Open Journal of Marine Science
Vol.2 No.4(2012), Article ID:23863,8 pages DOI:10.4236/ojms.2012.24022
The Distribution and Substrate Specificity of Extracellular Nuclease Activity in Marine Fungi
G.B. Elyakova Pacific Institute of Bioorganic Chemistry, Far-Eastern Branch of Russian Academy of Sciences, Vladivostok, Russia
Email: *balaban@piboc.dvo.ru
Received July 14, 2012; revised August 21, 2012; accepted September 6, 2012
Keywords: Marine Fungi; Marine Environment; Single-Strand-Specific Nuclease; DNAse; RNAse; ssDNA; Thymidine Uptake
ABSTRACT
The distribution and specificity of extracellular nucleases produced by marine fungi belonging to eleven genera, namely: Alternaria, Aspergillus, Aureobasidium, Chaetomium, Fusarium, Gliomastix, Humicola, Penicillium, Scopulariopsis, Wardomyces, Periconia, have implied its important function in the organic phosphorus and nitrogen circle in the Ocean. The fungal nucleases of 64 isolates tested were more or less specific for single-stranded DNA with a high preferential specificity towards poly-U substrate with forming of 5’-phosphate mononucleotides. A couple of the nucleases were capable of RNA digesting. The highest level of extracellular nucleolytic ability was observed in Penicillium spp. isolates. The tight correlation found between extracellular nuclease activity and the rate of thymidine uptake by actively growing and sporulating marine fungus Penicillium melinii suggests that this nuclease is required for fulfilling the nucleotide pool of precursors of DNA biosynthesis during transformation of hyphae into the aerial mycelium and conidia in stressful environmental conditions.
1. Introduction
The main portion of the organic carbon, phosphorus and nitrogen is a direct result of enzymatic processing of dissolved macromolecules of DNA, RNA and proteins in aquatic environments due to their wide occurrence [1,2]. Aquatic microorganism populations utilize nucleic acids, enabling efficiently scavenging of organic matter from the dissolved pool, rapidly cycling the nucleic acids [3, 4]. Therefore, there is considerable interest in molecular utilization mechanism of nucleic acids in marine ecosystem. Some fields of fungal community studies have been described [5]. However, the contribution of marine mycobiota as a source and consumer of the dissolved organic matter such as nucleic acids has not been yet shown.
It is known that a large group of fungi and filamentous bacteria generate and disseminate spores for implement of survival program in stressful environmental conditions. These events call forth increase of nuclease activity dramatically that coincides with lysis of the substrate hyphae and formation of aerial mycelium [6]. It has analogy to the late steps of apoptosis of other eukaryotic cells [7]. Indeed, these programs may be activated by environmental stresses or by natural processes of senescence [8,9]. Similarly, plant cells undergoing programmed cell death during tissue differentiation at the end of mitosis or under the influence of pathogens exhibit DNA degradation [10].
Only a few species of terrestrial fungi producing extracellular nucleolytic enzymes have been reported [11- 13]. No information is available on the genus or species specificity of nucleolytic enzymes in marine fungi. However, the previous data have shown the substantial difference between the marine fungi and their terrestrial counterparts [14,15]. Moreover, most of marine fungi were found to be facultative with the potential for longdistance dispersal that might enable unusually rapid propagation of epidemics in marine systems [16].
The purpose of the present study was to evaluate extracellular nucleolytic ability in marine fungi using different natural and synthetic substrates and its possible biological and ecological role in marine environment.
2. Materials and Methods
2.1. Fungal Isolates and Cultural Conditions
The fungal strains were isolated from marine animals and sea bottom sediments collected in the shore water of the Kuril Islands and Primorskyi krai (Table 1) and identified as described previously [17,18]. Fungi cultures were
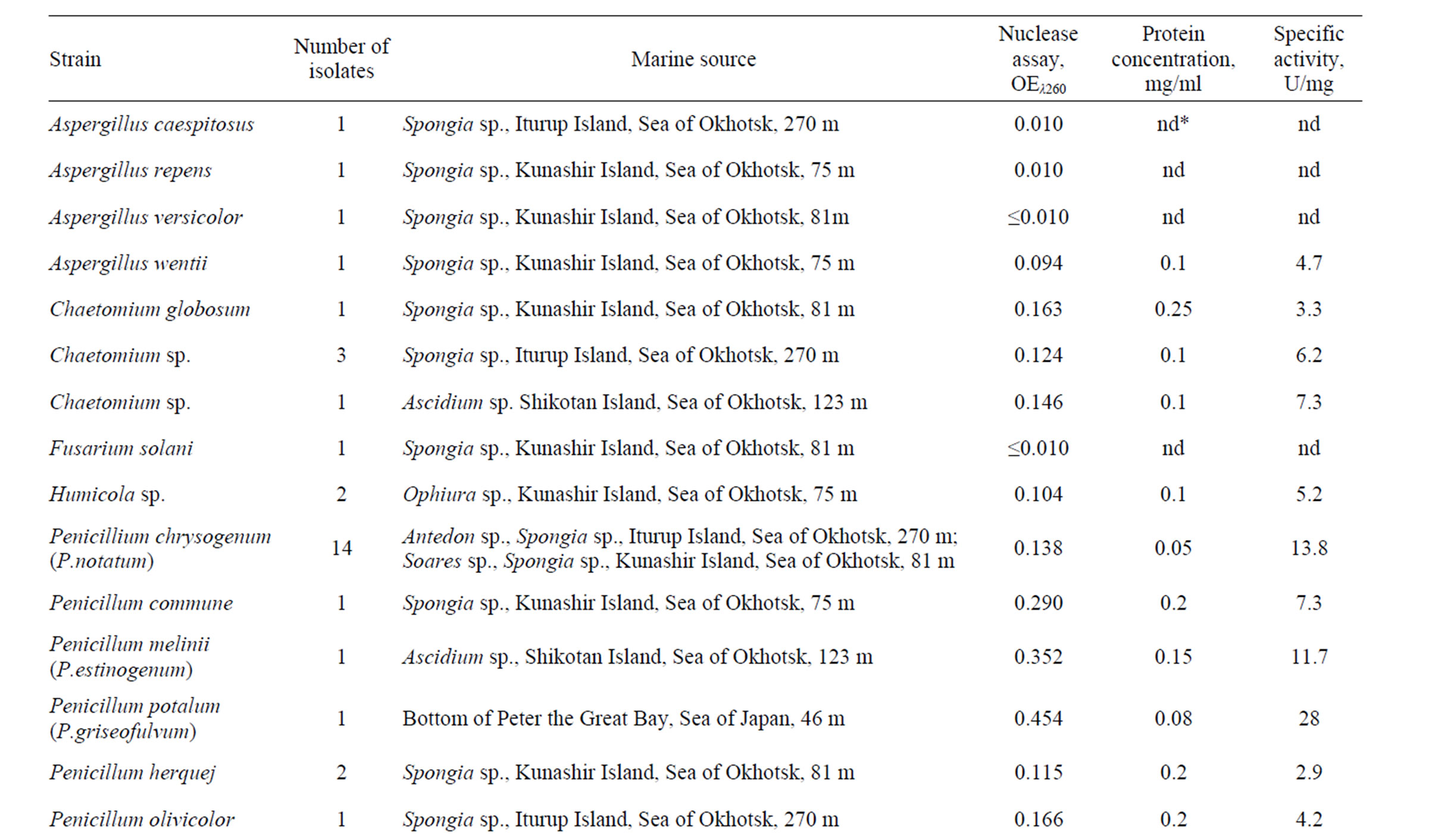

Table 1. The distribution of extracellular nucleolytic activity in marine fungi.
maintained on sterile seawater with Wort agar [19]. Each isolate was stationary grown in 100 ml of modified Ogata medium consisting of glucose 5%, pepton 1%, soybean flour 0.5%, MgSO4 × 7H2O – 0.05%, CaCl2 × 2H2O – 0.01%, KNO3 – 0.2% and natural seawater without phosphates at 22˚C for 7 days.
2.2. Extracellular Nuclease Activity Assay
Extracellular nucleolytic enzyme activity was determined in the fungal culture during the cells growth. For extracellular nucleolytic enzyme activity detection, cell-free supernatant after culture filtration was inoculated onto the agar plate. The agar plates were prepared with 3 g of liquefied agarose (“Hemapol”), 200 ml of 0.03 M sodium acetate buffer, pH 4.6, containing 1 mM ZnSO4 and 0.05 M NaCl, and 2 ml heat-denatured DNA (ssDNA) at concentration of 1 - 2 mg/ml. The resulting mixture was immediately poured onto marked horizontal plates with the final volume about 20 ml (8 × 12 cm and 2 mm thick). Wells of 3 mm in diameter were cut in the agar plates. Each of two-microliter samples of fungal culture in equal protein concentrations was inoculated into the wells. After incubation at 37˚C for 20 min, the result of the nuclease activity was observed at λ260. The more active fungal isolates were selected by comparison of the dark circle zones produced by enzymatic cleavage of the substrate for the further quantitative analysis.
2.3. Nuclease Specificity
Nuclease specificity was evaluated in a mixture of 0.5 ml 0.03 M sodium acetate buffer, pH 4.6, containing 1 mM ZnSO4 and 0.05 M NaCl, substrate in concentration of 10 OE260 and 0.02 ml of filtered fungal culture sample. The reaction mixture was incubated at 37˚C for 20 min and terminated by adding 2.0 ml of 0.5 M HCLO4 and 0.1% La(NO3)3. The mixture was centrifuged at 2000 g for 10 min. The absorbance of supernatant at λ260 was measured. One unit of the enzyme was defined as the amount that catalyzes the production of acid-soluble nucleotides increased by 1.0 absorbance unit for 20 min. Protein concentration was defined by Bredford method [20]. ssDNA was prepared by incubating bovine spleen DNA (Biolar, Russia) solution in boiling water for 30 min and then in the ice bath for 10 min. Poly (A), Poly (C), and poly (U), Poly (A:U), Poly (C:G), Poly (I:C) were from “Reanal”. Poly (G), Poly (I), yeast RNA was from “Vector” (Russia).
2.4. [H3]Thymidine Uptake and Conidia Productivity Assay
Penicillium melinii isolates were examined for the ability to take up [H3]thymidine during cell growth. [H3]Thymidine uptake was determined as a total cell-associated radioactivity by adding 1-ml samples to tubes containing 1 ml of 2 µCi/ml [H3]-labeled thymidine in nutrient-free artificial seawater. Subsamples were filtered within 5 min onto 0.2-mm-pore-diameter GFC filters (Whatman) and were washed with 5 ml of 5% trichloroacetic acid with 1% sodium pyrophosphate and ethanol. Radioactivity in the samples was measured by liquid scintillation counting (Mark III). The conidia were direct counted immediately and again after every two days of incubation. Counting of conidia was carried out as described by Bilai [21]. An aliquot of 10 ml of cultural fluid with fungus was shaken for 30 min and centrifuged at 3000 g for 20 min. Sediment was weighed and 0.3 g of fungal biomass was suspended in 1 ml of distilled water. Concentration of conidia was calculated five times in Thom’s cell.
2.5. Statistical Analysis
Extracellular nucleolytic ability, [H3]thymidine uptake and conidia productivity of fungi were evaluated as the average of three independent experiments. Statistical significance was analyzed by the Student’s t-test, the critical level of significance was 5%.
2.6. Molecular Cloning and Sequence Analysis
Recombinant DNA technique was performed using Ins T/Aclonetm PCR Product Cloning Kit, restriction endonucleases, T4 DNA ligase, Long PCR mix (Fermentas), Smart Taq Polymerase (Topotili), automatic amplifier (Eppendorf). Two pair of primers (For-1 and Rev, sequences: 5’-CAYTTYATHGGNGAYATGAC-3’ and 5’- GCCCANCKNGTNGCNGT-3’, respectively; For-2 and Rev, sequences: 5’-GAYTGGGAYACNTAYATGCC-3’ and 5’-GCCCANCKNGTNGCNGT-3’, respectively) were constructed on the base of S1-like nuclease homologue sequence alignments to amplify the active site region of S1 type nuclease using chromosomal DNA of P. melinii, P. patulum and P. chrysogenum as templates. Nand C-termini region coding sequences were determined using two pair of primers Nuc-for1 and Nuc-rev, sequences: 5’-ATGGTCTCTCTATCCAAGATTG-3’ and 5’-ATGGCTATGCTCACGTCCTTCTGAGG-3’, respectively. PCR products were cloned and sequenced using the automated PE/ABI 310 DNA sequencer and PE/ABIABI PRISM BigDye Terminator cycle sequencing Ready Reaction Kit (PE Applied Biosystems). E. coli strains TOP10 or XL1 (Evrogen) were used for standard cloning procedures. Nucleotide and amino acid sequence analysis were performed with Chromas, GenRunner, SMART programs. Nucleotide and amino acid sequences homology and similarity searches and alignments were carried out by using BLAST and ClustalW, МОЕ 9.10 facilities.
3. Results
3.1. Screening of Extracellular Nuclease Activities
Fifty percent from more than 60 isolates tested was found to produce a different level of extracellular nuclease activity catalyzing the cleavage of ssDNA at pH 4.6 (Table 1). Of the isolates with the high level of extracellular nuclease activity (about 10.0 - 30.0 U/mg), 14 isolates belonged to the species P. chrysogenum (formerly P. notatum) isolated from various species of sponges collected near Iturup and Kunashir Islands at the depth of 270 m and 81 m, respectively. The isolates of P. melinii (formerly P. estinogenum) and P. patulum (formerly P. griseofulvum) isolated from colonial ascidium near Shikotan Island at the depth of 123 m and P. patulum isolated from Peter the Greate Bay bottom sediments, Sea of Japan, at the depth of 46 m also showed a high level of extracellular nuclease activity (Table 1). Apparent nucleolytic activity was found in the isolates of Chaetomium sp., A. wentii, Humicola sp., P. commune, P. herquej, P. olivicolor, and Dematiaceae isolated from the bottom sediment of Sea of Okhotsk from 551 m depth (Table 1). The number of isolates belonging to genus Penicillium exhibited a highest level of extracellular nuclease activity accounted for 53% of the total. The majority of these isolates were identified as populations of P. chrysogenum. As shown in Table 1, Penicillium spp. coexists with the largest amount of the deep-sea habitants irrespectively of geographic area. Extracellular nucleolytic activity was not detected in any isolates belonging to the species A. alternata, A. сandidus, A. amilovorus, A. amstelodami, A. amstelodami, A. varians, Aureobasidium pullupans, F. sporotrichiella, G. cereales, P. citrinum, P. jensenii, P. namyslowskii, W. anomalus, Periconia sp. (Table 1).
3.2. Extracellular Nucleases Specificity
The marine fungi were found to produce non-specific extracellular ssDNA/RNA nucleases especially those, which was been habitable under the deep-sea conditions (Table 1). As shown in Figure 1, all of the strains, which were capable of ssDNA digestion, have a preference for Poly (U). The rate of Poly (U) hydrolysis was in 10 - 100-fold higher in comparison with the other substrates. The fungal extracellular nucleases acted both ssDNA and RNA at the same rate. Nuclease from a terrestrial strain of A. oryzae showed the similar specificity under the same reaction conditions. Nucleases from A. wentii (sponge, Kunashir Island, 75 m depth) and Chatomium spp. isolates (colonial ascidium, Shikotan Island, 123 m depth) were found to be much more active with RNA than DNA as a substrate (Figure 1).
3.3. [H3]Thymidine Uptake versus the Extracellular Nuclease Activity and Conidia Productivity
As shown in Figure 2, the maximal nuclease productivity achieved when fungus colony started actively growing after 3 days of incubation on a nutrient-free medium and it was highest at 10 days. Since the fungus cells were capable of [H3]thymidine inclusion (Figure 2). The tight correlation (r = 0.93) found between extracellular nuclease activity and the rate of thymidine uptake. As ex-
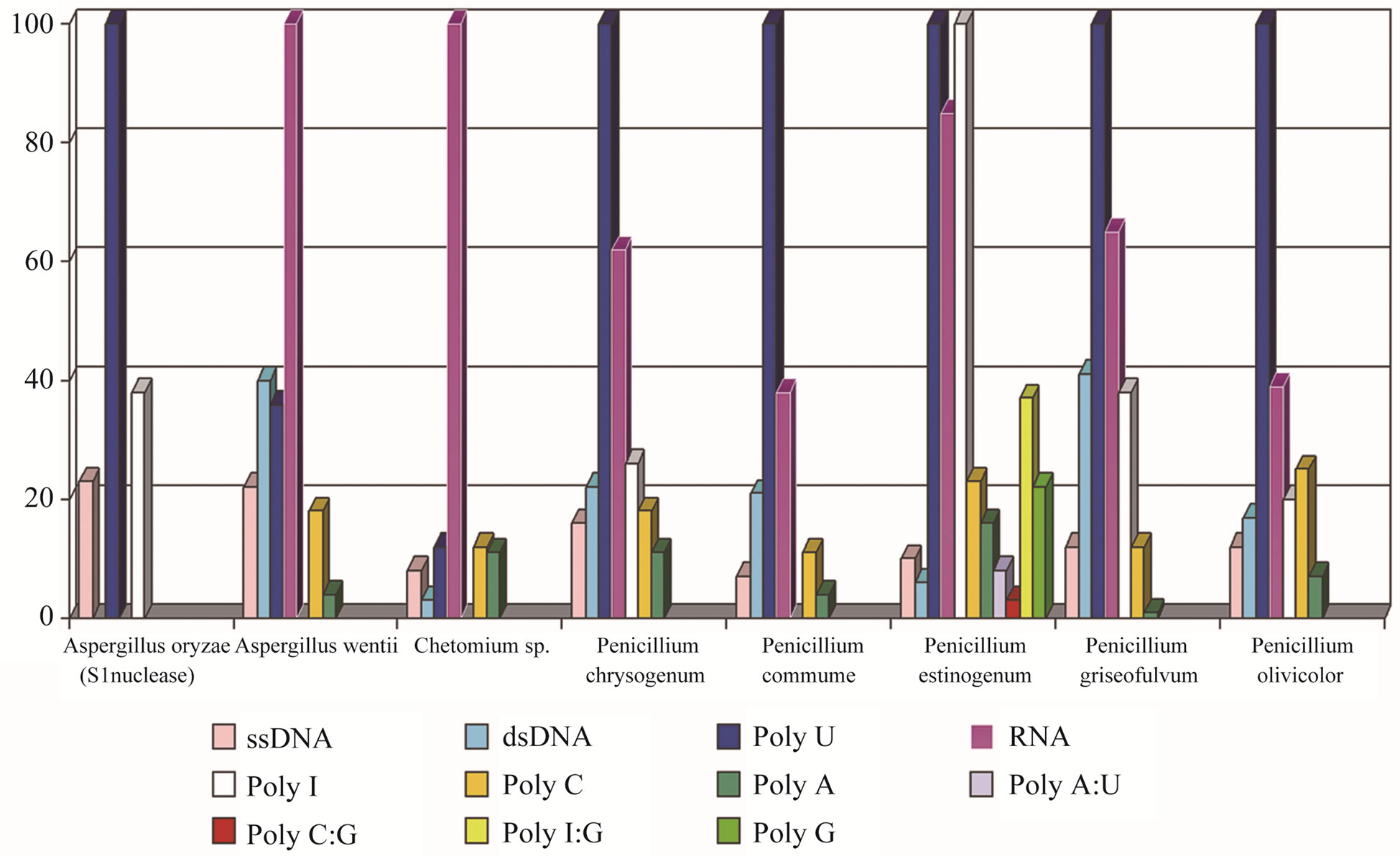
Figure 1. Specificity of extracellular nucleases of marine fungi.
pected, the amount of green conidia was highest at 11 days of incubation (Figure 3). The extracellular nucleolytic enzyme activity and [H3]thymidine uptake decreased when conidia productivity reached an exponential-stationary phase after 12 days of incubation (Figures 2 and 3). Shaking the fungal culture during the growth strongly repressed nuclease productivity, however, it was 2 - 5-fold higher when the fungus was cultivated without shaking (unpublished results).
3.4. Sequence Analysis
The structural and biochemical details of some ssDNApreferential nucleases of class 3.1.30.1 isolated from the terrestrial sources are known [13,22,23]. We have cloned and analyzed ssDNA-preferential highly active extracellular nucleases from marine fungi, namely: P. melinii (GU331890), P. patulum (formerly P. griseofulvum) (GU331891), and P. chrysogenum (GU33182).
The presence of putative signal peptide indicates that these proteins are destined for secretion to the periplasm or excretion into the environment. Gene Bank databases searching by BLAST software disclosed a significant sequence homology of these enzymes with terrestrial P. citrinum (70% identity), A. oyzae (52% identity), and N. crassa (47% identity) S1 type nucleases that suggested the same mechanism of action (Figure 4).
4. Discussion
Our investigation demonstrated a large number of the fungal strains collected from the sea deepwater sources, producing highly active extracellular nucleases into marine environment. Extracellular nucleolytic ability was detected in the strains of the genera Aspergillus, Chaetomium, Fusarium, Humicola, Penicillium, Scopulariopsis that were found prolifically in the sea bottom sediments and marine animals inhabiting of the shelf waters of Japan Sea. The species of the genera Penicillium, Aspergillus Chaetomium, Fusarium were of more frequent occurrence (Table 1). Some authors suggested the presence of interference competition among the fungi in mixed cultures on substrates, especially in congeneric species [24]. The ecological factor influences on the fungal population densities as well as their quality have also been reported [25]. The extracellular nuclease productivity among the marine fungi appears to be dependent on origin and depth as well as kind of host (Table 1). It might be related to taxonomic and ecological groups or/and biogeographic zones [26].
Of 64 isolates tested, two were capable of RNA digesting (Figure 1). Other fungal nucleases are more or less specific for ssDNA, preferentially cleaving Poly (U) substrate to leave 5’-phosphate mononucleotides. The high extracellular ability to cleave RNA indicates that
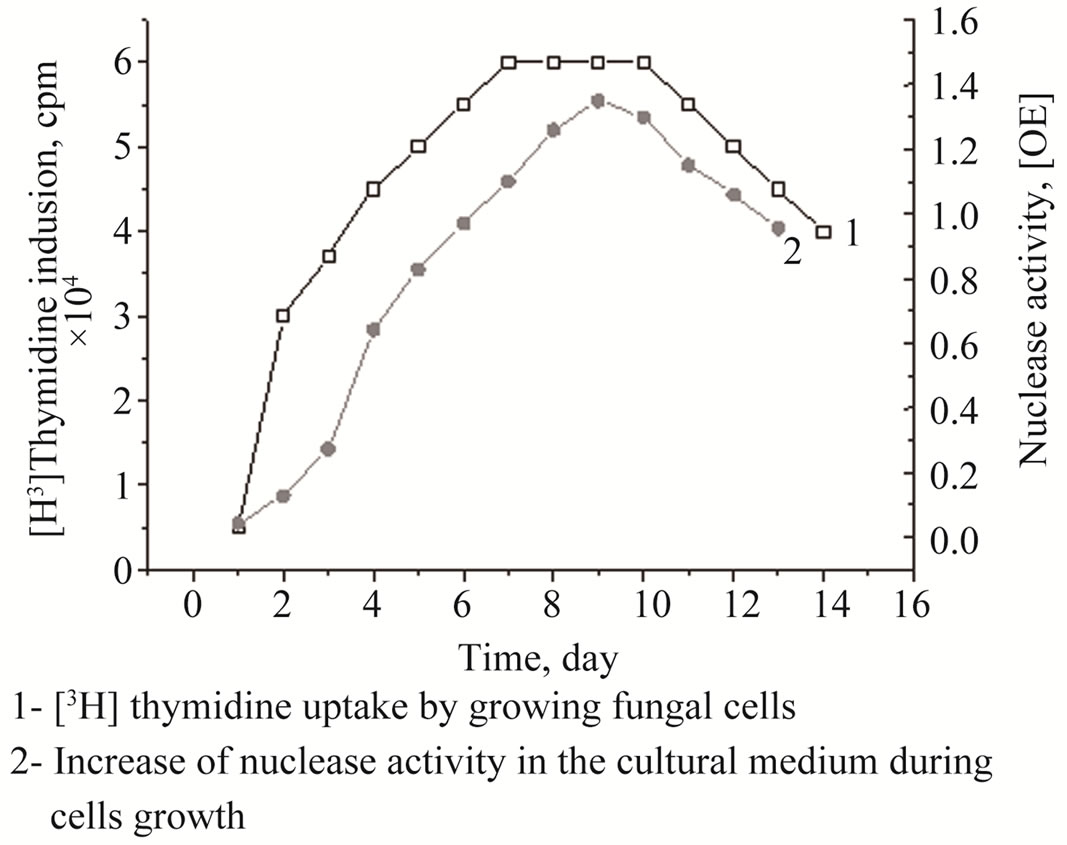
Figure 2. [H3]Thymidine uptake versus extracellular ssDNApreferential nuclease production by P. melinii.

Figure 3. Conidia formation intensity in marine fungus P. melinii during incubation.
some marine fungi species have a potential to safe genetic information from attacking carrier of viruses resides providing participation in host defense [27].
Actively growing marine fungus P. melinii (formerly P. estinogenum) collected from colonial Ascidium spp. at the depth of 270 m was examined for [H3]thymidine uptake into the cells (Figure 2). The results obtained indicated that the extracellular nucleolytic enzyme activities in marine fungi were required for nucleotide inclusions into DNA of the newly developing cells. Its function proposed to have a key role in complete DNA degradation of exogenous substrates as well as putative apoptotic destruction of the fungal hyphae to supply building blocks for macromolecular biosynthesis in the aerial mycelium and conidia as the result of senescence or stressful environmental conditions [8,9]. The inhibition of either hydrolytic activity at least strongly impaired aerial mycelium formation [6]. In the other hand, we have found that
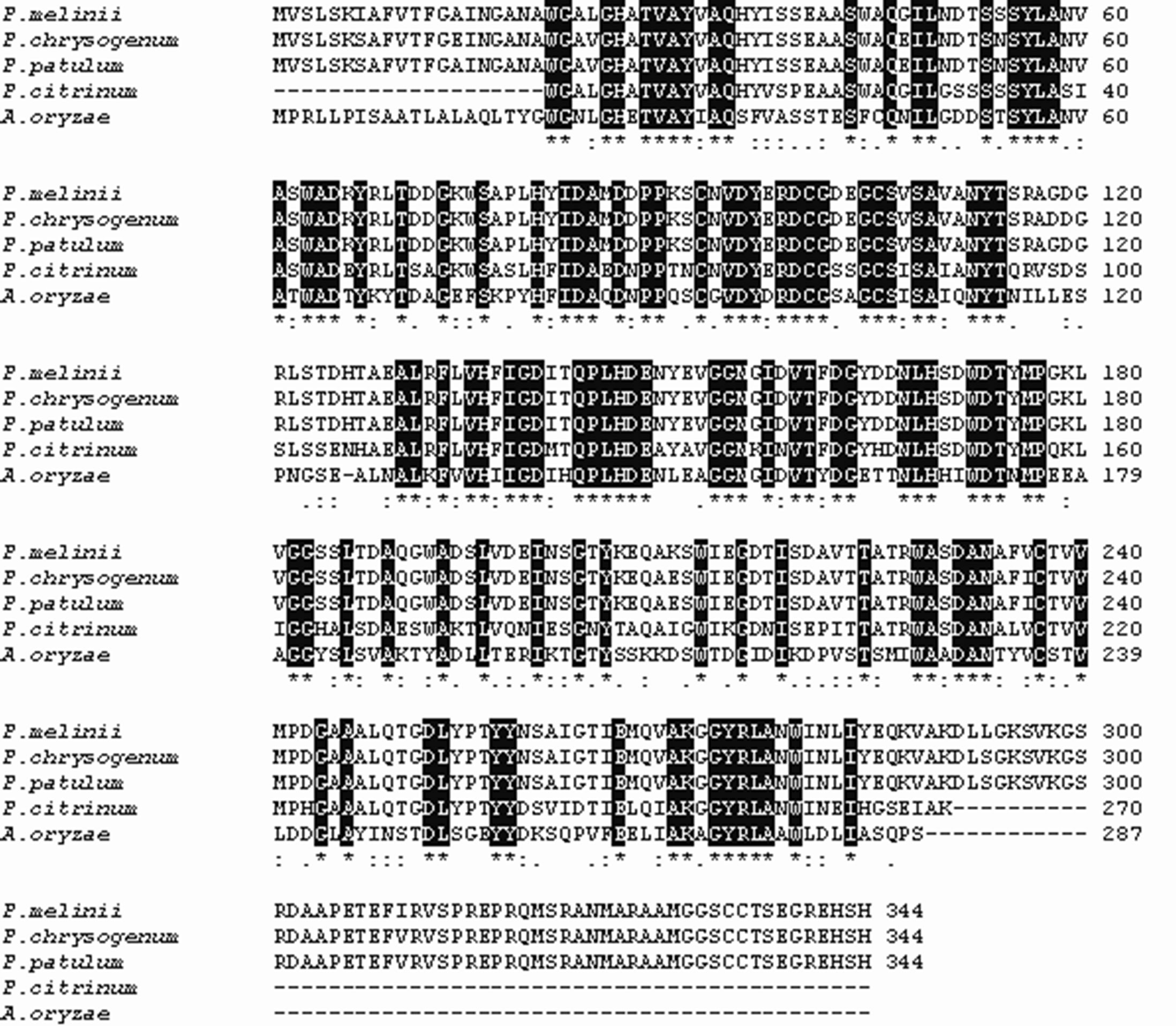
Figure 4. A ClustalW alignment of extracellular ssDNA-prefential nucleases from marine fungi P. melinii, P. chrysogenum, P. patulum, with homologous sequences of terrestrial counterparts S1 and P1. Identical residues are boxed.
the certain conditions of cultivation are required for the continuous synthesis of the enzyme (unpublished results). The aerification deficiency appears to provide the active conidia formation.
The extacellular nucleolytic ability in marine fungi, as demonstrated in this study, indicated that their enzymes, probably, are the major deliver of the nucleotide matter to the symbiothrophic organisms in the condition of the organic matter deficient (Table 1). An experiment in [H3]thymidine uptake by marine fungus demonstrated the capability for fungal communities to active participate in the cycling dissolved DNA and RNA in marine environment. Marine fungi are the major decomposer of living and/or dead substrates as well as participator in mutualistic symbioses with the most marine habitants [17, 28]. In fact, to provide the self-cleaning capacity of a habitant from pollutants the degradation of organic matter is archived by the action of an exoenzyme complex [17,29]. It seems to be simultaneous with programmed cell death associated with the different aspects of fungi development (Figures 2 and 3). Moreover significant similarities were found among the primary structures of S1 type nucleases of marine fungi and nuclease from barley secreted from the aleuron layer into the endosperm during germination and may be responsible for the digestion of nuclear DNA in the course of degradation of endosperm [9], and nuclease from zinnia, which activity appeared transiently at the time of autolysis associated with transdifferentiation from mesophyl cells into tracheary elements [30]. They also resemble enzymes from A. oryzae, P. citrinum and some other terrestrial S1-type nucleases [7,12,23]. However, some differences in the structure of the marine fungi ssDNA-preferential nucleases are considerable depending on species within a given group or genus that can be used as a unique molecular marker for providing useful information about genotypic variation (Figure 4). The largest amounts of negatively charged Glu and Asp comparatively with the terrestrial counterparts together with doubling of Arg content are essential in the marine fungi enzymes due to additional C-terminal peptide from 270 to 344 amino acid residues. An increase in polar intrinsic flexible Gly as well as non-polar small Ala residues and additional C-terminal sequence suggest that it is a functional adaptation molded by evolution under the deep-sea conditions [31].
In summary, we have shown that the marine fungi possess important extracellular nucleolytic enzymes, which are required for fulfilling the nucleotide pool of precursors of DNA biosynthesis and maintaining ecological balance. Their distribution in the deep-sea water and unique specificity confirmed the important function in the organic phosphorus and nitrogen circle in the Ocean. The potential of extracellular nucleolytic ability in marine fungi to estimate the marine environmental conditions as well as the biotechnology potential of the marine fungi nucleases will attract much attention.
5. Acknowledgements
Financial support was provided by grants from RFBR 08-08-00975-a and the RAS Presidium under the project “Molecular and Cellular Biology” 09-I-P22-05.
REFERENCES
- J. H. Paul, W. H. Jeffrey and J. P. Cannon, “Production of Dissolved DNA, RNA and Protein by Mycrobial Populations in a Florida Reservoir,” Applied and Environmental Microbiology, Vol. 56, No. 10, 1990, pp. 2957-2962.
- A. Dell’Anno and R. Danavaro, “Extracellular DNA Plays a Key Role in Deep-Sea Ecosystem Functioning,” Science, Vol. 309, No. 5744, 2005, p. 2179. doi:10.1126/science.1117475
- J. H. Paul, A. W. Jeffrey and M. F. DeFlaun, “Dynamics of Extracellular DNA in the Marine Environment,” Applied and Environmental Microbiology, Vol. 53, No. 1, 1987, pp. 170-179.
- J. H. Paul, M. F. DeFlaun and W. H. Jeffrey, “Mechanisms of DNA Utilization by Esturine Microbial Populations,” Applied and Environmental Microbiology, Vol. 54, No. 7, 1988, pp. 1682-1688.
- C. A. Shearer, “Fungal Competition,” Canadian Journal of Botany-Revue, Canadienne de Botanique, Vol. 73, 1995, pp. S1259-S1264. doi:10.1139/b95-386
- R. G. Nicieza, J. Huergo, B. A. Connolly and J. Sánchez, “Purification, Characterization and Role of Nucleases and Serine Proteases in Streptomyces Differentiation: Analogies with the Biochemical Processes Described in LateSteps of Eukaryotic Apoptosis,” Journal of Biological Chemistry, Vol. 274, No. 29, 1999, pp. 20366-20375. doi:10.1074/jbc.274.29.20366
- S. Aoyagia, M. Sugiyamaa and H. Fukudac, “BEN1 and ZEN1 cDNAs Encoding S1-Type DNases That Are Associated with Programmed Cell Death in Plants,” FEBS Letters, Vol. 429, 1998, pp. 134-138. doi:10.1016/S0014-5793(98)00563-8
- T. Panavas and B. Rubinstein, “Oxidative Events during Programmed Cell Death of Daylily (Hemerocallis Hybrid) Petals,” Plant Science, Vol. 133, No. 2, 1998, pp. 125- 138. doi:10.1016/S0168-9452(98)00034-X
- Y. Muramoto, A. Watanabe, T. Nakamura and T. Takabe, “Enhanced Expression of a Nuclease Gene in Leaves of Barley Plants under Salt Stress,” Gene, Vol. 234, No. 2, 1999, pp. 315-321. doi:10.1016/S0378-1119(99)00193-6
- M. Sugiyama, J. Ito, S. Aoyagi and H. Fukuda, “Endonucleases,” Plant Molecular Biology, Vol. 44, No. 3, 2000, pp. 387-397. doi:10.1023/A:1026504911786
- K. Shishido and T. Ando, “Single-Strand Specific Nucleases,” In: S. M. Linn and R. J. Roberts, Eds., Nucleases, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1982, pp. 155-185.
- V. Vogt, “Purification and Future Properties of SingleStrand-Specific Nuclease from Aspergillus oryzae,” FEBS Journal, Vol. 33, 2005, pp. 192-200.
- N. A. Desai and V. Shankar, “Single-Strand-Specific Nuclease from Basidiobolus haptosporus Nuclease Bh1),” Scientific Research and Essay, Vol. 2, No. 5, 2007, pp. 139-146.
- C. Osterhage, M. Schwibbe, G. M. Konig and A. D. Wright, “Difference between Marine and Terrestrial Phoma Species as Determined by HPLC-DAD and HPLC-MS,” Phytochemical Analysis, Vol. 11, No. 5, 2000, pp. 288- 294. doi:10.1002/1099-1565(200009/10)11:5<288::AID-PCA528>3.0.CO;2-G
- A. Baakza, B. P. Yala and H. C. Dube, “A Comparative Study of Siderophore Production by Fungi from Marine and Terrestrial Habitats,” Journal of Experimental Marine Biology and Ecology, Vol. 311, No. 1, 2004, pp. 1-9. doi:10.1016/j.jembe.2003.12.028
- H. McCallum, C. D. Harvell and A. Dobson, “Rates of Spread of Marine Pathogens,” Ecology Letters, Vol. 6, No. 12, 2003, pp. 1062-1067. doi:10.1046/j.1461-0248.2003.00545.x
- K. D. Hyde, C. A. Farrant and E. B. G. Jones, “Isolation of Marine Fungi,” Botanica Marina, Vol. 30, 1987, pp. 291-303. doi:10.1515/botm.1987.30.4.291
- Y. V. Khudyakova, M. V. Pivkin, T. A. Kuznetsova and V. I. Svetashev, “Fungi in Sediments of the Sea of Japan and Their Biologically Active Metabolites,” Microbiology, Vol. 69, No. 5, 2000, pp. 608-611. doi:10.1007/BF02756817
- N. Y. Artemchuk, “Microflora of USSR’s Seas,” Nauka, Moscow, 1981.
- M. M. Bradford, “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein-Dye Binding,” Analytical Biochemistry, Vol. 72, No. 1-2, 1976, pp. 248- 254. doi:10.1016/0003-2697(76)90527-3
- V. I. Bilai, “Definition of the Growth and Biosynthetic Activity of Fungi,” In: V. I. Bilay, Ed., Method of Experimental Mycology, Naukova Dumka, Kiev, 1982, pp. 146-147.
- S. A. Martin, R. S. Ullrich and W. L. Meyer, “A Comparative Study of Nucleases Exhibiting Preference for Single-Stranded Nucleic Acid,” Biochimica et Biophysica Acta, Vol. 867, No. 1-2, 1986, pp. 76-80. doi:10.1016/0167-4781(86)90031-X
- M. Pimkin, C. Miller, L. Blakesley, C. Oleykowski, N. Kodali and A. Yeung, “Characterization of a Periplasmic S1-Like Nuclease Coded by the Mesorhizobium Loti Symbiosis Island,” Biochemical and Biophysical Research Communications, Vol. 343, No. 1, 2006, pp. 77-84. doi:10.1016/j.bbrc.2006.02.117
- T. K. Tan, C. L. Teng and B. G. Jones, “Substrate Type and Microbial Interaction as Factors Affecting Ascocarp Formation by Mangroove Fungi,” Hydrobiologia, Vol. 295, 1995, pp. 127-134. doi:10.1007/BF00029119
- M. Muntanola-Cvetkovic, “A Mycological Survey of the South Adriatic Sea,” Journal of Experimental Marine Biology and Ecology, Vol. 43, No. 3, 1980, pp. 193-206. doi:10.1016/0022-0981(80)90047-7
- Y. Vishwakiran, N. L. Thakur, S. Raghukumar, P. L. Yennawar and A. C. Anil, “Spatial and Temporal Distribution of Fungi and Wood-Borers in the Coastal Tropical Waters of Goa, India,” Botanica Marina, Vol. 44, 2001, pp. 47-56. doi:10.1515/BOT.2001.007
- K. C. Hsia , C. L. Li and H. S. Yuan, “Structural and Functional Insight into Sugar-Nonspecific Nucleases in Host Defense,” Current Opinion in Structural Biology, Vol. 15, No. 1, 2005, pp. 193-206. doi:10.1016/j.sbi.2005.01.015
- S. Y. Newell, “Established and Potential Impacts of Eukariotic Mycelial Decomposers in Marine/Terrestrial Ecotones,” Journal of Experimental Marine Biology and Ecology, Vol. 200, 1996, pp. 187-206. doi:10.1016/S0022-0981(96)02643-3
- K. E. Gonga, D. Jendrossek and H. P. Molitoris, “Fungal Degradation of the Thermoplastic Polymer Poly-BetaHydroxybutyric Acid (PHB) under Simulated Deep Sea Pressure,” Hydrobiologia, Vol. 426, 2000, pp. 173-183. doi:10.1023/A:1003971925285
- J. Ito and H. Fukuda, “ZEN1 Is a Key Enzyme in the Degradation of Nuclear DNA during Programmed Cell Death of Tracheary Elements,” Plant Cell, Vol. 14, No. 12, 2002, pp. 3201-3211. doi:10.1105/tpc.006411
- L. A. Balabanova, Y. M. Gafurov, M. V. Pivkin, N. A. Terentyeva, G. N. Likhatskaya and V. A. Rasskazov, “An Extracellular S1-Type Nuclease of Marine Fungus Penicillium melinii,” Marine Biotechnology, Vol. 14, No. 1, 2012, pp. 87-95. doi:10.1007/s10126-011-9392-5
NOTES
*Corresponding author.

