quinolone-Resistant Clinical Isolates of Pseudomonas aeruginosa from Nini Hospital in North Lebanon,” Journal of Infection and Chemotherapy, Vol. 19, No. 1, 2013, pp. 77-81.
- J. Kriengkauykiat, E. Porter, O. Lomovskaya and A. Wong-Beringer, “Use of an Efflux Pump Inhibitor to Determine the Prevalence of Efflux Pump-Mediated Fluoroquinolone Resistance and Multidrug Resistance in Pseudomonas aeruginosa,” Antimicrobial Agents and Chemotherapy, Vol. 49, No. 2, 2005, pp. 565-570. doi:10.1128/AAC.49.2.565-570.2005
- M. Nakano, T. Deguchi, T. Kawamura, M. Yasuda, M. Kimura, Y. Okano and Y. Kawada, “Mutations in the gyrA and parC Genes in Fluoroquinolone-Resistant Clinical Isolates of Pseudomonas aeruginosa,” Antimicrobial Agents and Chemotherapy, Vol. 41, No. 10, 1997, pp. 2289-2291.
- H. Mouneimné, J. Robert, V. Jarlier and E. Cambau, “Type II Topoisomerase Mutations in Ciprofloxacin-Resistant Strains of Pseudomonas aeruginosa,” Antimicrobial Agents and Chemotherapy, Vol. 43, No. 1, 1999, pp. 62-66.
- P. G. Higgins, A. C. Fluit, D. Milatovic, J. Verhoef and F.-J. Schmitz, “Mutations in GyrA, ParC, MexR and NfxB in Clinical Isolates of Pseudomonas aeruginosa,” International Journal of Antimicrobial Agents, Vol. 21, No. 5, 2003, pp. 409-413. doi:10.1016/S0924-8579(03)00009-8
- G. G. Zhanel, J. A. Karlowsky, M. H. Saunders, et al., “Development of Multiple Antibiotic-Resistant (MAR) Mutants of Pseudomonas aeruginosa after Serial Exposure to Fluoroquinolones,” Antimicrobial Agents and Chemotherapy, Vol. 39, No. 2, 1995, pp. 489-495. doi:10.1128/AAC.39.2.489
NOTES
*Corresponding author.

