Natural Resources
Vol.4 No.1(2013), Article ID:29176,6 pages DOI:10.4236/nr.2013.41009
Molecular Relationships among Different Seryian Aegilops Species (Poaceae)
![]()
1Department of Biology, Faculty of Science, Taif University, Taif, Saudi Arabia; 2Department of Zoology, Faculty of Science, Cairo University, Cairo, Egypt.
Email: *yasser92us@yahoo.com
Received December 10th, 2012; revised January 17th, 2013; accepted January 28th, 2013
Keywords: Aegilops; Molecular Relationship; Syria; Biodiversity
ABSTRACT
Aegilops has been considered a complex genus with as many as 22 species in Syria. The current study has used 585 nucleotides from 5.8S nuclear ribosomal DNA gene and internal transcribed spacer 2 for these different species. These data were aligned manually and subjected to bioinformatics manipulation in order to construct the genetic relationship among these species. Three statistical methods (maximum-parsimony-MP, maximum-likelihood-ML and neighborjoining-NJ) were used to execute the most likely relationship. The constructed genetic relationship showed homogeneinty in clustering of the species of the same plant type (A, B or C) with each other. A single NJ tree and a single ML tree were obtained with slight difference in topology within each plant type. Both trees disagreed with our previous finding in that A. searsii, speltoides and A. longissima clustered in one group and the first two species were sisters while A. caudata was out. Therefore, A. speltoides was not the oldest among them and these differences could be related to the difference in taxon sampling size. This study, however, supported our previous molecular finding and did not support the previous karyotypic study in that A. searsii was not the oldest, A. caudata was not recently originated and both A. longissima and A. speltoides were not intermediate. The molecular markers and taxon sampling size are therefore mandatory in clarifying the genetic diversity of closely related species, particularly, those which possess an economic importance like Aegilops.
1. Introduction
The Mediterranean region, especially Syria, has been considered the main origin of plant genetic rescores [1] which constitutes the pillars of sustainable development. Aegilops is considered the main compound of the current cultivated wheat [2]. It is thought that the genome of this genus was incorporated in the genetic structure of other related taxa that have diploid, tetraploid and hexaploid chromosome numbers 2n = 14, 2n = 28 and 2n = 42, respectively [2].
Aegilops is distributed in a continuous landscape and is adapted with the climatic conditions of the Mediterranean basin. Syria, Palestine and other west Asian countries are considered as one of the main countries in which the different species of the genus are found with high density [1]. The author has recorded seventeen species in a very restricted ecological area of the region.
It is very difficult to classify the species of Aegilops morphologically because its species possess very similar morphological characteristics. Many phenotypes are found naturally which have been considered as hybrids or subspecies [3]. Because of these difficulties BoubesHammoud [3] and Silai et al. [4] conducted more deep studies on Aegilops karyotypically. Other molecular studies [5-10] were recently conducted for the same purpose.
Based on the abovementioned arguments, the present study aimed to revise the molecular relationship among different Syrian Aegilops species. We used the molecular data of 5.8S nuclear ribosomal DNA gene and internal transcribed spacer 2 (ITS2) for these different species to construct such relationship.
2. Materials and Methods
The sequences of 5.8S nuclear ribosomal DNA gene and internal transcriped spacer 2 for these different species were obtained from GenBank for 22 Aegilops species. The respective sequences for Bymus tsukushiensis, Leymus cappadociocus and Leymus racemosus from the same family (Poaceae) were used as outgroup due to their close relationship. Table 1 shows the details of all these sequences including the GenBank accession numbers.
Table 1. Aegilops species and their accession numbers for the genome data used in this study.

The obtained sequences were aligned manually using DNASIS v.3 and MacClade v.4 programs. The unalienable and gap-containing sites were deleted and the unambiguous data were then concatenated so that 585 bp were left for the analyses. The aligned nucleotide sequences can be obtained from author for correspondence upon request. The tree analyses were done by NeighborJoining (NJ) method with PAUP*4.0b10 [11]. We set the bootstrapping replicates to 1000 with simple additions. For the ML analysis, the general reversible model (GTR + I + G) and parameters optimized by Modeltest 3.0 [12] were used.
3. Results and Discussion
In the present study, the dataset of 5.8S rRNA gene and ITS2 were aligned for 22 Aegilops species and three outgroups. Both ambiguousand gap-containing sites were deleted from the concatenated alignment and therfore 585 unambiguous sites were left and used to construct the relationship. The base frequencies of these datset were A = 20.80%, C = 33.10%, G = 27.90% and T = 18.20%. Of the 585 nucleotides, 505 were constant and 80 were variable. Thirty five of the variable sites were parsimony-uninformative and 45 were informative under parsimony criterion. Maximum-likelihood and neighborjoining methods gave two trees with slight differences in the topology (Figures 1 and 2). With respect to the ML tree, the best-fit model that explained the dataset was GTR + G + I. Model parameters were as follows: substitution rate matrix R(a) = 1.000; R(b) = 1.685; R(c) = 1.000; R(d) = 3.479; R(e) = 1.000, assumed proportion of invariable sites = 0.675 and gamma shape parameter (alpha) = 0.749. This ML tree was found with a negative log likelihood score −lnL = 1,620,269. With respect to the NJ tree, the used distance measure was that of TamuraNei, branch-swapping algorithm was nearest-neighborjoining (NNI) and the bootstrapping was 1000 replicates. The same algorithm and bootstrapping replicates were also applied for the maximum-parsimony method.
The studied species are belonging to three Aegilops clusters A, B and C. Clusters A and C constitue sections Siptosis and Vertebrata, respectively while cluster B constitues three sections (Cylindropyrum, Comopyrum and Polycides) [13]. The ML tree (Figure 1) and the NL tree (Figure 2) agreed in clustering the same plant type with each other. Both tree showed that Aegilops species of cluster A have been grouped together. Similarly, the species of cluster B have been grouped together and those of type C also. Using genome analysis, the tree topologies in the present study showed approximately general agreement with several recent molecular studies [10,14-17] for the species that have been encompassed by each cluster.
The cluster A species showed similar genome type as they have S genome (Table 2) and all of them possess similar chromosomal (7) and microsatellite (2) numbers. The difference in their positions in the trees could be attributed to the difference in their ecological habitats or to the type of soil in which they have been cultivated. This result can be supported by that the cluster A contains 6 species five of which represent Sitopsis section. This section contains five species: Aegilops bicornis (SbSb, 2n = 2x = 14), Ae. longissima (SlSl, 2n = 2x = 14), Ae. sharonensis (SshSsh, 2n = 2x = 14), Ae. searsii, (SsSs, 2n = 2x = 14) and Ae. speltoides (SS, 2n = 2x = 14) [18]. Previous reports on cytogenetic and genetic investigations indicated that Aegilops genomes from this section are closely related [19-22]. Our study therefore approved the homogeniety of this section on molecular basis. Contradictions of the relationship among the species of Sitopsis section still be found as our results disagreed with previous recent studies [9,10] regarding the position of Ae. searsii, Ae. longissima and Ae. spiltiodes. This discripancy could be attributed to that the molecular data used by the autours were not enough to resolve this contradiction. Ae. tsuaschii was found within this cluster in
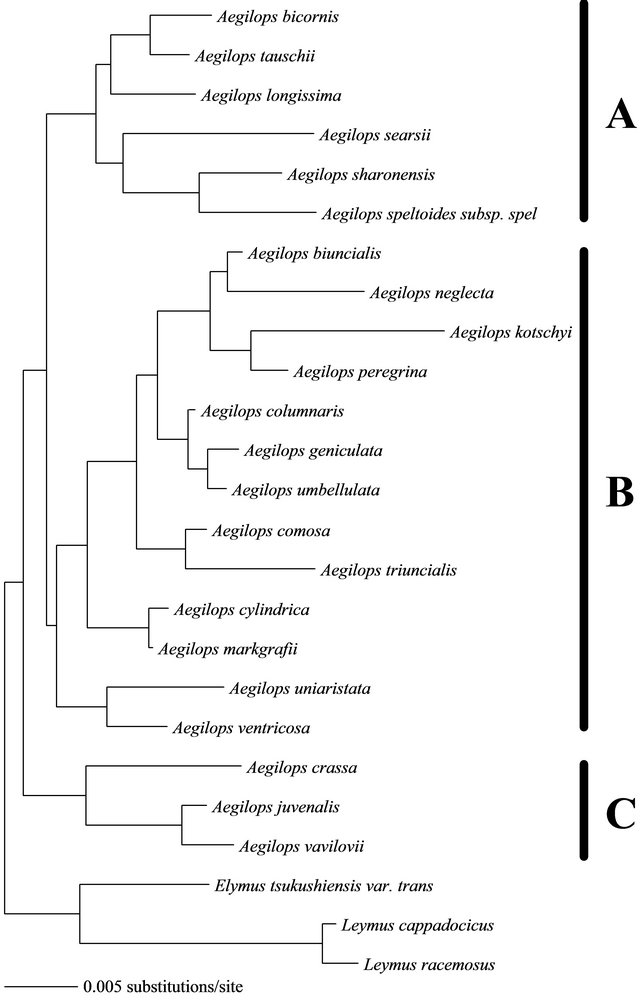
Figure 1. Neighbor-joining relationship between species belonging to the genus Aegilops, determined by aligning 585 sequences of the 5.8S gene and ITS2. Elymus and Leymus were used as outgroup.
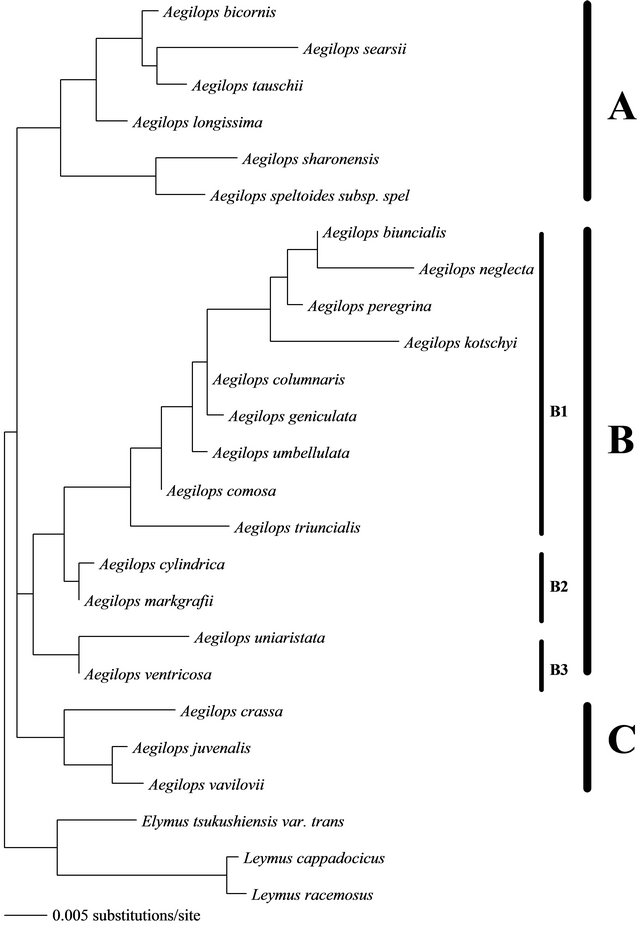
Figure 2. Maximum-likelihood relationship between species belonging to the genus Aegilops, determined by aligning 585 sequences of the 5.8S gene and ITS2. Elymus and Leymus were used as outgroup.
Table 2. Plant type, chromosomal number, genome constitution and origin of wild Aegilops species used in this study.
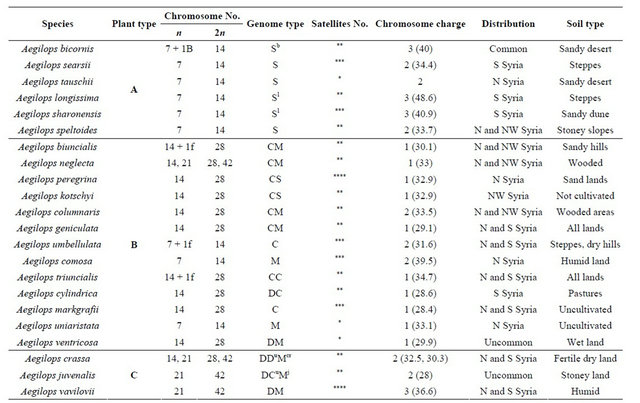
*1 pair equipped with satellites; **2 pair equipped with satellites; ***1 pair equipped with large satellite 1 pair with small satellite; ****3 pair equipped with satellites. N = north; S = south; W = west.
spite of being belong to section Vertebrata. The molecular analysis of Petersen et al. [23] showed section Sitopsis as the sister to Ae. tauschii (and the D genome of T. aestivum).
The cluster B showed a group of Aegilops species which possess M, D, C and S genomes or mixures of these types. The karyotypic number of Syrian taxa within this cluster was 14 chromosomes. Because of the complicated genome type and the difference in both number of microsatellites (2 or 3) and habitat, both trees showed great difference within this cluster. Konstantinos and Bebeli [17] observed Ae. triuncialis (genome UC) grouped in the same subgroup with Ae. markgrafii (C), which is its progenitor male parent. These evidences have been observed in our study by clustering of both species in the same group. In section B in which Aegilops contains Ae. ovata = Ae. geniculata, Ae. neglecta, Ae. umbellulata, Ae.triuncialis, Ae. Kotschyi, Alnaddaf et al. [10] found these species have identical restriction profiles. Konstantinos and Bebeli [17] noticed that Ae. kotschyi-SU, Ae. peregrina-SU) grouped closer to the male parent (Ae. umbellulata-U) than to Ae. searsii-S. This is agree with the study of Alnaddaf et al. [10] where Ae. kotschyi, Ae. umbellulata have identical restriction profiles.
Aegilops species of cluster C showed identical topology in both ML and NJ trees. The three species of this cluster poccess karyotypic number of 42 chromosome, DCM genome and 2 or 3 microsatellites. Because they did not show clear differences in these data, their topology was fixed in both trees. The section Vertebrata consists of five species which are Aegilops tauschii, Aegilops ventricosa, Aegilops crassa, Aegilops juvenalis and Aegilops vavilovii. The chromosomic numbers for these species are commonly 42 chromosomes with exceptions for Aegilops tauschii (2n = 14 chromosome) and Aegilops ventricosa (28 chromosome). Aegilops tauschii clustered with section Siptosis while Aegilops ventricosa clustered with cluster B. Molecular evidence for the sister relationship of Aegilops tauschii to section Siptosis [23] and clustering of Aegilops ventricosa with taxa of cluster B were possible.
4. Conclusion
The constructed genetic relationship showed homogeneinty in clustering of the species of the same plant type (either A, B or C) with each other. Using genome analysis, the constructed trees showed approximately general topological agreement with several molecular investigations for the species that have been encompassed by each cluster. The present study raised the significance of the size of both molecular data and taxon sampling in clarifying the genetic diversity of closely related species, particularly, those which possess an economic importance like Aegilops.
REFERENCES
- P. Mouterde, “Nouvelle Flore du Liban et de la Syrie,” Vol. 1, No. l, Impiemerie Catholique, Beyrouth, 1966.
- R. Riley and V. Chapman, “Evidence on the Origin of the B-Genome of Wheat,” Journal of Heredity, Vol. 49, No. 3, 1958, pp. 91-98.
- S. Boubes-Hammoud, “Contribution de l’etude Caryosystematique des Especes d’Aegilops et de Leurs Raports avec les Bles Cultives,” Doct. d’Etat, Université Louis Pasteur, de Strasbourg, 1986.
- A. Sliai, S. Boubes-Hammoud and G. H. Ayash, “Contribution l’etude de la Diversite Chromosomique d’Aegilops en Syrie, ”Bassel Al Assad Journal, Vol. 7, 1999, pp. 135-156.
- H. Meimberg, K. J. Rice, N. F. Milan, C. C. Njoku, J. K. Mckay, “Multiple Origins Promote the Ecological Amplitude of Allopolyploid Aegilops (Poaceae), ”American Journal of Botany, Vol. 96, No. 7, 2009, pp. 1262-1273. doi:10.3732/ajb.0800345
- N. Arrigo, F. Felber, C. Parisod, S. Buerki, N. Alvarez, J. David and R. Guadagnuolo, “Origin and Expansion of the Allotetraploid Aegilops geniculata, a Wild Relative of Wheat,” New Phytologist, Vol. 187, No. 4, 2010, pp. 1170-1180. doi:10.1111/j.1469-8137.2010.03328.x
- A. I. Sepsi, “Molecular Cytogenetic Characterisation of a Leaf-Rust Resistant Wheat-Thinopyrum ponticum Partial Amphiploid,” Doctoral Dissertation, Eotvos Lorand University, Budapest, 2010.
- K. G. Thomas and P. J. Bebeli, “Genetic Diversity of Greek Aegilops Species Using Different Types of Nuclear Genome Markers,” Molecular Phylogenetics and Evolution, Vol. 56, No. 3, 2010, pp. 951-961. doi:10.1016/j.ympev.2010.04.041
- A. M. Sliai and S. A. M. Amer, “Contribution of Chloroplast DNA in the Biodiversity of Some Aegilops Species,” African Journal of Biotechnology, Vol. 10, No. 12, 2011, pp. 2212-2215.
- L. M. Alnaddaf, M. Y. Moualla, N. Haider, “The Genetic Relationships among Aegilops L. and Triticum L. Species, ”Asian Journal of Agricultural Sciences, Vol. 4, No. 5, 2012, pp. 352-367.
- D. L. Swofford, “PAUP*. Phylogenetic Analysis Using Parsimony and Other Methods,” Version 4, Sinauer, Sunderland, 2003.
- D. Posada and K. A. Crandall, “MODELTEST: Testing the Model of DNA Substitution,” Bioinformatics, Vol. 14, No. 9, 1998, pp. 817-818. doi:10.1093/bioinformatics/14.9.817
- J. R. Witcombe, “A Guide to the Species of Aegilops L. Their Taxonomy, Morphology and Distribution,” IBPGR Secretariat, Rome, 1983.
- T. Terachi and K. Tsunewaki, “The Molecular Basis of Genetic Diversity among Cytoplasms of Triticum and Aegilops. V. Mitochondrial Genome Diversity among Aegilops Species Having Identical Chloroplast Genomes,” Theoretical Applied Genetics, Vol. 73, No. 2, 1986, pp. 175-181. doi:10.1007/BF00289272
- R. A. Queen, B. M. Gribbon, C. James, P. Jack and A. J. Flavell, “Retrotransposon-Based Molecular Markers for Linkage and Genetic Diversity Analysis in Wheat,” Molecular Genetics and Genomics, Vol. 271, No. 1, 2004, pp. 91-97. doi:10.1007/s00438-003-0960-x
- E. Al-Ahmar, N. Haider and H. Azzam, “Genetic Relationships among Aegilops L. Species Using DNA Molecular Markers,” General Commission for Scientific Agricultural Research, Gene Bank Division, Faculty of Agriculture, Damascus University, Syria, 2010.
- G. T. Konstantinos and P. J. Bebeli, “Genetic Diversity of Greek Aegilops Species Using Different Types of Nuclear Genome Markers,” Molecular Phylogenetics and Evolution, Vol. 56, No. 3, 2010, pp. 951-961. doi:10.1016/j.ympev.2010.04.041
- M. W. van Slageren, “Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae),” Wageningen Agricultural University Press, Wageningen, 1994.
- H. Kihara, “Consideration on the Evolution and Distribution of Aegilops Species Based on the Analyser-Method,” Cytologia, Vol. 19, No. 4, 1954, pp. 336-357. doi:10.1508/cytologia.19.336
- Y. Yen and G. Kimber, “Genomic Relationships of Triticum searsii to Other S-Genome Diploid Triticum Species,” Genome, Vol. 33, No. 3, 1990, pp. 369-373. doi:10.1139/g90-056
- T. Sasanuma, K. Chabane, T. R. Endo and J. Valkoun, “Characterization of Genetic Variation in and Phylogenetic Relationships among Diploid Aegilops Species by AFLP: Incongruity of Chloroplast and Nuclear Data,” Theoretical Applied Genetics, Vol. 108, No. 4, 2004, pp. 612-618. doi:10.1007/s00122-003-1485-8
- E. A. Salina, I. G. Adonina, T. Y. Vatolina and N. Kurata, “A Comparative Analysis of the Composition and Organization of Two Subtelomeric Repeat Families in Aegilops speltoides Tausch and Related Species, ”Genetica, Vol. 122, No. 3, 2004, pp. 227-237. doi:10.1007/s10709-004-5602-7
- G. Petersen, O. Seberg, M. Yde and K. Berthelsen, “Phylogenetic Relationships of Triticum and Aegilops and Evidence for the Origin of the A, B, and D Genomes of Common Wheat (Triticum aestivum),” Molecular Phylogenetics and Evolution, Vol. 39, No. 1, 2006, pp. 70-82. doi:10.1016/j.ympev.2006.01.023
NOTES
*Corresponding author.

