Advances in Biological Chemistry
Vol.4 No.1(2014), Article ID:43301,8 pages DOI:10.4236/abc.2014.41009
Interaction of calciumand integrin-binding protein 1 with integrin α11 and its possible involvement in pulmonary fibrosis
![]()
1Department of Biochemistry, Kinki University Faculty of Medicine, Osaka-Sayama, Japan
2Department of Microbiology, Kinki University Faculty of Medicine, Osaka-Sayama, Japan
Email: *kojiy@med.kindai.ac.jp
Copyright © 2014 Koji Yoshida et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Koji Yoshida et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 17 January 2014; revised 16 February 2014; accepted 22 February 2014
KEYWORDS:Calcium- and Integrin-Binding Protein 1; Fibrosis; Integrin α11; Yeast Two-Hybrid
ABSTRACT
Integrin α11 (ITGA11) is one of the collagen-binding integrin α chains; however, its biological significance remains unknown. To determine the functions of ITGA11, we performed a yeast two-hybrid screen using the cytoplasmic domain of ITGA11 as bait and transformed an EGY48 yeast strain with the baitcontaining plasmid using the plasmid from a human lung fibroblast cDNA library. This screen identified calciumand integrin-binding protein 1 (CIB1) as prey. Recombinant ITGA11 and CIB1 were expressed in mammalian cells and used in coimmunoprecipitation experiments, which showed that full-length ITGA11 and CIB1 are also associated in vivo. Overexpression of CIB1 in the human lung myofibroblast MRC-5 cells decreased the expression of α-smooth muscle actin and fibronectin. Using a mouse model of pulmonary fibrosis (bleomycin-treatment), we detected elevated expression of CIB1 in lung tissues compared with controls. These data suggest that CIB1 may regulate pulmonary fibrosis in concert with ITGA11.
1. INTRODUCTION
We have previously shown that the expression of integrin α11 (ITGA11), collagen type I, and fibronectin (FN) was upregulated in MRC-5 human myofibroblasts stimulated with transforming growth factor-β (TGF-β) [1]. TGF-β is considered to be the strongest inducer of tissue fibrosis [2-4]. Organ fibrosis occurs when normal wound healing fails to terminate, causing the deposition of excess amounts of extracellular matrix composed of collagen type I and FN in the tissue and resulting in loss of normal function [5-7].
ITGA11 is one of the collagen-binding integrin α- chains [8,9] that is overexpressed in non-small-cell lung carcinoma and regulates insulin-like growth factor-2 expression in fibroblasts [10]. In addition, ITGA11 expresses on mesenchymal stem cells and integrin α11β1- mediated interactions to collagen I are involved in its cell death [11]. It has been shown that integrin α11β1 is the major receptor for collagen type I on mouse embryonic fibroblasts, and that ITGA11 deficiency affects tooth eruption [12]. Furthermore, it has been reported that ITGA11 plays an important role in collagen deposition during corneal development [13], and that it stimulates myofibroblast differentiation in diabetic cardiomyopathy [14] and coordinates collagen proteolysis [15]. However, the precise physiological and pathological roles of ITGA11 are not fully understood.
In the present study, to investigate the functions of ITGA11, we performed a yeast two-hybrid assay using the cytoplasmic domain of ITGA11 as bait. Moreover, here, we describe the interaction between ITGA11 and a prey protein obtained by two-hybrid screening. Further, we report the behavior of the prey protein in bleomycin-induced pulmonary fibrosis.
2. MATERIALS AND METHODS
2.1. Yeast Two-Hybrid Screening
The LexA Yeast Two-Hybrid System (OriGene Technologies, Rockville, MD, USA) was used to screen for clones interacting with the cytoplasmic domain of human integrin α11 as previously described [16]. The pEG202- NLS (nuclear localization signal) vector is a standard bait plasmid for constitutive expression of LexA-NLSbait fusion protein in yeast, and it contains yeast selective marker HIS3. The pJG4-5 vector was used as library plasmid. Library plasmids express cDNAs inserted into EcoRI and XhoI sites as a translational fusion to a cassette consisting of the NLS. The pJG4-5 plasmid contains the TRP1 selectable marker in yeast. A cDNA encoding the transmembrane-cytoplasmic domain (Leu 1156-Glu 1189) of human ITGA11 (GenBank accession no. AF109681) was amplified from a human lung fibroblast cDNA library (OriGene) using the polymerase chain reaction (PCR) with KOD Plus DNA polymerase (Toyobo, Osaka, Japan) and the primers P1 and P2 (Table 1). The bait, pEG202-NLS/ITGA11, generated by inserting the cDNA encoding the transmembrane-cytoplasmic domain of integrin α11 into the EcoRI site of pEG202-NLS, was used to screen the human lung fibroblast cDNA library cloned into the pJG4-5 vector (OriGene). Before performing the large-scale library transformation, we confirmed that the bait had no autoactivation potential using the reporter genes of lacZ and LEU2, and could enter the nucleus and bind to the LexA operators. Positive clones were selected by simultaneously monitoring growth on a drop-out minimal medium (lacking His, Trp, Ura, and Leu) and the activity of the β-galactosidase gene. The cDNA clones from the positive yeast colonies were recovered and were sequenced using an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems, Foster, CA, USA).
2.2. Yeast Two-Hybrid One-On-One Interactions
To identify the interacting regions between the ITGA11 and calciumand integrin-binding protein 1 (CIB1), truncated constructs of CIB1 cDNA were prepared. DNA fragments corresponding to the N-terminal (Met 1-Ser 99) and C-terminal (Asp 100-Leu 191) domains of CIB1 were generated using PCR with human CIB1 cDNA as the template and the following sets of primers: P3 and P4 for the N-terminal domain of CIB1 and P5 and P6 for the C-terminal domain of CIB1 (Table 1). The PCR products were subcloned into the EcoRI and XhoI sites of the prey vector pJG4-5.
Similarly, to investigate the interaction between CIB1 and various ITGAs, the following sets of primers were used: P7 and P8 for ITGA1, P9 and P10 for ITGA2, P11
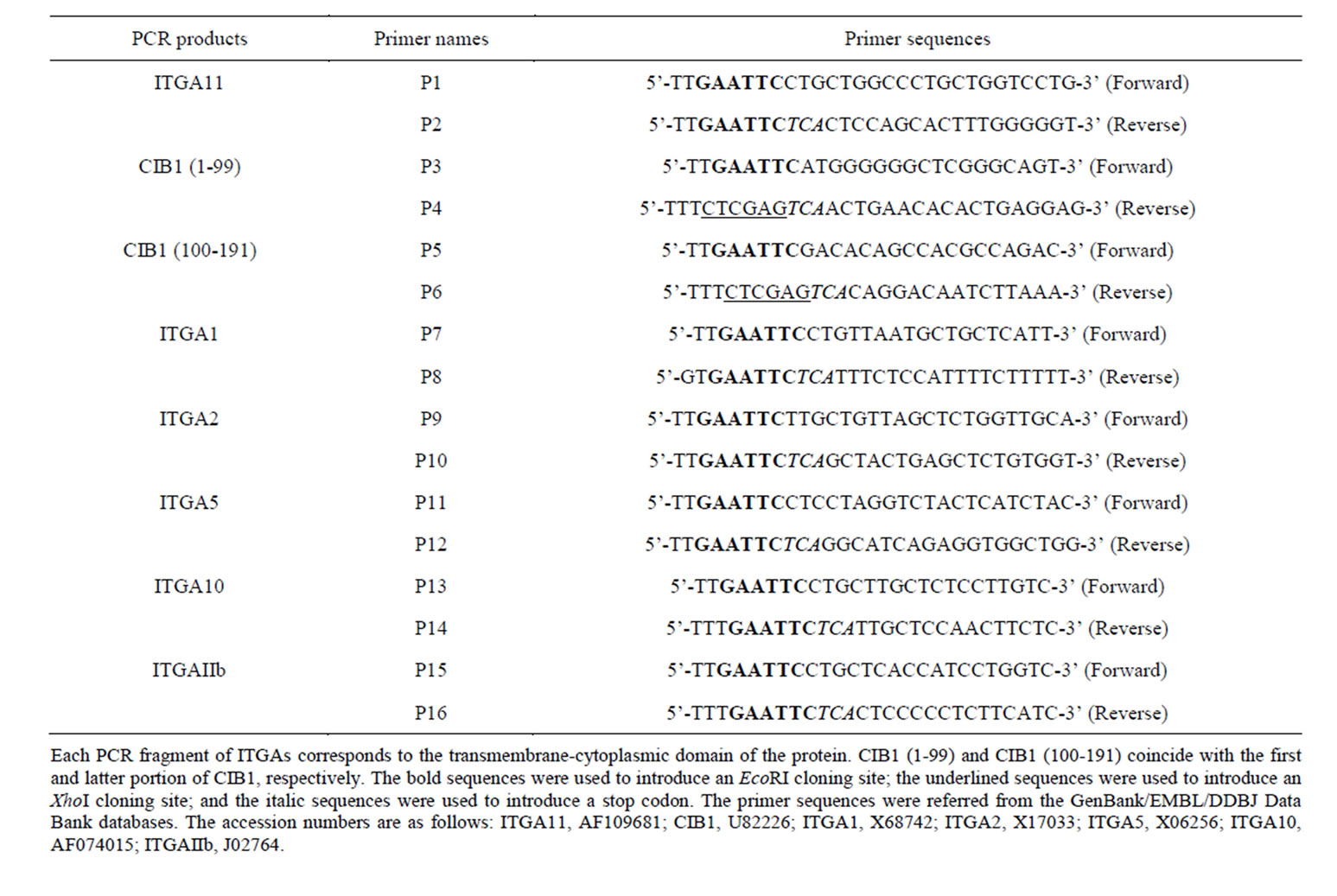
Table 1. Primers for generating PCR fragments used in two-hybrid assay.
and P12 for ITGA5, P13 and P14 for ITGA10, and P15 and P16 for ITGAIIb (Table 1). Each amplified cDNA fragments of ITGAs were subcloned into the EcoRI site of the bait vector pEG202-NLS. Plasmid DNA was isolated using a plasmid DNA purification kit (Promega, Madison, WI, USA), and the integrity of each cDNA sequence was confirmed by DNA sequencing.
2.3. Construction of Expression Vector for ITGA11 with FLAG
Plasmids were constructed according to standard recombinant DNA techniques. PCR-amplified fragments of human ITGA11 cDNA (Met 1-Glu 1189) with a C-terminal DYKDDDDK (FLAG) tag were cloned inframe into the EcoRI-XhoI sites of the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA, USA). The following PCR primers were used: 5’-TTGAATTCATGGACCTGCCCAGGGGCCTG-3’ (forward) and 5’-ATTCTCGAGCTACTTGTCATCGTCGTCCTTGTAATCCTCCAGCACTTTGGGGGTGGG-3’ (reverse). The respective restriction sites for EcoRI and XhoI are underlined.
2.4. Construction of Expression Vector for CIB1 with HA
This construct was prepared by cloning a fragment of full-length human CIB1 cDNA (Met 1-Leu 191, GenBank accession no. U82226) from the human lung fibroblast cDNA library (OriGene) using PCR. The CIB1 cDNA fragment with a C-terminal YPYDVPDYA (HA) tag was introduced into the EcoRI-XhoI sites of the pcDNA3.1(+) vector. The following PCR primers were used: 5’-TTGAATTCATGGGGGGCTCGGGCAGT-3’ (forward) and 5’-GGCTCGAGTCAAGCGTAGTCTGGGACGTCGTATGGGTACAGGACAATCTTAAAGGA-3’ (reverse). The respective restriction sites for EcoRI and XhoI are underlined. All constructs listed here were verified by DNA sequencing as described above.
2.5. Antibodies
Rat anti-human ITGA11 antibody was purchased from R&D Systems (Minneapolis, MN, USA). Mouse antiFLAG tag monoclonal antibody was obtained from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Rabbit anti-human FN, mouse anti-α-smooth muscle actin (α- SMA), and mouse anti-β-actin monoclonal anti-bodies were purchased from Sigma (St. Louis, MO, USA). A rabbit anti-HA tag polyclonal antibody was obtained from MBL (Nagoya, Japan). Normal rabbit IgG was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Rabbit CIB1 polyclonal antibody was obtained from Proteintech Group (Chicago, IL, USA). Horseradish peroxidase-conjugated donkey anti-rabbit or sheep anti-mouse IgG antibodies were purchased from GE Healthcare Biosciences (Piscataway, NJ, USA). A horseradish peroxidase-conjugated goat anti-rat IgG was obtained from Invitrogen.
2.6. Cell Culture and Transfection
The human lung fibroblast cell line MRC-5 and African green monkey cell line COS-7 were purchased from the RIKEN Cell Bank (Tsukuba, Japan). Cells were maintained in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37˚C in a humidified 5% CO2 and 95% air atmosphere. Cells were grown in tissue culture plates (Sumitomo Bakelite, Tokyo, Japan). Cells were transiently transfected using Lipofectamine® 2000 (Invitrogen) according to the manufacturer’s instructions. In brief, cells were seeded in cell culture dishes for 24 h at 90% confluence before transfection with the indicated plasmids. After 48 h, the cells were analyzed.
2.7. Coimmunoprecipitation
Anti-HA antibody was incubated with protein G-Sepharose (GE Healthcare) for 2 h at 4˚C and washed three times with phosphate-buffered saline (PBS; 147 mM NaCl, 2.7 mM KCl, 7 mM K2HPO4, and 1.5 mM KH2HPO4). After transient cotransfection of the ITGA11-FLAG and CIB1-HA expression constructs, COS- 7 cells in a 9-cm diameter tissue culture dish were washed three times with 10 ml of ice cold PBS. One milliliter of ice cold cell lysis buffer (50 mM Tris-HCl, pH 8.0; 1% NP-40; and 150 mM NaCl), which contained a cocktail of protease inhibitors (EDTA-free) (Roche Molecular Biochemicals, Mannheim, Germany), was added to the dish. The dish was placed on ice for 1 min, and the cells were recovered by scraping and vortexing and were then centrifuged at 18,000 g for 15 min at 4˚C. The supernatant was reacted with anti-HA antibody and bound to protein G-Sepharose beads (20 μl), as described above, for 5 h at 4˚C. After the reaction, the protein G-Sepharose beads were collected by centrifugation at 200 g for 2 min and washed three times with PBS. Immunoprecipitated proteins were then removed from the beads by boiling in 2× Laemmli sample buffer (0.125 M Tris-HCl, pH 6.8; 4% sodium dodecyl sulfate (SDS); 20% glycerol; and 10% 2-mercaptoethanol) (Wako) for 5 min, resolved using 10% SDS-polyacrylamide gel electrophoresis, and analyzed using western blotting.
2.8. Pulmonary Fibrosis Induction in Mice
CD1 mice were obtained from Charles River Laboratories Japan (Yokohama, Japan) and used in accordance with our institutional guidelines. Bleomycin (Wako) was dissolved in PBS and instilled intratracheally at a dose of 1.5 mg/kg body weight, and the lungs were then removed on day 14 after treatment. For western blot analysis, lung tissue lysates were prepared using CelLytic MT (Sigma) containing a cocktail of protease inhibitors.
This study was performed in compliance with the Rules and Regulations of the Animal Care and Use Committee of Kinki University Faculty of Medicine and according to the Guide for the Care and Use of Laboratory Animals of Kinki University Faculty of Medicine.
3. RESULTS
3.1. Detection of Interaction between ITGA11 and CIB1 in a Two-Hybrid Screen
To identify proteins that interact with ITGA11, a twohybrid screen was performed using the transmembranecytoplasmic domain of ITGA11 fused to the LexA DNAbinding domain. Using this bait, we screened 1 × 106 independent transformants obtained from the human lung fibroblast cDNA library and identified CIB1 (GenBank accession no. U82226) as one of the binding partners of ITGA11. Two independent clones of full-length CIB1 were isolated, namely, both CIB1 inserts in the prey vector pJG4-5 consisted of 191 amino acids. The interaction between the bait and prey was also confirmed by an interchange experiment (Figure 1).
The yeast two-hybrid system was used to identify specific protein regions of CIB1 that interact with ITGA11. Two prey constructs were generated by individually cloning cDNA fragments corresponding to the N-terminal (Met 1-Ser 99) and C-terminal (Asp 100-Leu 191) domains of CIB1 protein into the pJG4-5 vector. The C-terminal domain of CIB1 bound to the cytoplasmic domain of ITGA11; however, the N-terminal domain of CIB1 did not (Figure 2).
3.2. Interaction between ITGA11 and CIB1 in Vivo
To further investigate the relevance of the interaction between ITGA11 and CIB1, we performed coimmunoprecipitation experiments. Full-length ITGA11 and CIB1 were coexpressed in COS-7 cells. The CIB1 construct incorporated an HA-tag epitope fused to its C-terminus. Thus, we used the anti-HA antibody to immunoprecipitate CIB1 protein. As shown in Figure 3, anti-HA antibody was able to coimmunoprecipitate ITGA11.
3.3. The Analysis of the Interaction between ITGA Isoforms and CIB1
To examine the interaction between different ITGA iso-
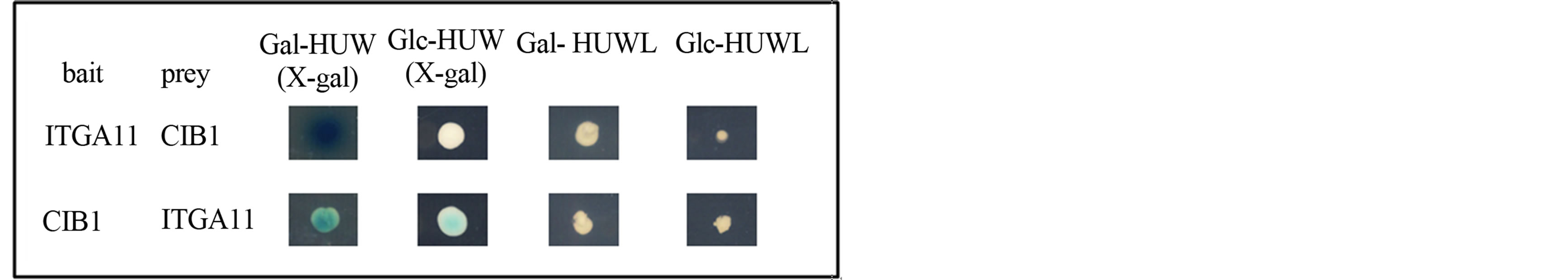
Figure 1. Yeast two-hybrid interactions between ITGA11 and CIB1. The binding also occurred when the bait and prey were swapped over in the two-hybrid assay. In the bait-prey interchange experiments, we used a cDNA encoding full-length CIB1 and a cDNA encoding the amino acid sequence of the carboxyl-terminal 34 residues of ITGA11 as bait and prey, respectively. Interaction is indicated by growth of the transformants on a His−Ura−Trp−Leu− galactose (Gal-HUWL) plate but not on Glc-HUWL plate, with blue colonies on the X-gal (Gal), but not the X-gal (Glc), plate.
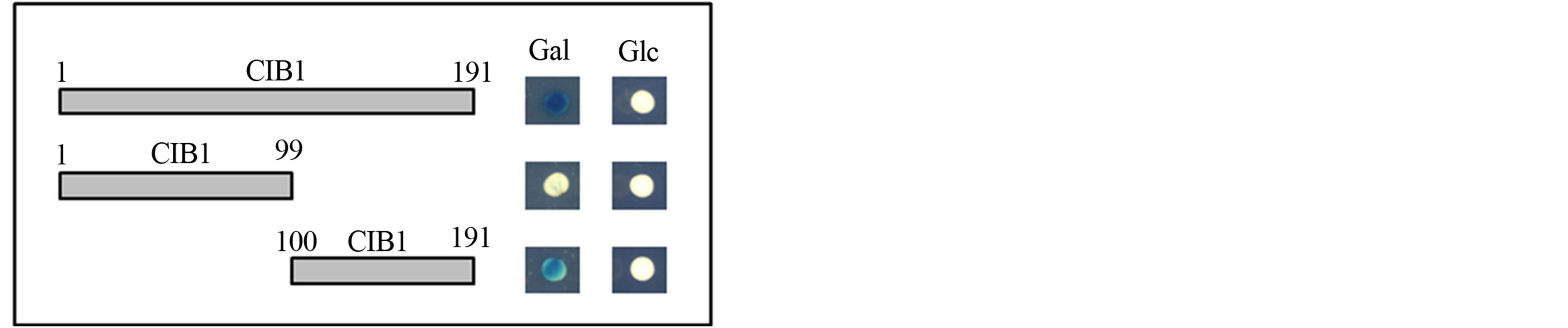
Figure 2. Interactions between the ITGA11 transmembrane-cytoplasmic domain and CIB1 fragments. CIB1 truncated constructs (size indicated by amino acid residues) were individually analyzed for interaction with the transmembrane-cytoplasmic domain of ITGA11. The CIB1 prey constructs corresponding to amino acid residues 1-191 (full-length) and 100-191 (C-terminal domain) showed β-galactosidase expression when grown with Gal, but not Glc, as carbon source. The CIB1 construct corresponding to amino acids 1-99 (N-terminal domain) had no β-galactosidase activity on both Gal and Glc.

Figure 3. Immunoprecipitation analysis of the interaction between ITGA11 and CIB1. HAtagged CIB1 was cotransfected with ITGA11 into COS-7 cells and immunoprecipitated with anti-HA antibody (HA) or normal rabbit IgG (control). The rat anti-human ITGA11 antibody detected coprecipitated ITGA11 at the band of 160 kDa. IP, immunoprecipitation; WB, western blot; WCL, whole cell lysates.
forms related to collagen binding and CIB1, a yeast twohybrid assay was performed using each transmembrane– cytoplasmic domain of ITGA as bait and full-length CIB1 as prey. The interactions are summarized in Figure 4. ITGA1, 11, and IIb bound to CIB1; however, ITGA2, 5, and 10 did not.
3.4. The Effects of CIB1 on α-SMA and FN Expression in MRC-5 Cells
To investigate the influence of CIB1 on fibrotic change in MRC-5 cells, CIB1 was overexpressed in the cells. After transfection treatments, the cells were cultured for 48 h and collected for immunoblot assay. As shown in Figure 5, CIB1 overexpression decreased α-SMA and FN expression.
3.5. CIB1 Protein Expression in Bleomycin-induced Lung Fibrosis in Mice
To determine CIB1 protein expression in normal and fibrotic lungs, we examined the lung tissue lysates of bleomycin-treated mice using western blotting. Western blot analysis revealed that CIB1 expression was readily detectable in lysates from bleomycin-treated mice but barely detectable in lysates prepared from untreated mice (Figure 6).

Figure 4. Yeast two-hybrid interactions between CIB1 and selected ITGA isoforms. The assay was performed as previously described in the legend of Figure 1, and the results obtained for growth on GalHUW + X-gal media (Gal) and Glc-HUW + X-gal media (Glc) are shown. ITGA1, 11, and IIb bound to CIB1 but ITGA2, 5, and 10 did not.
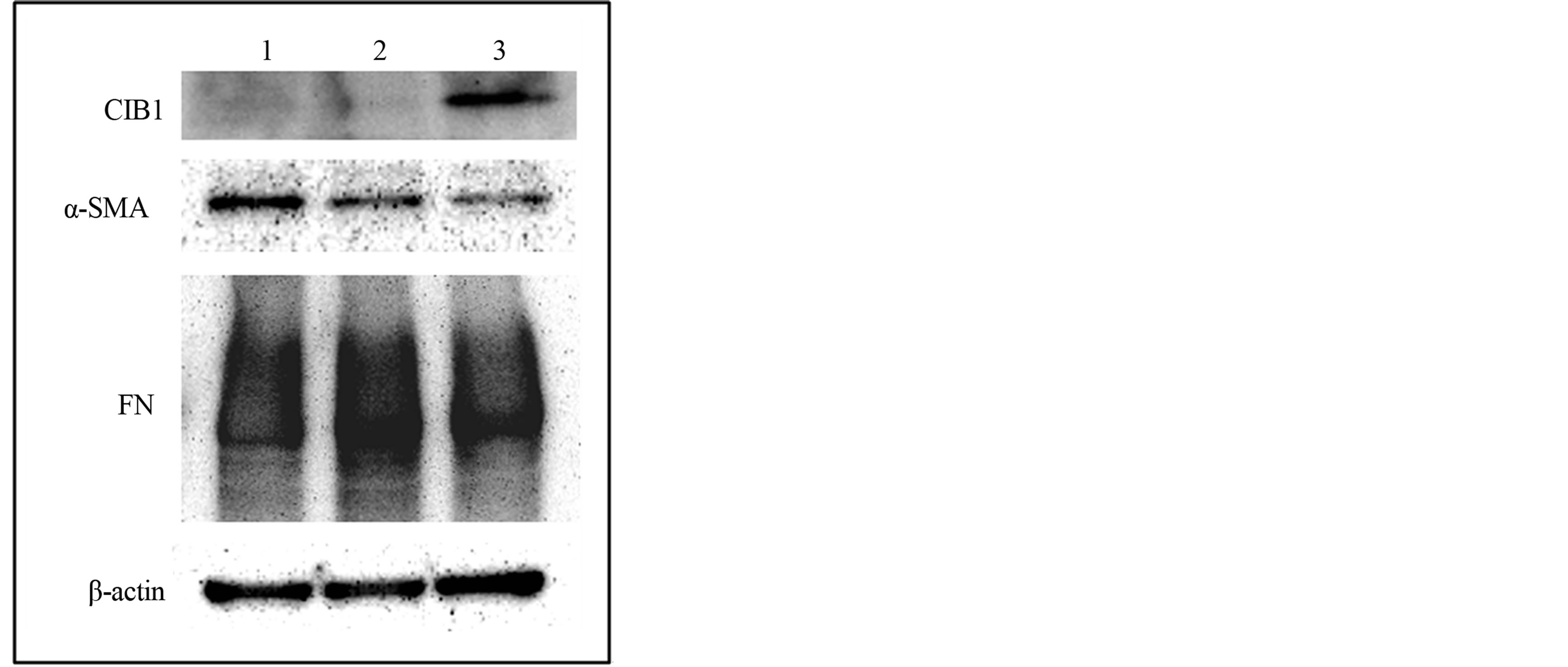
Figure 5. Effects of CIB1 on α-SMA and fibronectin (FN) expression in MRC-5 cells. Western blot analysis of intact (lane 1) or empty (lane 2) vector or CIB1 expression (lane 3) vector-transfected MRC-5 cells was performed with primary antibodies against CIB1, α-SMA, FN, and β-actin. When CIB1 was overexpressed in MRC-5 cells, α-SMA and FN expression decreased.

Figure 6. Western blot analysis of CIB1 and β-actin expression in lung tissue lysates from control and bleomycin-treated mice. Con, lung tissue lysates from control mice; BM, lung tissue lysates from bleomycin-treated mice. CIB1 expression in lung tissue was increased in bleomycin-treated mice. Similar results were obtained for two experiments.
4. DISCUSSION
We demonstrate here that ITGA11 binds to CIB1 (Figure 1), which is a 22-kDa protein present in various tissues [17,18]. CIB1 belongs to the calcium-binding protein family and contains two EF-hand domains in its C-terminal domain [19]. Using the yeast two-hybrid system, we demonstrate that ITGA11 interacts with the Cterminal domain of CIB1 but not with the N-terminal domain (Figure 2). These results suggest that the Cterminal domain of CIB1 has an amino acid sequence for essential binding to ITGA11, and that the calcium-binding domain of CIB1 may be necessary for this interaction. Although CIB1 is myristoylated at its N-terminus [20], the two-hybrid analysis presented here shows that this modification is not required for binding to ITGA11. Moreover, the results obtained from the yeast two-hybrid system were corroborated by immunoprecipitation experiments using full-length proteins (Figure 3). ITGA11 and CIB1 are distributed among various organs and are expressed together in tissues such as heart, lung, small intestine, testis, and prostate [18,21]. Taken together, these findings indicate that ITGA11 and CIB1 may be associated in vivo.
Integrins are heterodimeric cell surface and transmembrane proteins consisting of α and β subunits. Four different collagen-binding integrins, i.e., α1β1, α2β1, α10β1, and α11β1, are known [22,23]. To determine whether CIB1 interacts with other collagen-binding integrins, except for ITGA11, we conducted a two-hybrid assay between various ITGAs and CIB1. Consistent with the studies of Naik et al. [19], we observed binding of ITGAIIb to CIB1 (Figure 4). In addition, we have identified an interaction between ITGA1 and CIB1 (Figure 4). ITGA1, 11 and IIb contain the common amino acid sequence motif K-x (I or L or V)-G-F-F-x (K or R) in their membrane-proximal cytoplasmic region; moreover, this sequence is also observed in ITGA2 and 5 (Figure 7). Nevertheless, these ITGAs showed no interaction with CIB1. In contrast, the amino acid sequences of the overall cytoplasmic domain of ITGA1, 11, IIb share little similarity, and each cytoplasmic domain completely differs in length. These findings indicate that there are variations among the amino acid sequences of the CIB1 that mediate binding to ITGA1, 11, and IIb.
As for the interaction between CIB1 and integrins αvβ3 and αIIbβ3, β3 activation motifs are not involved in the binding [24]. Our study also showed that the interaction between CIB1 and ITGAs was independent of β subunit. Freeman, Jr. et al. predicted that CIB1 could bind all 24 known integrin heterodimers from competitive inhibition solid-phase binding assays [25]. However, obtained results from two-hybrid analysis in our study revealed that CIB1 does not bind to ITGA2, 5 and 10 (Figure 4). This apparent difference between two studies seems to be due to the sensitivity of the methods used.
We next evaluated the role of CIB1 in MRC-5 myofibroblasts by overexpressing CIB1 and assessed the effect of the overexpressing by measuring the levels of α-SMA and FN expression. When CIB1 was overexpressed, the expression of α-SMA and FN was reduced compared with the controls; however, β-actin expression was not significantly changed (Figure 5). Increased α-SMA expression is sufficient to enhance fibroblast contractile activity, and the level of α-SMA expression is enhanced by fibrotic progression (e.g. TGF-β treatment) [1,26]. FN is an extracellular matrix protein that is present with collagen in fibrotic tissues [27,28]. Thus, the results obtained from the CIB1 overexpressing experiments suggest that CIB1 may be involved in suppressing fibrotic changes in MRC-5 myofibroblasts.
It has been reported that Cib1 null mice show a marked reduction in myocardial hypertrophy, fibrosis, and cardiac dysfunction [29]. In contrast, perivascular fibrosis was increased in melanoma tumors that developed in CIB1-KO mice [30]. To assess the role of CIB1 in tissue fibrosis, we analyzed CIB1 expression in the lung tissues of bleomycin-treated mice. As shown in Figure 6, the level of CIB1 expression was significantly higher in bleomycin-treated mice compared with controls. Bleomycin is an antitumor agent that can cause lung damage as a side effect by inducing pulmonary fibrosis [31]. Collectively, these results imply that CIB1 may be induced to suppress the bleomycin-induced pulmonary fibrosis.
In conclusion, using the yeast two-hybrid system, we indicated that the cytoplasmic domain of ITGA11 binds to CIB1 and demonstrated that the C-terminal domain of CIB1 is required for this interaction. This interaction was also confirmed by a coimmunoprecipitation analysis. Our present study indicates that CIB1 may regulate fibrosis in concert with ITGA11. Therefore, CIB1 may serve as a
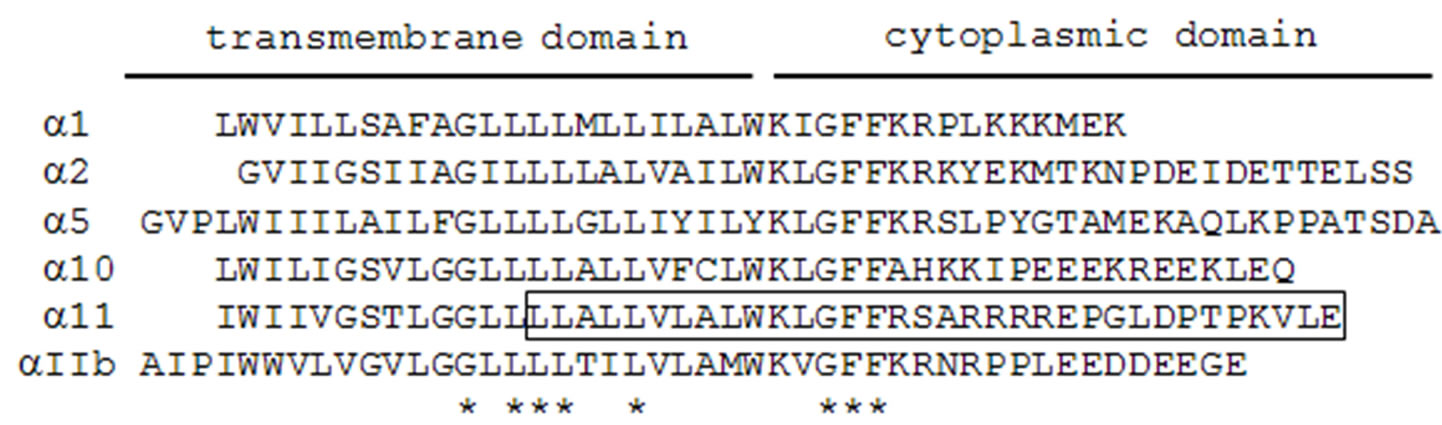
Figure 7. Alignment of the amino acid sequences of integrin α subunits. The peptide sequence in ITGA11 used as bait for two-hybrid screening is boxed. Asterisks indicate identical residues in all ITGA isoforms.
new target for fibrosis therapy.
ACKNOWLEDGEMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research (C) (23592596) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- Honda, E., Yoshida, K. and Munakata, H. (2010) Transforming growth factor-β upregulates the expression of integrin and related proteins in MRC-5 human myofibroblasts. Tohoku Journal of Experimental Medicine, 220, 319-327. http://dx.doi.org/10.1620/tjem.220.319
- Border, W.A. and Noble, N.A. (1994) Transforming growth factor β in tissue fibrosis. New England Journal of Medicine, 331, 1286-1292. http://dx.doi.org/10.1056/NEJM199411103311907
- Schuppan, D., Krebs, A., Bauer, M. and Hahn, E.G. (2003) Hepatitis C and liver fibrosis. Cell Death & Differentiation, 10, S59-S67. http://dx.doi.org/10.1038/sj.cdd.4401163
- Akhmetshina, A., Palumbo, K., Dees, C., Bergmann, C., Venalis, P., Zerr, P., Horn, A., Kireva, T., Beyer, C., Zwerina, J., Schneider, H., Sadowski, A., Riener, M.O., MacDougald, O.A., Distler, O., Schett, G. and Distler, J.H. (2012) Activation of canonical Wnt signalling is required for TGF-β-mediated fibrosis. Nature Communications, 3, 735. http://dx.doi.org/10.1038/ncomms1734
- Chung, H.J., Steplewski, A., Chung, K.Y., Uitto, J. and Fertala, A. (2008) Collagen fibril formation. A new target to limit fibrosis. Journal of Biological Chemistry, 283, 25879-25886. http://dx.doi.org/10.1038/ncomms1734
- Iredale, J.P., Thompson, A. and Henderson, N.C. (2013) Extracellular matrix degradation in liver fibrosis: Biochemistry and regulation. Biochimica et Biophysica Acta, 1832, 876-883. http://dx.doi.org/10.1016/j.bbadis.2012.11.002
- Honda, E., Park, A.M., Yoshida, K., Tabuchi, M. and Munakata, H. (2013) Myofibroblasts: Biochemical and proteomic approaches to fibrosis. Tohoku Journal of Experimental Medicine, 230, 67-73. http://dx.doi.org/10.1620/tjem.230.67
- Gullberg, D., Velling, T., Sjoberg, G. and Sejersen, T. (1995) Up-regulation of a novel integrin α-chain (αmt) on human fetal myotubes. Developmental Dynamics, 204, 57-65. http://dx.doi.org/10.1002/aja.1002040108
- Velling, T., Kusche-Gullberg, M., Sejersen, T. and Gullberg, D. (1999) cDNA cloning and chromosomal localization of human α11 integrin. A collagen-binding, I domain-containing, β1-associated integrin α-chain present in muscle tissues. Journal of Biological Chemistry, 274, 25735-25742. http://dx.doi.org/10.1074/jbc.274.36.25735
- Zhu, C.Q., Popova, S.N., Brown, E.R., Barsyte-Lovejoy, D., Navab, R., Shih, W., Li, M., Lu, M., Jurisica, I., Penn, L.Z., Gullberg, D. and Tsao, M.S. (2007) Integrin α11 regulates IGF2 expression in fibroblasts to enhance tumorigenicity of human non-small-cell lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America, 104, 11754-11759. http://dx.doi.org/10.1073/pnas.0703040104
- Popov, C., Radic, T., Haasters, F., Prall, W.C., Aszodi, A., Gullberg, D., Schieker, M. and Docheva, D. (2011) Integrins α2β1 and α11β1 regulate the survival of mesenchymal stem cells on collagen I. Cell Death and Disease, 2, e186. http://dx.doi.org/10.1038/cddis.2011.71
- Popova, S.N., Barczyk, M., Tiger, C.F., Beertsen, W., Zigrino, P., Aszodi, A., Miosge, N., Forsberg, E. and Gullberg, D. (2007) α11β1 integrin-dependent regulation of periodontal ligament function in the erupting mouse incisor. Molecular and Cellular Biology, 27, 4306-4316. http://dx.doi.org/10.1128/MCB.00041-07
- Bystrom, B., Carracedo, S., Behndig, A., Gullberg, D. and Pedrosa-Domellof, F. (2009) α11 integrin in the human cornea: Importance in development and disease. Investigative Ophthalmology & Visual Science, 50, 5044- 5053. http://dx.doi.org/10.1167/iovs.08-3261
- Talior-Volodarsky, I., Connelly, K.A., Arora, P.D., Gullberg, D. and McCulloch, C.A. (2012) α11 integrin stimulates myofibroblast differentiation in diabetic cardiomyopathy. Cardiovascular Research, 96, 265-275. http://dx.doi.org/10.1093/cvr/cvs259
- Barczyk, M.M., Lu, N., Popova, S.N., Bolstad, A.I. and Gullberg, D. (2013) α11β1 integrin-mediated MMP-13- dependent collagen lattice contraction by fibroblasts: Evidence for integrin-coordinated collagen proteolysis. Journal of Cellular Physiology, 228, 1108-1119. http://dx.doi.org/10.1002/jcp.24261
- Yoshida, K., Suzuki, Y., Saito, A., Fukuda, K., Hamanishi, C. and Munakata, H. (2005) Aggrecanase-1 (ADAMTS- 4) interacts with α1-antitrypsin. Biochimica et Biophysica Acta, 1725, 152-159. http://dx.doi.org/10.1016/j.bbagen.2005.06.009
- Shock, D.D., Naik, U.P., Brittain, J.E., Alahari, S.K., Sondek, J. and Parise, L.V. (1999) Calcium-dependent properties of CIB binding to the integrin αIIb cytoplasmic domain and translocation to the platelet cytoskeleton. Biochemical Journal, 342, 729-735. http://dx.doi.org/10.1016/j.bbagen.2005.06.009
- Whitehouse, C., Chambers, J., Howe, K., Cobourne, M., Sharpe, P. and Solomon, E. (2002) NBR1 interacts with fasciculation and elongation protein zeta-1 (FEZ1) and calcium and integrin binding protein (CIB) and shows developmentally restricted expression in the neural tube. European Journal of Biochemistry, 269, 538-545. http://dx.doi.org/10.1046/j.0014-2956.2001.02681.x
- Naik, U.P., Patel, P.M. and Parise, L.V. (1997) Identification of a novel calcium-binding protein that interacts with the integrin αIIb cytoplasmic domain. Journal of Biological Chemistry, 272, 4651-4654. http://dx.doi.org/10.1074/jbc.272.8.4651
- Stabler, S.M., Ostrowski, L.L., Janicki, S.M. and Monteiro, M.J. (1999) A myristoylated calcium-binding protein that preferentially interacts with the Alzheimer’s disease presenilin 2 protein. Journal of Cell Biology, 145, 1277-1292. http://dx.doi.org/10.1083/jcb.145.6.1277
- Lehnert, K., Ni, J., Leung, E., Gough, S.M., Weaver, A., Yao, W.P., Liu, D., Wang, S.X., Morris, C.M. and Krissansen, G.W. (1999) Cloning, sequence analysis, and chromosomal localization of the novel human integrin α11 subunit (ITGA11). Genomics, 60, 179-187. http://dx.doi.org/10.1006/geno.1999.5909
- Leitinger, B. and Hohenester, E. (2007) Mammalian collagen receptors. Matrix Biology, 26, 146-155. http://dx.doi.org/10.1016/j.matbio.2006.10.007
- Barczyk, M., Carracedo, S. and Gullberg, D. (2010) Integrins. Cell and Tissue Research, 339, 269-280. http://dx.doi.org/10.1007/s00441-009-0834-6
- Yamodo, I.H. and Blystone, S.D. (2012) Calcium integrin binding protein associates with integrins αVβ3 and αIIbβ3 independent of β3 activation motifs. CellBio (Irvine, Calif), 1, 30-37. http://dx.doi.org/10.4236/cellbio.2012.12004
- Freeman Jr., T.C., Black, J.L., Bray, H.G., Dagliyan, O., Wu, Y.I., Tripathy, A., Dokholyan, N.V., Leisner, T.M. and Parise, L.V. (2013) Identification of novel integrin binding partners for calcium and integrin binding protein 1 (CIB1): Structural and thermodynamic basis of CIB1 promiscuity. Biochemistry, 52, 7082-7090. http://dx.doi.org/10.1021/bi400678y
- Hinz, B., Celetta, G., Tomasek, J.J., Gabbiani, G. and Chaponnier, C. (2001) α-smooth muscle actin expression upregulates fibroblast contractile activity. Molecular Biology of the Cell, 12, 2730-2741. http://dx.doi.org/10.1091/mbc.12.9.2730
- Kadler, K.E., Hill, A. and Canty-Laird, E.G. (2008) Collagen fibrillogenesis: Fibronectin, integrins, and minor collagens as organizers and nucleators. Current Opinion in Cell Biology, 20, 495-501. http://dx.doi.org/10.1016/j.ceb.2008.06.008
- Klingberg, F., Hinz, B. and White, E.S. (2013) The myofibroblast matrix: Implications for tissue repair and fibrosis. Journal of Pathology, 229, 298-309. http://dx.doi.org/10.1002/path.4104
- Heineke, J., Auger-Messier, M., Correll, R.N., Xu, J., Benard, M.J., Yuan, W., Drexler, H., Parise, L.V. and Molkentin, J.D. (2010) CIB1 is a regulator of pathological cardiac hypertrophy. Nature Medicine, 16, 872-879. http://dx.doi.org/10.1038/nm.2181
- Zayed, M.A., Yuan, W., Chalothorn, D., Faber, J.E. and Parise, L.V. (2010) Tumor growth and angiogenesis is impaired in CIB1 knockout mice. Journal of Angiogenesis Research, 2, 17. http://dx.doi.org/10.1186/2040-2384-2-17
- Moeller, A., Ask, K., Warburton, D., Gauldie, J. and Kolb, M. (2008) The bleomycin animal model: A useful tool to investigate treatment options for idiopathic pulmonary fibrosis? International Journal of Biochemistry & Cell Biology, 40, 362-382. http://dx.doi.org/10.1016/j.biocel.2007.08.011
LIST OF ABBREVIATIONS
α-SMA, α-smooth muscle actin;
CIB, calciumand integrin-binding protein;
FN, fibronectin;
Gal, galactose;
Glc, glucose;
ITGA, integrin α;
PBS, phosphate-buffered saline;
SDS, sodium dodecyl sulfate;
TGF-β, transforming growth factor-β;
X-gal, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
NOTES
*Corresponding author.

