Advances in Infectious Diseases
Vol.2 No.4(2012), Article ID:25361,8 pages DOI:10.4236/aid.2012.24022
Characterization of Virulence Factors in Enteroaggregative and Atypical Enteropathogenic Escherichia coli Strains Isolated from Children with Diarrhea
![]()
1Research Group of Endemic and Parasitic Diseases, CEUMA University, São Luis, Brazil; 2Federal Institute of Maranhão, São Luis, Brazil; 3Health and Biological Sciences Center, Federal University of Maranhão, São Luís, Brazil.
Email: *figueiredo.patricia@gmail.com
Received August 17th, 2012; revised September 18th, 2012; accepted October 2nd, 2012
Keywords: E. coli; Diarrhea; Virulence Factors
ABSTRACT
Enteroaggregative (EAEC) and atypical enteropathogenic (EPEC) Escherichia coli are important bacterial etiologic agents causing diarrhea among children. The aim of the present study was to examine the impact of virulence factors predisposes to diarrhea. In this study some virulence properties were examined on 11 EAEC and 8 EPEC strains identified by Polymerase Chain Reaction (PCR), isolated from stool samples of children were analyzed genotypically and phenoltypically for the prevalence of virulence factors. The most frequently detected factor was resistance to serum (94%), followed by curli fimbriae (78%), biofilm production (73%), and gene coding for Extended-Spectrum Beta-Lactamase (ESBL) (68%). EPEC isolates showed at least three of the evaluated properties, while EAEC isolates showed at least two. The prevalence of these virulence factors between the two strains showed no statistical difference. This study showed the heterogeneity of the virulence profile of the isolates of EAEC and atypical EPEC strains and suggests that this diversity may influence in the disease severity.
1. Introduction
A survey by the World Health Organization (WHO), from 2002 to 2003, found that 73% of the 10.6 million annual deaths of children under five years old were related to five causes. Diarrheal disease is the second most common (18%), after pneumonia (19%), which evidences the importance of this illness to public health, since it causes deep impacts on the infant morbidity and mortality rates, especially in the less developed regions in the world [1].
Despite recent studies showing that there was a significant reduction in infant mortality rates due to diarrhea from 4.6 million per year to approximately 2.5 million per year worldwide, such values are still considered high and the morbidity rates remain as high as 30 years ago [2] .
Several pathogens such as bacteria, viruses and intestinal parasites are related to diarrhea in humans. Among them, Escherichia coli are the most important etiologic agent in childhood and represent the biggest public health problem in developing countries [3].
Diarrheagenic strains of E. coli (DEC) are classified in 6 subtypes, with differences among themselves and between them and non-pathogenic microorganisms, mem bers of the normal human flora, according to their virulence properties [4].
Among these properties, we highlight those which enable the bacterium to recognize and colonize the surface of the host’s cells (fimbrial and non-fimbrial adhesins); production of toxins and hemolysins; expression of iron uptake systems (siderophores); and resistance to antibiotics and to the immune system.
Currently, the two most important diarrhea agents that affect children under five years old among the isolates of E. coli, both in developed and developing countries, are enteroaggregative (EAEC) and enteropathogenic (EPEC) [5].
EPEC causes a histopathological lesion known as “attaching and effacing (A/E)”. Strains of A/E genotype, which do not possess the plasmid Enteropathogenic E.
coli Adherence Factor (EAF), are classified as atypical EPEC [6]. Enteroaggregative E. coli (EAEC) are defined as E. coli strains that adhere in vitro to HEp-2 cells in a pattern known as autoaggregative [7]. Some studies have suggested targets for molecular characterization of EAEC, how genes setBA, aaiA, astA and aat. However, it was observed that some of these markers are not unique to this categoria [7,8]. Genes afa, dra and daa, encoding a family of adhesins have been used to characterize DAEC, though they may be present in no pathogenic E. coli. Therefore, due to these peculiarities, it is recommended to include tests of adhesions in cultured Hep-2 or HeLa method as the gold standard for defining categories and EAEC and DAEC [8,9].
Although a large number of virulence factors have been identified in EAEC and EPEC, not even a single factor seems to be present in all the pathogenic strains. Despite its high prevalence, the enteropathogenicity in these two groups is not completely established yet [8,9]. This happens due to: few systematic studies on the virulence and heterogeneity properties of these characteristics.
This study aimed to investigate the virulence factors prevalence, some of which have been previously investigated in EAEC and EPEC, isolated from children under 5 years with diarrhea.
2. Materials and Methods
Sampling: The studied sample was comprised by 162 children divided in two groups (case and control), with 81 children each other. Acute diarrhea was considered for 4 or more liquid voiding with evolution until 7 days.
Female and male children under 5 years old with a diarrheic episode 72 hours before the survey, with rising frequency of voidings and/or reduction in the fecal consistency, were considered belonging to the case group. The control group was formed by children in the same age group and had not presented gastrointestinal disturbances 72 hours before the survey. This study was ap proved by the Committee on Ethics in Research of UNICEUMA (License No. 00559/09).
Collection of samples: For each child one fecal sample was collected which was stored into a sterile plastic container, and one fecal sewab transported thorough Cary-Blair. The fecal samples were transported through an isothermal container to the Medical Microbiology Laboratory of UNICEUMA. Further, the fecal samples were examined and the remaining Cary-blair sample was used for bacteriological culture and isolation.
Fecal culture: For isolation of enteropathogenic bacteria, the Cary Blair fecal samples were added into the MacConkey agar, Salmonella-Shigella agar, and the Selenito-Cystine preparation. The incubation was accomplished at 37˚C for 24 hours. The suspicious colonies of E. coli got under preliminary identification in the Enterokit B (Probac) for biochemical identification and in some confirmed cases by the VITEK automatic system.
Molecular identification of pathogenic categories of Escherichia coli and virulence markers: DNA extraction was obtained from E. coli cultures grown during the night they were resuspended in sterile distilled deionized water and boiled for 5 to 10 minutes. The survey included the detection of genes or DNA conserved regions for analysis of the diarrheagenic E. coli strain type: eae (for EPEC and EHEC), stx 1 and stx 2 (for EHEC), elt and est (for ETEC), ipaH (for EIEC) and aggR (for EAEC) [10].
The colonies eae+ and stx– were researched for the presence of bfpA gene (which encodes the pilin of the Bundle Forming Pilus fimbriae) to discriminate sub-groups of EPEC (typical, bfpA carriers and atypical, lacking this gene).
It was also included in this study the investigation of other virulence markers, which encode: extended-spectrum beta-lactamase (ESBL), cytolethal distending toxin (cdt), enterohemolysin (hlyAehec), and the siderophore yersiniabactin (irp), homologue adhesion irgA (iha) and adherence factor of enterohemorrhagic Escherichia coli (EHEC). The primers used are listed in Table 1.
Detection of hemolytic activity: To search for the hemolysin on blood agar, the Escherichia coli strains were grown in Brain and Heart Infusion (BHI) at 37˚C for 24 h in stationary culture. After bacterial growth, strains were grown on the surface an in depth in agar Muller Hinton, with 5% of sheep erythrocytes washed with Phosphate Buffered Saline (PBS) at pH 7.4 [11].
Identification of type I fimbriae: Performed by growth in BHI for 24 h at 37˚C. Right after, a portion of each culture was centrifuged three times (12,000 rpm for 15 minutes), always discarding the supernatant and putting PBS pH 7.4. About 500 µl of the isolates were mixed, on glass slides, with 500 µl of blood washed three times with PBS, with and without addition of mannose. The samples that produced hemagglutination only in the absence of mannose were considered positive for type I fimbriae [12].
Identification of curli fimbriae: the culture was grown at 37˚C in Luria Bertani medium (LB), with pH 8, for 24 h. Afterwards, the culture was sown on LB plates (without salt) with 0.004% of Congo Red and 0.002% of Bright Blue. The plates were incubated for 24 to 48 hours at room temperature. Red or pink colonies indicate curliproducing bacteria [13].
Biofilm production: Performed by the cultures in BHI,
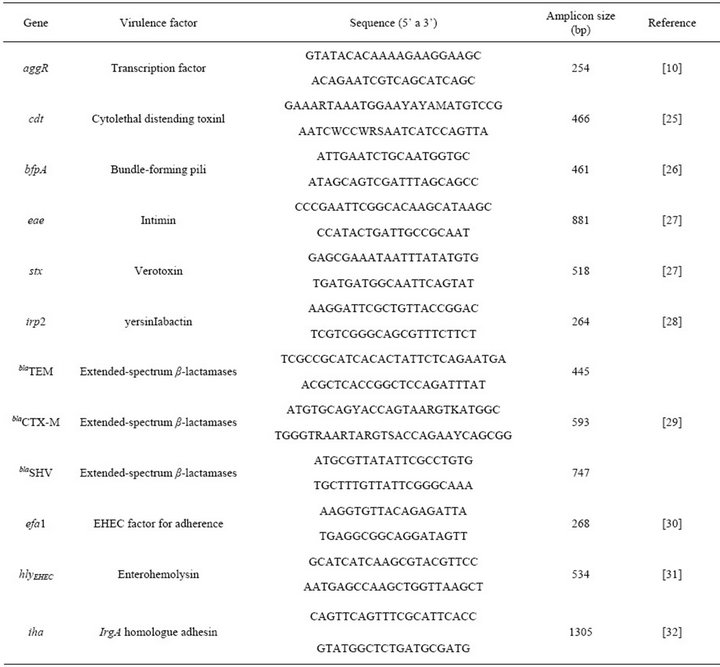
Table 1. Primers used to identify different Escherichia coli virulence factors.
24 h after inoculation at 37˚C. Right after, a portion of each culture was diluted to 1:40 in BHI sterile medium. Each sample, already standardized, was added to tissue culture plates with 96 wells deep U-shaped, in triplicate, along with controls, 20 µL/well. Once completed, the plates were incubated at 37˚C for 24 h in microbiological oven. After the incubation, the plates were washed three times with PBS buffer with pH 7.4 and let to dry at room temperature. On the dry plates was added 200 µL of crystal violet in each well for 15 minutes at room temperature. Then, each well was washed three times with sterile distilled water and dried at room temperature. The plates were stained with crystal Violet and submitted to spectrophotometry with a 470 ηm filter to measure the respective absorbance for each well. Basing on the optical density (D.O.i) produced by the isolates, and taking as basis the negative control (D.O.c), isolates classified in the following categories [14]: Non-Producer: D.O.i < D.O.c; Poor Producer: D.O.c < D.O.i ≤ (2× D.O.c); Moderate Producer: (2× D.O.c) < D.O.i ≤ (4× D.O.c); Strong Producer: (4× D.O.c) < D.O.i.
Serum Resistance: The isolates were incubated in BHI at 37˚C for 24 hours. 500 µl of the culture was incubated in 4.5 ml of sterile BHI for 90 minutes at 37˚C. Then, the isolates were centrifuged for 15 minutes, to resuspend in PBS later. 120 µl of the resuspended isolates were incubated with 60 µl of normal human serum. The absorbance was analyzed at 0, 30, 60, 90 and 120 minutes. Three types of normal human serum were used for the test: 1) Inactivated serum at 56˚C for 30 minutes; 2) Serum treated with 10mM of EDTA; and 3) Normal Human Serum (NHS). The isolates which had reduced the absorbance in 50% were considered sensitive; in 50 to 90% of the initial absorbance, were considered intermediate resistant; and above 90, as resistant [15].
Statistical analysis: it was performed using ANOVA and multiple logistic regression, where p < 0.05 was considered statistically significant.
3. Results
The distribution of the virulence factors is shown in Table 2. All examined virulence factors were more prevalent in the EPEC group, except for the presence of blaTEM gene and serum resistance. Besides, all the isolates from this group show at least 3 virulence factors. The most prevalent property among the 11 EAEC isolates was serum resistance (100%), followed by curli fimbriae (63.3%), biofilm production (63.3%) and blaTEM gene (63.3%); while curli fimbriae (87.5%) and serum resistance (87.5%) were more prevalent in the EPEC group. However, there was no association between the presence of any of these factors and the two evaluated groups (p > 0.05).
Table 3 shows the combination of several virulence factors, according to the E. coli strain. The number of isolates with 3 or more factors was higher in the EPEC group (100%) than in the EAEC group (82%). However, the most complex combination of factors was found in the second group.
Notably, the isolates with hemolytic activity were more homogeneous and had a larger number of associated properties. The frequency of the virulence factors of these isolates was: 100% of the isolates showed biofilm production and serum resistance, 75% curli fimbriae and blaTEM.
Interestingly, only 2 isolates (10.5%) had the profile with blaTEM, biofilm production, curli fimbriae, serum resistance and hemolysis; and other 2 with blaCTX-M, biofilm, curli and serum resistance.
3.1. Serum Resistance Assays
The survival of both enteroaggregative and enteropathogenic E. coli strains were examined: in normal human serum (NHS); under conditions which inactivated the classical and the alternative pathways of the complement system; and in inactivated serum. Over 70% of the isolates survived to two hours of incubation in normal serum and about 21% decreased significantly after 30 minutes of incubation (Figure 1).
On the other hand, when the isolates were incubated in heat-inactivated or EDTA-treated serum, they did not decrease. The visualized growth was similar between these types of serum (p > 0.05), but different when compared to normal serum (p > 0.05), which evidences the importance of the alternative and the classical pathways, indicating that the intact complement system is essential for the bactericidal activity. It was not verified any association between the serum resistance and any other factors mentioned in this study (p > 0.05).
3.2. Biofilm Production Assays
In this study, about 73% of the isolates were biofilm producers (Figure 2). 60% of the poor producers were EAEC, while 56% of the moderate and strong producers were EPEC. However, there was no difference between the strains (p > 0.05). Among the EPEC isolates, 50% were strong producers and 12.5% were moderate producers.

Table 2. Virulence factors in EAEC and EPEC isolates.

Table 3. Combinations of putative virulence factors among EAEC and EPEC strains.
In relation to the difference virulence factors determined by producers versus non-producers, curli fimbriae was the most prevalent factor in the first group (85%), but there was no association between this or other factors with biofilm (p > 0.05).
4. Discussion
Several studies have reported the prevalent heterogeneity of different virulence factors among EAEC and atypical EPEC isolates, due to which the pathogenicity mechanism of the infection in these two strains is not completely understood yet. This study determined the distribution of 16 virulence factors in EAEC and atypical EPEC isolates. There was no association between these strains specific properties. There are few studies evaluating the prevalence of these properties in the isolates we studied, especially in São Luís and Macapá.
None of the studied properties was present in all the isolates, however this study identified serum resistancecurli fimbriae and ESBL as the most frequent among the isolates. All the evaluated properties were more prevalent in EPEC than in EAEC. The most present factor in EPEC was curli fimbriae (100%), biofilm production and serum resistance (87.5% each); while in EAEC, the most frequent was serum resistance (100%), biofilm, curli fimbriae and blaTEM (63.3% each). However, these factors were not specific for EAEC and EPEC.
The number of isolates carrying 4 or more virulence factors was expressive in all strains (63%). Some authors suggested that only strains presenting at least 2 virulence factors should be considered as potential pathogens [16].
Compared to other studies performed with EAEC and atypical EPEC isolated from children in developing countries, our data showed significant differences and a few similarities. Recent researches report high variation in the presence of yersiniabactin (22% to 60%), moderate prevalence of cdt (8% to 23%), efal (20%) and iha (20%) [8,17,18]. Most of them were not found in this study.
On the other hand, ESBL was found in most of the
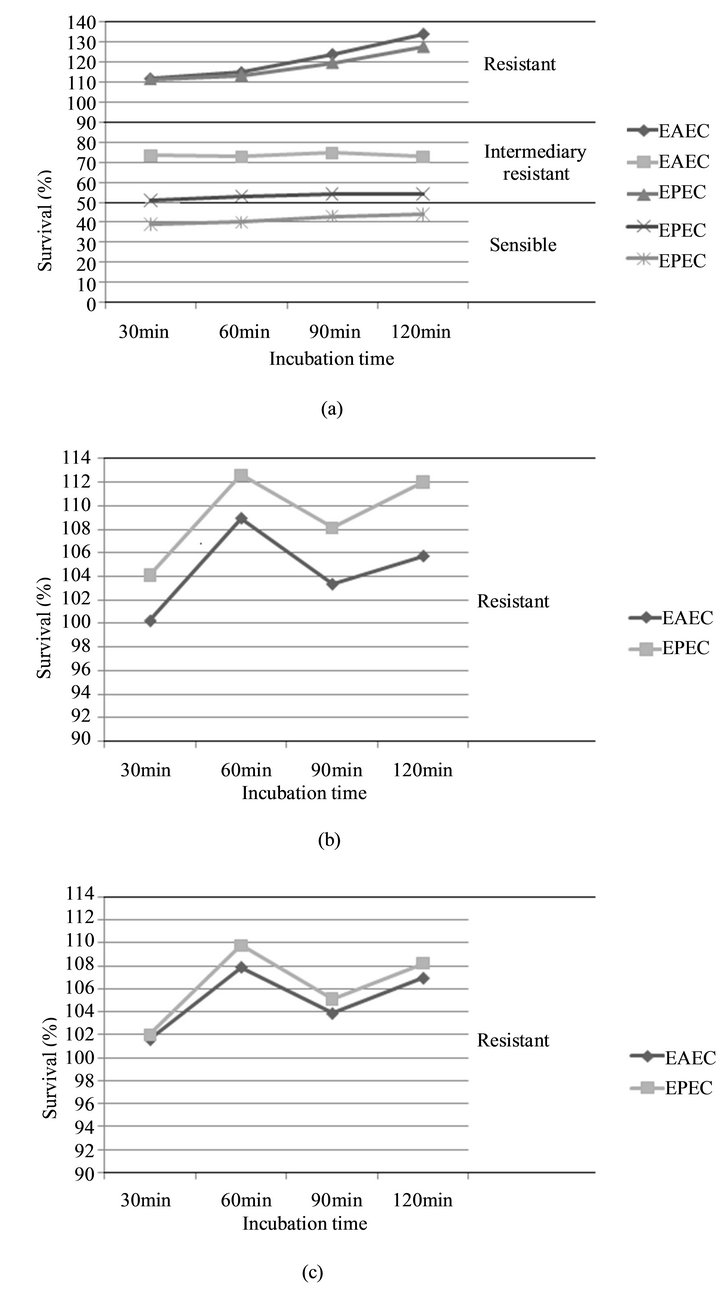
Figure 1. Survival of EAEC and EPEC strains in adult human sera. E. coli strains grown overnight (postexponential) were subjected to serum bactericidal assays using adult human sera either untreated. (a) EDTA-treated serum; and (b) heat-inactivated; (c) The results are presented as means of three category (sensible, intermediary and sensible) in individual experiments and are expressed as relative survival with the survival of E. coli at 0 min being taken as 100%.

Figure 2. Biofilm formation (BF) of EAEC and EPEC strains. The mean BF index was higher for EPEC than for EAEC strains, but there was not difference among two strains ANOVA (p > 0.05).
isolates: in our study, this factor was detected in 13 (68%). This result is similar to the other studies data, which show a big number of Escherichia coli samples with blaTEM and blaCTX-M genes [19-21]. One of the most important properties in pathogenic microorganisms is the resistance to bactericidal activity. To survive, microorganisms must avoid serum bactericidal activity to establish infection. The complement system is the first line of defense and the greatest weapon of innate immunity. E. coli has shown strategies to regulate several steps of the complement system cascade, such as ability to reduce or delay C3b deposition through the outer protein of the membrane A [22-24]. Interestingly, we demonstrated that only one isolate in this research was sensitive to the bactericidal activity, although growth in normal human serum (NHS) has been smaller than in EDTAtreated serum and heat-inactivated serum. This data suggests that the survival pattern of both EAEC and EPEC strains is the same and that the classical and the alterna tive pathways of the complement system contribute to control the isolates growth.
In conclusion, our data has clearly indicated that both EAEC and atypical EPEC present a heterogeneous combination of virulence factors. Thus, it’s not possible to associate any of these properties or combinations with the two groups.
5. Acknowledgements
Fundação de Amparo à Pesquisa e ao Desenvolvimento Científico e Tecnológico do Maranhão (FAPEMA) (APPUniversal-01176/09 and APP-Universal-011 80/09).
REFERENCES
- J. Bryce, C. Boschi-Pinto K. Shibuya and R. E. Black, “WHO Estimates of the Causes of Death in Children,” Lancet, Vol. 365, No. 9465, 2005, pp. 1147-1152. doi:10.1016/S0140-6736(05)71877-8
- M. Kosek, C. Bern and R. L. Guerrant, “The Global Burden of Diarrhoeal Disease, as Estimated from Studies Published between 1992 and 2000,” Bulletin of the World Health Organization, Vol. 81, No. 3 2003, pp. 197-204.
- S. J. Moyo, S. Y. Maselle, M. I. Matee, N Langeland and H. Mylvaganam, “Identification of Diarrheagenic Escherichia coli Isolated from Infants and Children in Dar Es Salaam, Tanzania,” BMC Infectious Diseases, Vol. 7, 2007, p. 92. doi:10.1186/1471-2334-7-92
- R. Naveen and E. Mathai, “Some Virulence Characteristics of Uropathogenic Escherichia coli in Different Patient Groups,” Indian Journal of Medical Research, Vol. 122, No. 2, 2005, pp. 143-147.
- V. Bueris, et al., “Detection of Diarrheagenic Escherichia coli from Children with and without Diarrhea in Salvador, Bahia, Brazil,” Memórias do Instituto Oswaldo Cruz, Vol. 102, No. 7, 2007, pp. 839-844. doi:10.1590/S0074-02762007005000116
- L. R. Trabulsi, R. Keller and T. A. Tardelli Gomes, “Typical and atypical enteropathogenic Escherichia coli,” Emerging Infectious Diseases, Vol. 8, No. 5, 2002, pp. 508-513. doi:10.3201/eid0805.010385
- J. Flores and P. C. Okhuysen, “Enteroaggregative Escherichia coli Infection,” Current Opinion in Gastroenterology, Vol. 25, No. 1, 2009, pp. 8-11. doi:10.1097/MOG.0b013e32831dac5e
- E. Mendez-Arancibia, M. Vargas, S. Soto, J. Ruiz, E. Kahigwa, D. Schellenberg, H. Urassa, J. Gascon and J. Vila, “Prevalence of Different Virulence Factors and Biofilm Production in Enteroaggregative Escherichia coli Isolates Causing Diarrhea in Children in Ifakara (Tanzania),” The American Journal of Tropical Medicine and Hygiene, Vol. 78, No. 6, 2008, pp. 985-989.
- J. H. Kim, J. C. Kim, Y. A. Choo, H. C. Jang, Y. H. Choi, J. K. Chung, S. H. Cho, M. S. Park and B. K. Lee, “Detection of Cytolethal Distending Toxin and Other Virulence Characteristics of Enteropathogenic Escherichia coli Isolates from Diarrheal Patients in Republic of Korea,” Journal of Microbiology and Biotechnology, Vol. 19, No. 5, 2009, pp. 525-529. doi:10.4014/jmb.0801.033
- C. Toma, Y. Lu, N. Higa, N. Nakasone, I, Chinen, A. Baschkier, M. Rivas and M. Iwanaga, “Multiplex PCR Assay for Identification of Human Diarrheagenic Escherichia coli,” Journal of Clinical Microbiology, Vol. 41, No. 6, 2003, pp. 2669-2671. doi:10.1128/JCM.41.6.2669-2671.2003
- L. Beutin, et al., “Enterohemolysin, a New Type of Hemolysin Produced by Some Strains of Enteropathogenic E. coli (EPEC),” Zentralbl Bakteriol, Vol. 267, No. 4, 1988, pp. 576-588.
- S. Clegg and D. C. Old, “Fimbriae of Escherichia coli K-12 Strain AW405 and Related Bacteria,” Journal of Bacteriology, Vol. 137, No. 2, 1979, pp. 1008-1012.
- S. Da Re and J. M. Ghigo, “A CsgD-Independent Pathway for Cellulose Production and Biofilm Formation in Escherichia coli,” Journal of Bacteriology, Vol. 188, No. 8, 2006, pp. 3073-3087. doi:10.1128/JB.188.8.3073-3087.2006
- S. Stepanovic, I. Cirkovic, L. Ranin and M. Svabic-Vlahovic, “Biofilm Formation by Salmonella spp. and Listeria monocytogenes on Plastic Surface,” Letters in Applied Microbiology, Vol. 38, No. 5, 2004, pp. 428-432. doi:10.1111/j.1472-765X.2004.01513.x
- S. Pelkonen and J. A. Finne, “A Rapid Turbidimetric Assay for the Study of Serum Sensitivity of Escherichia coli,” FEMS Microbiology Letters, Vol. 42, No. 1, 1987, pp. 53-57. doi:10.1111/j.1574-6968.1987.tb02298.x
- I. N. Okeke, A. Lamikanra, F. Dubovsky, J. B. Kaper and J. P. Nataro, “Heterogeneous Virulence of Enteroaggregative Escherichia coli Strains Isolated from Children in Southwest Nigeria,” Journal of Infectious Diseases, Vol. 181, No. 1, 2000, pp. 252-260. doi:10.1086/315204
- D. B. Huang, J. A. Mohamed, J. P. Nataro, H. L. DuPont, Z. D. Jiang and P. C. Okhuysen, “Virulence Characteristics and the Molecular Epidemiology of Enteroaggregative Escherichia coli Isolates from Travelers to Developing Countries,” Journal of Medical Microbiology, Vol. 56, 2007, pp. 1386-1392. doi:10.1099/jmm.0.47161-0
- S. M. Tennant, M. Tauschek, K. Azzopardi, A. Bigham, V. Bennett-Wood, E. L. Hartland, W. Qi, T. S. Whittam and R. M. Robins-Browne, “Characterisation of Atypical Enteropathogenic E. coli Strains of Clinical Origin,” BMC Microbiology, Vol. 9, 2009, p. 117. doi:10.1186/1471-2180-9-117
- T. M. Coque, F. Baquero and R. Canton “Increasing Prevalence of ESBL-Producing Enterobacteriaceae in Europe,” Eurosurveillance, Vol. 13, No. 47, 2008, p. 19051.
- C. F. Oliveira, N. L. Dal Forno, I. A. Alves, J. A. Horta, A. Rieger and S. H. Alves, “Prevalence of the TEM, SHV and CTX-M Families of Extended-Spectrum Beta-Lactamases in Escherichia coli and Klebsiella spp at the University Hospital of Santa Maria, State of Rio Grande do Sul,” Revista da Sociedade Brasileira de Medicina Tropical, Vol. 42, No. 5, 2009. pp. 556-560. doi:10.1590/S0037-86822009000500014
- A. Lago, S. R. Fuentefria and D. B. Fuentefria, “ESBLProducing Enterobacteria in Passo Fundo, State of Rio Grande do Sul, Brazil,” Revista da Sociedade Brasileira de Medicina Tropical, Vol. 43, No. 4, 2010, pp. 430-434. doi:10.1590/S0037-86822010000400019
- N. V. Prasadarao, A. M. Blom, B. O. Villoutreix and L. C. Linsangan, “A Novel Interaction of Outer Membrane Protein A with C4b Binding Protein Mediates Serum-Resistance of Escherichia coli K1,” The Journal of Immunology, Vol. 169, No. 11, 2002, pp. 6352-6360.
- D. G. Wooster, R. Maruvada, A. M. Blom and N. V. Prasadarao, “Logarithmic Phase Escherichia coli K1 Efficiently Avoids Serum Killing by Promoting C4bp-Mediated C3b and C4b Degradation,” Immunology, Vol. 117, No. 4, 2006, pp. 482-493. doi:10.1111/j.1365-2567.2006.02323.x
- R. Maruvada, A. M. Blom and N. V. Prasadarao, “Effects of Complement Regulators Bound to Escherichia coli K1 and Group B Streptococcus on the Interaction with Host Cells,” Immunology, Vol. 124, No. 2, 2008, pp. 265-276. doi:10.1111/j.1365-2567.2007.02764.x
- I. Toth, F. Herault, L. Beutin and E. Oswald, “Production of Cytolethal Distending Toxins by Pathogenic Escherichia coli Strains Isolated from Human and Animal Sources: Establishment of the Existence of a New cdt Variant (Type IV),” Journal of Clinical Microbiology, Vol. 41, No. 9, 2003, pp. 4285-4291. doi:10.1128/JCM.41.9.4285-4291.2003
- R. M. Robins-Browne, A.-M. Bordun, M. Tauschek, V. Bennett-Wood, J. Russell, F. Oppedisano, N. A. Lister, K. A. Bettelheim, C. K. Fairley, M. I. Sinclair and M. E. Hellard, “Atypical Enteropathogenic Escherichia coli: A Leading Cause of Community-Acquired Gastroenteritis in Melbourne, Australia,” Emerging Infectious Diseases, Vol. 10, No. 10, 2004, pp. 1797-1805. doi:10.3201/eid1010.031086
- C. Toma, et al., “Multiplex PCR Assay for Identification of Human Diarrheagenic Escherichia coli,” Journal of Clinical Microbiology, Vol. 41, No. 6, 2003, pp. 2669- 2671. doi:10.1128/JCM.41.6.2669-2671.2003
- J. R. Czeczulin, T. S. Whittam, I. R. Henderson, F. NavarroGarcia and J. P. Nataro, “Phylogenetic Analysis of Enteroaggregative and Diffusely Adherent Escherichia coli,” Infection and Immunity, Vol. 67, No. 6, 1999, pp. 2692- 2699.
- H.-J. Monstein, Å. Östholm-Balkhed, M. Nilsson, K. Dornbusch and L. Nilsson, “Multiplex PCR Amplifi- cation Assay for the Detection of blaSHV, blaTEM and blaCTX-M Genes in Enterobacteriaceae,” Acta Pathologica, Microbiologica et Imunologica Scandinavica, Vol. 115, No. 12, 2007, pp. 1400-1408. doi:10.1111/j.1600-0463.2007.00722.x
- L. Nicholls, T. H. Grant and R. M. Robins-Browne, “Identification of a Novel Locus That Is Required for in vitro Adhesion of a Clinical Isolate of Enterohaemorrhagic Escherichia coli to Epithelial Cells,” Molecular Microbiology, Vol. 35, No. 2, 2000, pp. 275-288. doi:10.1046/j.1365-2958.2000.01690.x
- A. W. Paton and J. C. Paton, “Detection and Characterization of Shiga Toxigenic Escherichia coli by Using Multiplex PCR Assays for stx1, stx2, eaeA, Enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157,” Journal of Clinical Microbiology, Vol. 36, No. 2, 1998, pp. 598-602.
- C. Toma, E. E. Martinez, T. Song, E. Miliwebsky, I. Chinen, S. Iyoda, M. Iwanaga and M. Rivas, “Distribution of Putative Adhesins in Different Seropathotypes of Shiga Toxin-Producing Escherichia coli,” Journal of Clinical Microbiology, Vol. 42, No. 11, 2004, pp. 4937-4946. doi:10.1128/JCM.42.11.4937-4946.2004
NOTES
*Corresponding author.

