American Journal of Molecular Biology
Vol.3 No.2(2013), Article ID:30852,8 pages DOI:10.4236/ajmb.2013.32017
Clinical applications of molecular profiling in colorectal cancer: Review of the literature
![]()
Department of Surgery, National University of Ireland, Galway, Ireland
Email: rashmed1111@gmail.com
Copyright © 2013 Elrasheid A. H. Kheirelseid et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 10 February 2013; revised 27 March 2013; accepted 25 April 2013
Keywords: Molecular Profiling in Colorectal Cancer; Gene Expression in Colorectal Cancer; Colorectal Cancer Genetics; Colorectal Cancer
ABSTRACT
Despite the developments in the diagnostic and management strategies, a considerable number of colorectal cancer (CRC) patients present with disease recurrence after curative surgery. Moreover; there are no reliable indicators to determine the prognosis and response of CRC patients to therapy. By harnessing recent technological advances in molecular profiling techniques, it is anticipated that greater insight to the various combinations of genetic events or alternative pathways underlying carcinogenesis will be gained. By carrying out literature search, we were able to identify a comprehensive list of genes with high differential expression patterns in colorectal cancer that could serve as molecular markers to complement existing histopathological factors in diagnosis, follow up and therapeutic strategies for individualized care of patients.
1. INTRODUCTION
At the molecular level, activation of oncogenes and inactivation of tumour suppressor genes [1] are processes known to be involved in colorectal carcinogenesis. Additionally, abrogation of mismatch repair systems [2] contributes to some colorectal cancers. Nevertheless, exactly how those genetic alterations bring about the development and progression of colorectal carcinomas remains to be resolved. To complicate the picture, accumulations of mutant genes in neoplasms tend to be accompanied by other genetic and epigenetic changes including loss of heterozygosity, inactivation of important genes by methylation or loss of imprinting [3] or gene amplifications, all of which can alter gene expression profiles. Therefore, genome wide monitoring of gene expression is of great importance if we are to disclose the numerous and diverse events associated with carcinogenesis. Molecular profiling, a tool of genome monitoring, is an attempt to identify the different combinations of genetic events or alternative pathways that may be represented by cancers of a similar type. Examples of the type styles are provided throughout this document and are identified in italic type, within parentheses, following the example. Some components, such as multi-leveled equations, graphics, and tables are not prescribed, although the various table text styles are provided. The formatter will need to create these components, incorporating the applicable criteria that follow.
2. GENE EXPRESSION PROFILING IN CRC
Molecular biology represents one of the most interesting topics in medical oncology, because it provides a global and detailed view on the molecular changes involved in tumour progression, leading to a better understanding of the carcinogenesis process, to discovering new prognostic markers and novel therapeutic targets. Despite of clinical and pathological parameters are available for the classification and prognostic stratification of cancer, they may be inadequate in everyday practice due to the great biologic and genetic heterogeneity of this multiform disease.
CRC represents an interesting field of molecular profiling research for several reasons: CRC is considered a biological model of tumourigenesis, because clinical progression from adenoma to early stage carcinoma until advanced stage carcinoma seems to parallel distinctive molecular alterations [4]. In addition; traditional clinical and pathological parameters are not always sufficient to discriminate high risk from low risk CRC and validated molecular markers with prognostic value are still not available. The studies on molecular profiling in CRC have mainly focused on carcinogenesis process, disease prognosis prediction and therapeutic targets and response prediction. (Table 1)
2.1. Diagnostic and Prognostic Biomarkers
The application of gene expression profiling on carcinogenesis studies purposes to identify specific alterations on gene expression according to tumour development and to diagnose and classify tumours on the basis of molecular features. Studies comparing gene expression between normal mucosa, adenoma and carcinoma or between primary tumour and metastases, as well as between left-side and right-side tumours are performed, in order to discover distinctive genetic signatures belonging to each. Furthermore; studies on prognosis prediction aim to identify specific alterations to the gene expression profile that may be useful to discriminate high risk from low risk CRC, to provide a molecular stratification according to the clinical outcome and to predict the metastatic potential of the primary tumour.
Table 1. Genes that are consistently represented in CRC literature. Of many published studies on colorectal gene expression profiling, little correlation exists between validated candidate genes associating with disease status. Some candidate genes are consistently represented in the literature however, examples are shown below.
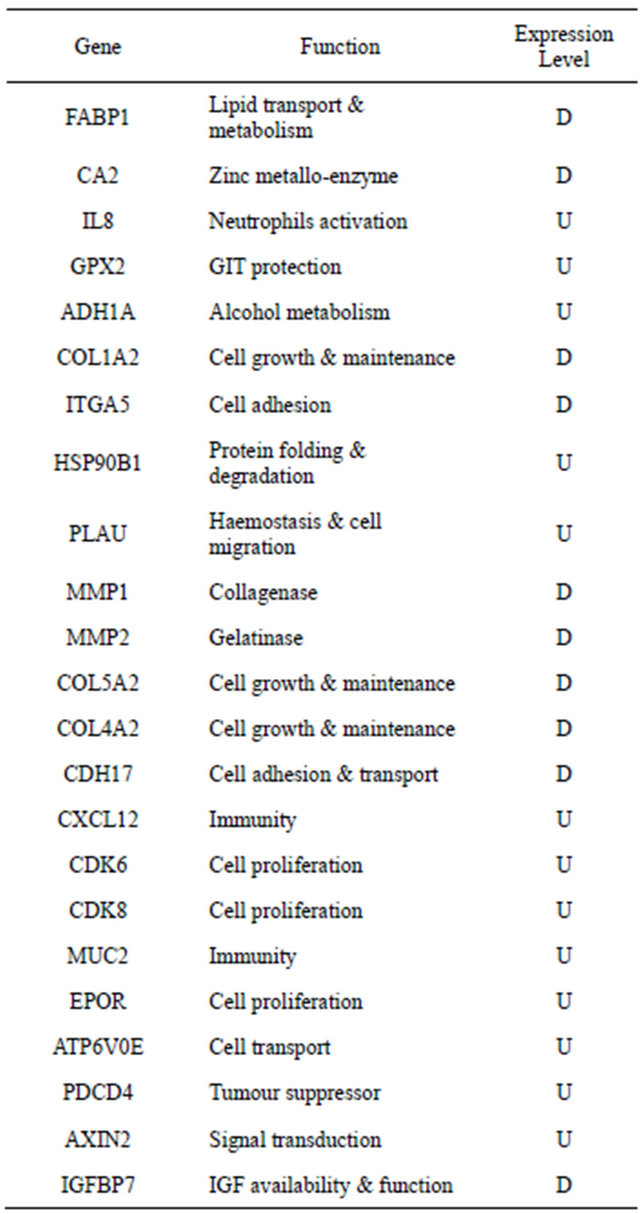
Several studies were set to investigate the difference in gene expression levels between tumour and normal colorectal tissues. In 1999, Alon and colleagues reported a clustered data set of 2000 genes able to separate 22 normal and 40 tumour colon tissues with the highest minimal intensity across samples. Subsequently many studies reported other sets of genes that were differentially expressed between cancer and normal tissue and therefore potentially involved in the development of colorectal carcinogenesis [5-9]. In addition, some studies reported significant differences in gene expression profile between adenoma and normal mucosa, suggesting that different mechanisms of development of these precancerous lesions may exist [10,11]. Furthermore, and in order to clarify the molecular modifications underlying the development of metastases, some studies compared the gene expression profile of primary tumours with their corresponding metastases [12-16]. Agrawal et al. reported that among all genes associated with disease progress, osteopontin expression seemed to be the leading candidate [12]. Moreover; Yanagawa et al. [14] analysed genome-wide expression profiles of 10 primary CRCs and their corresponding liver metastasis and identified 40 genes whose expression was commonly up-regulated in metastatic lesions, and 7 that were commonly down-regulated. On the other hand; Watanabe et al. studied 89 CRC patients to identify a set of discriminating genes that can be used for characterization and prediction of lymph node metastasis and identified 73 genes in which expression was significantly different between patients with and without lymph node metastasis [17]. Using this gene set, they established a model to predict the presence of lymph node metastasis with an accuracy of 88.4%. In addotion, the controversial data on the benefit of adjuvant chemotherapy in stage II CRC led to the identification of molecular prognostic factors that may identify stage II CRC patients who develop disease recurrence and may benefit by adjuvant treatment. Wang et al. studied the gene expression profile in this set of patients and, using two supervised class prediction approaches of analysis, they identified a 23-genes signature that may predict recurrence in stage II CRC with 78% accuracy [18].
Some studies have also investigated differences in gene expression between CRC of the right side and left side, due to their epidemiological, morphological and pathogenetic diversity and found distinct profiles according to the anatomical stratification. Birkenkamp-Demtroder et al. [19] investigated the difference in gene expression between the caecum vs sigmoid and rectosigmoid and identified 58 genes to be differentially expressed between the normal mucosa of caecum and the sigmoid and rectosigmoid.nd the sigmoid and rectosigmoid.
2.2. Therapeutic Targets and Treatment Response Prediction
While gene expression profiling has been widely applied to CRC for diagnosis, classification and prognosis prediction based on molecular patterns of expression, its application to response prediction to medical treatment is still lacking reliable results due to few currently available studies [20-24]. In a panel of 30 colon carcinoma cell lines Mariadason et al. identified 420 genes correlated with response to 5-fluorouracil (5-FU) and involved in two main biological processes, DNA replication and repair and protein processing/targeting [22]. The predictive value of 50 genes best correlated with 5-FU response was subsequently validated using a leave one out cross validation approach and it was higher than the traditional markers, such as thymidylate synthase, thymidine phosphorylase, mismatch repair and p53 status. Furthermore they also found that 149 genes best-correlated with CPT-11-induced apoptosis significantly predicted response of colon cancer cell lines to this agent. In addition; Del Rio et al. analyzed gene expression profile of 21 primary advanced CRC tissues, in order to identify an expression pattern that could predict response to leucovorin, fluorouracil and irinotecan as first-line treatment: 14 genes were found expressed differently between responders and non responders and were able to predict treatment response with 95% accuracy [23]. In the same year Khambata-Ford et al. investigated gene expression pattern of metastatic biopsies of 80 advanced CRC patients treated with cetuximab to identify genes whose expression correlates with best clinical response [24]. They found that, among 629 genes expressed differently between 25 patients with disease control and 55 non responders, the top candidate markers based on lowest p value were epiregulin and amphiregulin, both ligands for epidermal growth factor receptor (EGFr), suggesting that these markers could select patients for cetuximab therapy.
Some studies evaluated the ability of gene expression profiling for predicting response of advanced rectal cancer (RC) to preoperative chemoradiotherapy [20,25-27]. Ghadimi et al. analyzed gene expression signatures of biopsies from 30 locally advanced RC and found 54 genes differentially expressed between responders and non responders [20]. Kim et al. reported 261 genes differentiated between 20 partial response and 11 complete response patients affected by locally advanced RC treated with preoperative chemoradiotherapy. In their study 95 genes predicted complete and partial response with an 84% accuracy [25]. Similarly another study identified a gene expression signature of 42 genes that was able to distinguish responder from non responder locally advanced RC patients with a 71% accuracy [26]. Recently; Spitzner et al. were able to identify a gene expression signature for chemoradiosensitivity of colorectal cancer cells [27]. They exposed 12 colorectal cancer cell lines to of 5-fluorouracil and radiation therapy. The differences in treatment sensitivity were then correlated with the pretherapeutic gene expression profiles of these cell lines. Their data have suggested a potential relevance of the insulin and Wnt signalling pathways for treatment response, and they also identified STAT3, RASSF1, DOK3, and ERBB2 as potential therapeutic targets [27].
Although colorectal cancer (CRC) is still one of the leading causes of cancer related death, the introduction of new therapeutic options like oxaliplatin and irinotecan in addition to 5-fluorouracil, the standard therapeutic for CRC has increased the overall survival of affected patients from 10 to 18 - 24 months. Furthermore, the “biological” therapeutics cetuximab, an IgG1 chimeric monoclonal antibody against epidermal growth factor receptor (EGFR), and bevacizumab, a monoclonal antibody against vascular endothelial growth factor (VEGF), have augmented the course of the disease and brought in the new era of targeted therapy against cancer specific molecular pathways [28-31]. Although these biologicals have entered clinical routine due to their encouraging results, their effect has been shown to be limited due to adaptation or previously existing resistance of tumour cells. This has been clearly shown in the case of patients with mutations of K-ras, which lead to resistance against cetuximab. Therefore, several new pathways are currently investigated for therapeutic targeting in CRC. These include WNT-signaling, downstream mediators of EGFR as the mitogen-activated protein kinase (MAPK) or the phosphatidylinositol 3-kinase (PI3K)-pathway, the hypoxia response system involving hypoxia inducible factor-1 (HIF-1), mechanisms of tumour development following chronic inflammation, and many others [32].
3. MICRORNA EXPRESSION PROFILING IN CRC
MiRNAs have recently emerged as an exciting new class of disease biomarker with further potential as the next generation of cancer therapeutics. Although elucidating their mechanisms of action is still in its infancy, the discovery of miRNAs has uncovered an entirely new and exciting repertoire of molecular factors upstream of gene expression, with great potential for new developments in current diagnostic and therapeutic strategies in the management of cancer patients. MiRNAs are small 19 to 22 nucleotide sequences of RNA found in both prokaryotes and eukaryotes that are intimately involved in cell differentiation, cell cycle progression, and apoptosis. Hence, miRNAs may be useful tools for characterizing specific cancers and for determining patient prognosis and response to therapy. The study of miRNA has been extended into many types of cancer, including leukemias, lung, breast, and colon cancer. The first description of miRNA appeared in 1993 by Lee et al., who proved that lin-4 is involved in controlling the temporal progression of cell differentiation in C. elegans [33]. Discoveries of other miRNAs that regulate apoptosis, proliferation, and differentiation in Drosophila, mice, and humans soon followed [34,35]. Calin et al. [36] published the first study to link miRNAs to cancer in 2002. These authors demonstrated that miR-15 and miR-16 are located on chromosome 13 in a position where deletion of a putative tumour suppressor, known to be associated with greater than half of chronic lymphocytic leukemia cases, was identified. Researchers have proposed that specific miRNA expression patterns could help identify human solid tumours, suggest patient prognosis, and even represent a novel molecular target for cancer treatment.
3.1. miRNA Expression and Function in CRC
In 2003 Michael et al. published the first report to profile miRNAs in CRC. Using cloning technology followed by Northern blotting, they reported consistently reduced accumulation of the specific mature miR-143 and miR- 145 in the adenomatous and carcinoma stages of colorectal tumours [37]. Thereafter, several studies were set to investigate the role of miRNAs in colorectal cancer. MiRNAs with tumour suppressor properties which are under-expressed in CRC specimens, and thus potentially function as tumour suppressors, include miR-31, miR- 34a, miR-96, miR-143, miR-145, and let-7a [38,39]. MiR-34a is a well described tumour suppressor miRNA which regulates the p53 pathway and when overexpressed induces apoptosis and acute senescence. Conversely reduction of miR-34 expression and function attenuates p53-mediated cell death and is therefore implicated in tumourigenesis, including initiation of CRC [40, 41]. It is postulated therefore that loss of miR-34a expression in colorectal biopsy specimens may be an early biomarker of CRC. Other miRNAs like miR-31, miR-96, miR-135b, and miR-183 have been found to be upregulated in colorectal neoplasm. The transcription factor CHES1 which is involved in repressing apoptosis is a potential target of miR-96. Schetter et al. identified miRNAs which can distinguish cancerous from normal colon tissue, with miR-21 over-expressed in 87% of colon cancers [38]. Subsequent mechanistic investigations provide evidence for the oncogenic role of miR-21 in CRC by demonstrating how it suppresses the cell cycle regulator CDC25A [42], and can also target and repress the tumour suppressor gene PDCD4 thus inducing invasion, intravasation and metastatic potential [43]. MiR-21 may also target PTEN and TPM1. In addition; miR-135a and miR-135b are upregulated, and this upregulation correlates with reduced expression of the APC [44]. Moreover; miR-143 and miR-145 are both downregulated in colorectal cancer. The genes encoding these miRNAs are both located on 5q23, and these miRNAs possibly originate from the same primary miRNA [37, 39]. MiR-126 promotes cell proliferation through modulation of phosphatidylinositol 3-kinase signaling [45]. MiR-133b is also downregulated, and one of its putative targets is KRAS [46], which is a member of the Ras family of proteins, that regulates signaling pathways involved in cellular proliferation, differentiation, and survival. Moreover; over-expression of the oncogenic miR-17-92 cluster is also implicated in the etiology of CRC, specifically in adenoma to adenocarcinoma progression.
3.2. Clinical Value of Mirna Expression in CRC
Accumulating evidence shows that miRNA expression patterns are unique to certain cancers and may be used clinically as prognostic and diagnostic factors as well as therapeutic targets.
3.2.1. Diagnostic and Prognostic Value
To test the function of miRNAs in the pathogenesis of CRC, expression of 156 miRNAs was measured in both tumour and normal tissues from patients with CRC and cell lines [46]. Expression of 13 miRNAs was significantly altered, and the most significantly dysregulated miRNAs were miR-31, miR-96, miR-133b, miR-135b, miR-145 and miR-183. In addition, the expression level of miR-31 was significantly correlated with tumour stage. Xi et al. [47] analysed patients with adenocarcinoma of the colon and rectum and found that tumours expressing high levels of miR-200c are correlated with poorer prognosis, regardless of tumour stage: approximately 12 months decreased survival compared with patients whose tumour expresses low levels of miR-200c. Furthermore; Arndt et al. [48] identified 37 miRNAs that were differentially expressed between CRC and normal tissues. They also reported that loss of miR-133a and gain of miR-224 are associated with tumour progression. Overexpression of miR-21 was shown in many reports to be associated with worse prognosis, lymph node and distant metastasis and poor response to chemotherapy in CRC. Moreover, Asangani et al. [43] reported that overexpression of miR-21causes tumour cells to invade and metastasize more aggressively when implanted into mouse models. In addition; a study by Motoyama and colleagues [49] showed that expression of miR-31, miR-183, miR-17-5p, miR-18a and miR-92 were significantly higher in tumour tissues compared to normal, while expression of miR-143 and miR-145 in cancer were lower than in normal tissues. They also showed that miR-18a expression was associated with poor disease prognosis. Moreover, miR-31 expression was positively related to advanced TNM stage and tumour invasion suggesting its role in CRC initiation and progression [50]. Of further interest, Lanza et al. [51] identified a molecular signature consisting of 27 differentially expressed genes, inclusive of 8 miRNAs that can correctly distinguish high microsatellite instable (MSI-H) vs microsatellite stable (MSS) colon cancers.
3.2.2. Therapeutic Potential
The synthesis and functions of miRNAs can be manipulated with various oligonucleotides that encode the sequences complementary to mature miRNAs [52]. Overexpression of miRNAs can be induced either by using synthetic miRNA mimics or chemically modified oligonucleotides [53]. Conversely, miRNAs can be silenced by antisense oligonucleotides and synthetic analogues of miRNAs [54,55]. Cross-sensitivity with endogenous miRNAs and lack of specificity for target miRNAs can cause non-specific side-effects with miRNA modulation therapy. However, the use of an effective delivery system and less toxic synthetic anti-miRNA oligonucleotides may minimize such side-effects. The role of miRNAs in pathogenesis of cancer makes them important targets for therapeutic intervention. Gene therapies may be designed to treat colorectal cancers and to block the progression of precursor lesions by manipulating the tumour suppressor or promoter miRNAs [56]. Such manipulation may control the tumour growth rate and have potential as a new therapy for both early and advanced cancers.
Studies have revealed that inhibition of miR-21 and miR-17-92 activity is associated with reduced tumour growth, invasion, angiogenesis and metastasis [57,58]. Targeting such miRNAs may help to prevent the recurrence of disease in high-risk tumours and may control the growth of advanced metastatic tumours. Overexpression of miR-21 is associated with low sensitivity and poor response to chemotherapy [56]. Its inhibition may improve the response to chemotherapy. In addition; some drugs were found to alter the expression of miRNAs. Rossi et al. reported a suggestive pattern of miRNA rearrangement in HT-29 and HCT-116 human colon cancer cell lines after exposure to 5-FU [59]. It leads to downregulation of miR-200, which is a microRNA known to inhibit a tumour-suppressor gene, protein tyrosine phosphatase, non-receptor type 12 (PTPN12) [60]. 5-FU treatment also induces up-regulation of miR-133a, which is thought to inhibit the proto-oncogene K-ras. Strangely, treatment with 5-FU also causes up-regulation of microRNA known to be mitogenic. To this, Rossi suggests that the cytotoxic effect of 5-FU induces cells to express anti-apoptotic factors, of which are these abnormally up-regulated, and tumourigenic, miRNAs. Besides these, 5-FU treatment leads to significant elevation in expression of many other miRNAs, and it remains to be seen what genes these miRNAs target. 5-FU induces p53 protein expression at a posttranscriptional level without correspondingly elevated mRNA level in a pattern that has become a hallmark for microRNA involvement. When wild-type HCT-116 cells are treated with 5-FU, they express high levels of certain miRNAs, and a great majority of these affected microRNA have a binding site for p53 in the gene. When HCT-116 cells knocked out for p53 are treated with 5-FU, these miRNAs are not upregulated. These results suggest that 5-FU acts as a switch to turn on p53 and, through p53, a cascade of miRNAs that may act with or independently of p53.
4. ACKNOWLEDGEMENTS
We would like to thank the National Breast Cancer Research Institute (NBCRI) for their financial support of the study.
REFERENCES
- Forrester, K., Almoguera, C., Han, K., et al. (1987) Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature, 327, 298-303. doi:10.1038/327298a0
- Ionov, Y., Peinado, M.A., Malkhosyan, S., et al. (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature, 363, 558-561. doi:10.1038/363558a0
- Cui, H., Horon, I.L., Ohlsson, R., et al. (1998) Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nature Medicine, 4, 1276- 1280. doi:10.1038/3260
- Vogelstein, B., Fearon, E.R., Hamilton, S.R., et al. (1988) Genetic alterations during colorectal-tumor development. New England Journal of Medicine, 319, 525-532. doi:10.1056/NEJM198809013190901
- Kitahara, O., Furukawa, Y., Tanaka, T., et al. (2001) Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Research, 61, 3544-3549.
- Lin, Y.M., Furukawa, Y., Tsunoda, T., et al. (2002) Molecular diagnosis of colorectal tumors by expression profiles of 50 genes expressed differentially in adenomas and carcinomas. Oncogene, 21, 4120-4128. doi:10.1038/sj.onc.1205518
- Zou, T.T., Selaru, F.M., Xu, Y., et al. (2002) Application of cDNA microarrays to generate a molecular taxonomy capable of distinguishing between colon cancer and normal colon. Oncogene, 21, 4855-4862. doi:10.1038/sj.onc.1205613
- Kwon, H.C., Kim, S.H., Roh, M.S., et al. (2004) Gene expression profiling in lymph node-positive and lymph node-negative colorectal cancer. Diseases of the Colon & Rectum, 47, 141-152. doi:10.1007/s10350-003-0032-7
- Friederichs, J., Rosenberg, R., Mages, J., et al. (2005) Gene expression profiles of different clinical stages of colorectal carcinoma: Toward a molecular genetic understanding of tumor progression. International Journal of Colorectal Disease, 20, 391-402. doi:10.1007/s00384-004-0722-1
- Kim, K., Park, U., Wang, J., et al. (2008) Gene profiling of colonic serrated adenomas by using oligonucleotide microarray. International Journal of Colorectal Disease, 23, 569-580. doi:10.1007/s00384-008-0451-y
- Kita, H., Hikichi, Y., Hikami, K., et al. (2006) Differential gene expression between flat adenoma and normal mucosa in the colon in a microarray analysis. Journal of Gastroenterology, 41, 1053-1063. doi:10.1007/s00535-006-1894-y
- Agrawal, D., Chen, T.G., Irby, R., et al. (2002) Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. Journal of the National Cancer Institute, 94, 513-521. doi:10.1093/jnci/94.7.513
- Koehler, A., Bataille, F., Schmid, C., et al. (2004) Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinicopathological stage. Journal of Pathology, 204, 65-74. doi:10.1002/path.1606
- Yanagawa, R., Furukawa Y., Tsunoda T., et al. (2001) Genome-wide screening of genes showing altered expression in liver metastases of human colorectal cancers by cDNA microarray. Neoplasia, 3, 395-401. doi:10.1038/sj.neo.7900185
- Ki, D.H., Jeung, H.C., Park, C.H., et al. (2007) Whole genome analysis for liver metastasis gene signatures in colorectal cancer. International Journal of Cancer, 121, 2005-2012. doi:10.1002/ijc.22975
- Lin, H.M., Chatterjee, A., Lin, Y.H., et al. (2007) Genome wide expression profiling identifies genes associated with colorectal liver metastasis. Oncology Reports, 17, 1541-1549
- Watanabe, T., Kobunai, T., Tanaka, T., et al. (2009) Gene expression signature and the prediction of lymph node metastasis in colorectal cancer by DNA microarray. Diseases of the Colon & Rectum, 52, 1941-1948. doi:10.1007/DCR.0b013e3181b53684
- Wang, Y., Jatkoe, T., Zhang, Y., et al. (2004) Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. Journal of Clinical Oncology, 22, 1564-1571. doi:10.1200/JCO.2004.08.186
- Birkenkamp-Demtroder, K., Olesen, S.H., Sorensen, F.B. et al. (2005) Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut, 54, 374-384. doi:10.1136/gut.2003.036848
- Ghadimi, B.M., Grade, M., Difilippantonio, M.J., et al. (2005) Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. Journal of Clinical Oncology, 23, 1826-1838. doi:10.1200/JCO.2005.00.406
- Kim, I.J., Lim, S.B., Kang, H.C., et al. (2007) Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Diseases of the Colon & Rectum, 50, 1342-1353. doi:10.1007/s10350-007-277-7
- Mariadason, J.M., Arango, D., Shi, Q.H., et al. (2003) Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Research, 63, 8791-8812
- Del Rio, M., Molina, F., Mollevi, C.B., et al. (2007) Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. Journal of Clinical Oncology, 25, 773-780. doi:10.1200/JCO.2006.07.4187
- Khambata-Ford, S., Garrett C.R., Meropol N.J., et al. (2007) Expression of epiregulin and amphiregulin and Kras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal of Clinical Oncology, 25, 3230-3237. doi:10.1200/JCO.2006.10.5437
- Kim, I.J., Lim, S.B., Kang, H.C., et al. (2007) Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Diseases of the Colon & Rectum, 50, 1342-1353. doi:10.1007/s10350-007-277-7
- Rimkus, C., Friederichs, J., Boulesteix, A.L., et al. (2008) Microarray-based prediction of tumor response to neoadjuvant radiochernotherapy of patients with locally advanced rectal cancer. Clinical Gastroenterology and Hepatology, 6, 53-61. doi:10.1016/j.cgh.2007.10.022
- Spitzner, M., Emons, G., Kramer, F., et al. (2010) A gene expression signature for chemoradiosensitivity of colorectal cancer cells. International Journal of Radiation Oncology, Biology, Physics, 78, 1184-1192. doi:10.1016/j.ijrobp.2010.06.023
- Cunningham, D., Humblet, Y., Siena, S., et al. (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. New England Journal of Medicine, 351, 337-345. doi:10.1056/NEJMoa033025
- Hurwitz, H., Fehrenbacher, L., Novotny, W., et al. (2004) Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. New England Journal of Medicine, 350, 2335-2342. doi:10.1056/NEJMoa032691
- Mayer, R.J. (2009) Targeted therapy for advanced colorectal cancer-more is not always better. New England Journal of Medicine, 360, 623-625. doi:10.1056/NEJMe0809343
- Saltz, L.B., Meropol, N.J., Loehrer, P.J. et al. (2004) Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. Journal of Clinical Oncology, 22, 1201-1208. doi:10.1200/JCO.2004.10.182
- Waldner, M.J. and Neurath, M.F. (2010) The molecular therapy of colorectal cancer. Molecular Aspects of Medicine, 31, 171-178. doi:10.1016/j.mam.2010.02.005
- Lee, R.C., Feinbaum, R.L. and Ambros, V. (1993) The C-elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell, 75, 843- 854. doi:10.1016/0092-8674(93)90529-Y
- Brennecke, J., Hipfner, D.R. and Stark, A. et al. (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell, 113, 25-36. doi:10.1016/S0092-8674(03)00231-9
- Lagos-Quintana, M., Rauhut, R., Yalcin, A. et al. (2002) Identification of tissue-specific microRNAs from mouse. Current Biology, 12, 735-739. doi:10.1016/S0960-9822(02)00809-6
- Calin, G.A., Dumitru, C.D., Shimizu, M. et al. (2002) Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America, 99, 15524-15529. doi:10.1073/pnas.242606799
- Michael, M.Z., O’Connor, S.M., Pellekaan, N.G.V., et al. (2003) Reduced accumulation of specific microRNAs in colorectal neoplasia. Molecular Cancer Research, 1, 882- 891.
- Schetter, A.J., Leung, S.Y., Sohn, J.J. et al. (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JamaJournal of the American Medical Association, 299, 425- 436. doi:10.1001/jama.299.4.425
- Akao, Y., Nakagawa, Y. and Naoe T. (2007) MicroRNA- 143 and-145 in colon cancer. DNA and Cell Biology, 26, 311-320. doi:10.1089/dna.2006.0550
- Braun, C.J., Zhang, X., Savelyeva, I., et al. (2008) P53- responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Research, 68, 10094- 10104. doi:10.1158/0008-5472.CAN-08-1569
- He, X.Y., He, L. and Hannon, G.J. (2007) The guardian’s little helper: MicroRNAs in the p53 tumor suppressor network. Cancer Research, 67, 11099-11101. doi:10.1158/0008-5472.CAN-07-2672
- Wang, P., Zou, F., Zhang, X. et al. (2009) MicroRNA-21 negatively regulates cdc25A and cell cycle progression in colon cancer cells. Cancer Research, 69, 8157-8165. doi:10.1158/0008-5472.CAN-09-1996
- Asangani, I.A., Rasheed, S.A.K., Nikolova, D.A. et al. (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene, 27, 2128-2136. doi:10.1038/sj.onc.1210856
- Nagel, R., Le Sage, C., Diosdado, B. et al. (2008) Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Research, 68, 5795-5802. doi:10.1158/0008-5472.CAN-08-0951
- Guo, C.G., Sah, J.F., Beard, L., et al. (2008) The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes & Cancer, 47, 939-946. doi:10.1002/gcc.20596
- Bandres, E., Cubedo, E., Agirre, X., et al. (2006) Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and nontumoral tissues. Molecular Cancer, 5, 29.
- Xi Y., Formentini, A., Chien, M., et al. (2006) Prognostic values of microRNAs in colorectal cancer. Biomark Insights, 2, 113-121.
- Arndt, G.M., Dossey, L., Cullen, L.M. et al. (2009) Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer, 9, 374.
- Motoyama, K., Inoue, H., Takatsuno, Y., et al. (2009) Over-and under-expressed microRNAs in human colorectal cancer. International Journal of Oncology, 34, 1069-1075.
- Wang, C.J., Zhou, Z.G., Wang, L. et al. (2009) Clinicopathological significance of microRNA-31,-143 and-145 expression in colorectal cancer. Disease Markers, 26, 27- 34.
- Lanza, G., Ferracin, M., Gafa, R. et al. (2007) MRNA/ microRNA gene expression profile in microsatellite unstable colorectal cancer. Molecular Cancer, 6, 54.
- Kloosterman, W.P., Lagendijk, A.K., Ketting, R.F. et al. (2007) Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. Plos Biology, 5, 1738-1749. doi:10.1371/journal.pbio.0050203
- Hutvagner, G. and Zamore P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056-2060. doi:10.1126/science.1073827
- Elmen, J., Lindow, M., Silahtaroglu, A. et al. (2008) Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Research, 36, 1153-1162. doi:10.1093/nar/gkm1113
- Krutzfeldt, J., Rajewsky, N., Braich, R. et al. (2005) Silencing of microRNAs in vivo with “antagomirs”. Nature, 438, 685-689. doi:10.1038/nature04303
- Brosens, L.A.A., Van Hattem, W.A., Jansen, M. et al. (2007) Gastrointestinal polyposis syndromes. Current Molecular Medicine, 7, 29-46. doi:10.2174/156652407779940404
- Lu, Z., Liu, M., Stribinskis, V. et al. (2008) MicroRNA- 21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene, 27, 4373-4379. doi:10.1038/onc.2008.72
- Dews, M., Homayouni, A., Yu, D. et al. (2006) Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nature Genet, 38, 1060-1065. doi:10.1038/ng1855
- Rossi, L., Bonmassar, E. and Faraoni, I. (2007) Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacological Research, 56, 248-253. doi:10.1016/j.phrs.2007.07.001
- [61] Meng, F.Y., Henson, R., Lang, M. et al. (2006) Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology, 130, 2113-2129. doi:10.1053/j.gastro.2006.02.057

