American Journal of Molecular Biology
Vol.2 No.3(2012), Article ID:20982,7 pages DOI:10.4236/ajmb.2012.23027
The full length PtSRP (Pisolithus tinctorius symbiosis related protein) fungal mRNA encodes a potential marker of ectomycorrhiza formation*
![]()
1Departamento de Micologia, Universidade Federal de Pernambuco, Recife, Brazil
2Laboratório de Imunologia Celular e Molecular, Centro de Pesquisas René Rachou, Belo Horizonte, Brazil
3Centro de Pesquisas Aggeu Magalhães—Fundação Oswaldo Cruz, Laboratório de Virologia e Terapia Experimental, Universidade Federal de Pernambuco, Recife, Brazil
Email: #bartacioli@cpqam.fiocruz.br
Received 17 February 2012; revised 22 April 2012; accepted 19 May 2012
Keywords: Ectomycorrhiza; Symbiosis Related Genes/Proteins; RACE; EST
ABSTRACT
The Pisolithus tinctorius symbiosis related protein expressed sequence tag (EST PtSRP) was previously identified in the first hours of the interaction between the fungus Pisolithus tinctorius and sweet chestnut Castanea sativa, and partially characterized as a fungal marker gene of ectomycorrhizal symbiosis formation. We used the 5’ rapid amplification of cDNA ends (RACE) to obtain the PtSRP mRNA 5’ region, and together with our previously reported 3’ mRNA region, the full mRNA sequence was assembled by use of bioinformatics tools and deposited to GenBank (Accession: GU733439). The full-length mRNA sequence (636 bp) revealed the locations of the 5’ and 3’ untranslated regions (UTRs) and contained the Kozak sequence (ccc aag ATG A) in the 5’ UTR. The in silico translated PtSRP open reading frame (ORF) codes for a 127 amino acid protein and contained four putative post-translational modification sites (two Nglycosylation and two phosphorylation). The protein secondary structure is postulated to be composed of one N-terminal hydrophobic transmembrane alpha helix and at least six hydrophilic beta-strands spread across the protein. Sub-cellular localization prediction suggests that the protein is involved in cellular secretory pathway, supported by the presence of a cleavage site motif close to the membrane anchor. The data presented herein indicate the role of PtSRP as a fungal membrane secreted protein involved in early stages of ectomycorrhizal formation, with application as a possible marker for nascent ectomycorrhiza fungal development.
1. BACKGROUND
The establishment of ectomycorrhiza involves controlled, intense gene expression in both partners that leads to drastic morphological and physiological changes, crucial to the development of mutualism and symbiotic harmony [1-3] . The comparison of protein extracts from mycorrhizal and non-mycorrhizal mycelia in previous studies has shown differences that suggest specific gene activation during the symbiosis process [4-6] . These findings highlighted a new class of biomolecules thought to control the ectomycorrhiza symbiosis process: the ectomycorrhizins [7]. However, recent studies evaluating the fungal transcript pattern during symbiosis formation have demonstrated that mycorrhization also induces changes in the expression of genes normally expressed in free organisms [3,8-10] .
Among the ectomycorrhizins, SRAPs (Symbiosis Related Acid Proteins) and hydrophobins are the most investigated and discussed classes of proteins. However, these proteins were generally isolated from fully established mycorrhiza or those developing associations after several days of interaction [8,9] . The identification of new early stage ectomycorrhizal molecules could bring new insight to the molecular and functional understanding of the ectomycorrhiza formation process. The fungal PtSRP mRNA (previously called Pisolithus tinctorius symbiosis related receptor 1, accession number EL563703) was isolated [3] and partialy characterized [11] as a possible fungal membrane protein probably secreted in the first hours of fungus-root interaction. In this paper, we present the full-length PtSRP fungal mRNA sequence, supported by sequencing of the 5’ region and our previously reported 3’ region, followed by in silico characterization of the most probable ORF and its relationship with early stages of ectomycorrhiza.
2. RESULTS
2.1. The Full-Length PtSRP mRNA
The 5’ RACE technique generated a partial 355 bp fragment corresponding to the 5’ portion of PtSRP mature mRNA (deposited at NCBI as nucleotide record GU733- 439). The complete sequence, assembled by contig construction between the 5’ RACE fragment and a 3’ previously reported sequence (EL563703, [11]) resulted in a 636 bp sequence (Figure 1). Additionally, a unique 604 bp product was obtained by direct PCR of fungal cDNA samples. The nearly perfect alignment of the 604 bp consensus fragment to the 636 bp contig supported the reliability of the contig; only two nucleotide differences (291 C/T and 306 A/C, Figure 2) were observed and they were in the putative ORF region, with no changes to the amino acid (synonymous mutations). The PtSRP mRNA putative ORF is 384 bp long and codes for a protein of 127 amino acids (a.a.) (Figure 1), with untranslated regions (UTRs) upstream and downstream of the ORF. The Kozak motif sequence (ccc aag ATG A) was present in the 5’ UTR, albeit slightly variable from the original Kozak sequence (gcc Rcc AUG G) for three of the nucleotides: –6 (C), –2 (A), –1 (G) and 4 (A) (Figure 1).
2.2. In Silico Analysis of PtSRP
In silico analysis of PtSRP primary structure indicated theoretical molecular weight of 13,969 kDa and an iso-

Figure 1. The PtSRP mRNA. Complete nucleotide sequence (636 bp) and its probable ORF (the codons and corresponding 127 amino acids). The ORF represents the largest translation region for the sequence. Stop codon is represented by the word “End”. Full line = new fragment obtained by RACE 5’; Interrupted line = 3’ previously reported sequence (EL563703, Acioli-Santos et al., 2009); ← = 5’ UTR; → = 3’ UTR; □ = Kozak sequence with variation and respective base positions.
electric point of 3.92. Further, four targets of post-translation modifications were predicted: two N-glycosylation sites (residues 118 to 121 (NFSQ) and 122 to 125 (NFTI)) and two casein kinase II phosphorylation sites (residues 65 to 68 (TNSE) and 99 to 102 (TVPD), Figure 3).
The predicted secondary structure was an N-terminal hydrophobic transmembrane alpha-helix, from residues 10 to 20, followed by six beta-sheets interspersed by short loops (Figure 3). A signal peptide cleavage site was predicted close to the membrane, between the 23rd (serine) and 24th (valine) residues. Sub-cellular localization prediction indicated strong probability that the PtSRP protein is involved in cellular secretory pathway, which is also supported by the presence of the N-terminal signal peptide cleavage site. Hydrophobicity analysis demonstrated that the PtSRP initial region is strongly hydrophobic (Figure 4) supporting the secondary structure prediction analysis (i.e. an initial alpha-helix region).

Figure 2. Alignment between the RACE assembly (636 bp; A) and the directly amplified and cloned sequence (604 bp; B) indicating only two nucleotide changes in 291C/T and 306 A/C positions (#). ▬: PtSRP ORF.

Figure 3. Secondary structure prediction and transmembrane domain of the PtSRP. Legend: AA = amino acid sequence, PHD_htm = transmembrane helix prediction (M = transmembrane helix, blank spaces = non-membrane regions), Rel_htm = PHD_htm prediction reliability index (Reliable predictions are marked “*”), PROF_sec = secondary structure prediction (H = Helix, E = sheet, blank: “loop”), Rel_sec = PROF_sec prediction reliability index (0 = low to 9 = high), SUB_sec = all PROF_sec predictions subset, for all residues with an expected average accuracy > 82% (L = “loop”, “.” = no prediction for the residue), O_3_acc = observed relative solvent accessibility (b = 0% - 9%, i = 9% - 36%, e = 36% - 100%). P_3_acc = predicted relative solvent accessibility, Rel_acc: P_3_acc prediction reliability index, SUB_acc: P_3_acc prediction subset, for all residues with an average correlation > 0.69. Glycosylation sites NFSQ and NFTI = (─); phosphorylation sites TNSE and TVPD = (). The cleavage site near to membrane is represented by (↓). Analyses were carried out in http://www.predictprotein.org.
GLOBE prediction indicated that PtSRP is not a globular protein.
3. DISCUSSION
The studies of unknown ectomycorrhizal genes need to be carried out in several stages, as there are as yet none or few elements for comparison. Hydrophobins are fungal proteins usually found during the early stages of P. tinctorius-E. globulus interaction [12]. However, its expression is dubious in some cases, and its use as a symbiotic development marker is debatable [9]. SRAP (Symbiosis Related Acid Proteins) genes have been considered as robust ectomycorrhiza development marker after two days of contact [8,9,13] . To the best of our knowledge, there is no report of SRAPs expression during shorter intervals, such as within early hours of mycorrhizal establishment. PtSRP QRT-PCR data [11] confirmed the cDNA microarray analysis [3] of its high relative transcription at 12 h of ectomycorrhizal stimulus. Transcription of this gene apparently does not occur until 6 h of contact, suggesting that the period between 6 and 12 h can be crucial for the PtSRP expression [11]. The complete PtSRP mRNA sequence showed 78% identity with a Pisolithus microcarpus sequence (CB010071; [14]) in ectomycorrhizal association with Eucalyptus globulus. Further, 78% identity was detected when compared with a fungus cDNA from a four-day-old Pisolithus tinctorius-Eucalyptus globulus association (BF942- 674; [8]).
The Kozak consensus sequence was found in the 5’ UTR of the PtSRP mRNA. This regulatory element plays an important role during early processes of gene translation [15] through recognition by the ribosome, resulting in higher or lower protein synthesis [16]. Typical Kozak sequences are followed by the start codon and a guanine base, gccRccAUGG, where R is a purine base three positions before the AUG start codon [17]. In PtSRP mRNA, a variant Kozak sequence is observed, but these differ-
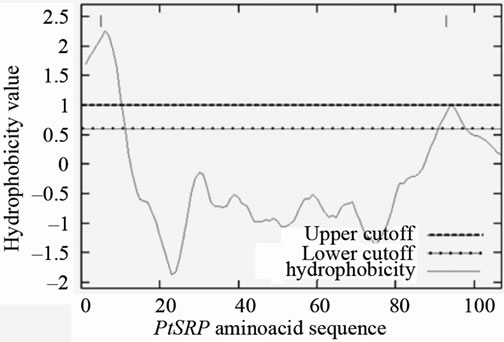
Figure 4. PtSRP hydrophobicity analysis. A strong hydrophobic initial region is observed in the protein (scaled by the kyte and doolittle, 1982).
ences are common, as variation between species [18-20] . In Saccharomyces cerevisiae, for example, adenine is commonly observed to precede the initial methionine codon [19].
The computationally translated protein ORF showed significant local sequence alignment with two previously reported proteins. The first (48% identity and 65% similarity) was isolated from the mycelia of the ectomycorrhizal Laccaria bicolor (accession XP_001876100.1). This protein is associated with a small-secreted protein (SSP) of unknown function, up regulated in symbiotic tissues [10] . The other homologous protein (45% identity and 61% similarity) was isolated from the non-ectomycorrhizal Schizophyllum commune (accession AF335537) and is highly expressed when the mycelium is growing under low nitrogen availability, an important environment aspect to mycorrhizal formation.
The presence of the integrin binding RGD domain (arginine-glycine-aspartic acid) in a protein has been related to cell adhesion [21], a key feature in the initial stages of ectomycorrhiza formation, as has been observed in some SRAPs [13] . The RGD domain and its variants [21] have not been found in the PtSRP protein. This, however, is not an isolated event. Other genes from ectomycorrhizal systems do not present this motif (e.g. SC13 and SC25 in [22] and Lbras in [23] ), suggesting it is not a critical ectomycorrhizal domain.
The PtSRP post-translation modification sites previously indicated [11] were confirmed after obtention of the full-length mRNA. In addition, the prediction of a transmembrane region (composed of an alpha-helix between the 10th and the 20th a.a. residues) in the protein with a probable cleavage site close to the membrane, and an external portion composed of beta-sheets interspersed with loops were observed. These data suggest that the PtSRP protein could act as a signalling secreted protein during early stages of symbiosis. Further studies of the PtSRP gene and its protein are required to confirm its function as a potential controller/marker of fungal development in ectomycorrhiza symbiosis.
4. MATERIALS AND METHODS
4.1. Biological Material and Culture Conditions
The fungal strain, culturing and ectomycorrhizal induction were done as described in [24]. P. tinctorius (isolate 289/Marx from the University of Tübingen) was maintained on modified Melin-Norkrans agar MNM [25]. Liquid cultures were obtained by transferring mycelia discs from solid cultures to 250 ml liquid MNM contained in Erlenmeyer flasks and kept in the dark at 25˚C until a dense mycelium was observed. Fungal biomass was washed in sterile water, immediately frozen in liquid nitrogen and stored at –80˚C.
4.2. Synthesis of PtSRP mRNA 5’ Portion
Total fungal RNA was extracted using the PureLinkTM Micro-to-Midi Total RNA Purification System (Invitrogen). The 5’ PtSRP was obtained using 5’ RACE technology (GeneRacer—full-length, RNA ligase-mediated rapid amplification of 5’ cDNA ends—RLM-RACE, Invitrogen) according to the manufacturer’s instructions. The 5’ phosphate free ends were linked to GeneRacerTM RNA Oligo and the cDNA was synthesised (Superscript II—Invitrogen). PCR reactions were preformed using the primers supplied in the kit, aiming at the GeneRacerTM RNA Oligo combined with gene specific primers targeted to the 3’ sequence (PT-1440 REV: 5’-AAATCGTTCAGAGAGATAAAGTTG-3’ and PCR 1R REV: 5’- CGTCCGGTACTGTGACCATC-3’). Cloning of the largest RACE fragment was performed using the pGEM-T Easy Vector System (Promega) and the insertion was confirmed by PCR using Promega’s specific primers (SP6 and T7) directed to the cloning vector.
4.3. PtSRP 5’ Fragment Sequencing and Obtention of Full-Length PtSRP mRNA
Cloned plasmids of recombinant bacteria (TG1) were extracted using Mini-prep. ABI PRISM BigDyeTM Terminator v3.1 Cycle sequencing Ready Reaction kit (Applied Biosystems) was used for sequencing the 5’ RACE fragment in an ABI PRISM 3100 Genetic Analyzer system (Applied Biosystems). Full-length mRNA was obtained by contig formation using SeqMan NGen v1.2 (DNASTAR Lasergene V8.0, Madison-US) after manual edition of sequences using ApE v1.15 (University of California-US) and Chromas Lite v2.01 (Technelysium Pty Ltd., Australia) software. This sequence will be referred in the text as 636 bp.
4.4. PtSRP Cloning and Sequencing
To confirm that the above contig sequence was correctly constructed, a primer pair (PtSRP FW: 5’-CCTCTCTCTCGAACACCTCCAC-3’ and PtSRP REV: 5’-ACGTACAGCAGAATGCGAAAG-3’), directed to the flanking regions of the gene ORF were designed (by use of ApE v1.15) for the direct PCR amplification of the gene from cDNA samples. The amplicons were cloned using the CloneJETTM PCR Cloning Kit (Fermentas). Cloned plasmids were extracted from recombinant DH10B using QIAprep® spin Miniprep kit and sequenced on an ABI PRISM 3100 as described above. Twelve experimental sequences were aligned giving a 604 bp consensus gene sequence which was further aligned to the 636 bp sequence described above.
4.5. PtSRP mRNA Nucleotide Sequence Analysis and PtSRP Protein Prediction
The most probable ORF definition of the 636 bp was achieved using the “Find ORF” routine in the ApE v1.15, with identification of initial methionine and termination codons. Untranslated regions (UTRs) of possible ORFs were compared to the original Kozak sequence [17]. Additionally, online BLASTx searches [26] were carried out to detect significant similarity between the new ORF and those previously deposited in the NCBI Entrez Protein Database.
Structural analysis of the putative ORF was carried out using the Predict Protein Web-server (http://www.predictprotein.org) [27]. Functional databases were searched for conserved domains by use of the Inter Pro Web-server (http://www.ebi.ac.uk/interpro/) [28] . The subcellular location prediction of the PtSRP protein was obtained using the TargetP 1.1 Web-server [29] and [30] and potential cleavage site predicted using ChloroP 1.1 [31] and SignalP 3.0 servers [32] . TargetP (http://www.cbs.dtu.dk/services/TargetP/) incorporates prediction of cleavage sites and signal/non-signal peptide based on combination of artificial neural networks and hidden Markov models. The following cutoff levels were used: 0.780 for mitochondrial targeting peptide (mTP), 0.000 for secretory pathway signal peptide (SP) and 0.730 for the other features. Further, analyses were performed to study the hydrophobicity (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=toppred) and globular shape of the protein (http://www.predictprotein.org; GLOBE function used). Prediction of a three-dimensional protein model based on homology modelling was not performed due to lack of significant similarity crystallized template structures in RCSB Protein Databank (PDB) (http://www.rcsb.org/pdb) [33].
5. ACKNOWLEDGEMENTS
This work was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brazil) and the Post-Graduation Program on Fungal Biology (PPG-BF, Federal University of Pernambuco—Brazil).
REFERENCES
- Tagu, D., Lapeyrie, F. and Martin, F. (2002) The ectomycorrhizal symbiosis: genetics and development. Plant Soil, 244, 97-105. doi:10.1023/A:1020235916345
- Le Quere, A., Wright, D.P., Soderstrom, B., Tunlid, A. and Johansson, T. (2005) Global patterns of gene regulation associated with the development of ectomycorrhiza between birch (Betula pendula Roth.) and Paxillus involutus (Batsch) Fr. Molecular Plant-Microbe Interactions, 18, 659-673. doi:10.1094/MPMI-18-0659
- Acioli-Santos, B., Sebastiana, M., Pessoa, F., Sousa, L., Figueiredo, A., Fortes, A.M., Balde, A., Maia, L.C and Pais, M.S. (2008) Fungal transcript pattern during the preinfection stage (12 h) of ectomycorrhiza formed between Pisolithus tinctorius and Castanea sativa roots, identified using cDNA microarrays. Current Microbiology, 57, 620-625. doi:10.1007/s00284-008-9253-2
- Hilbert, J.L., Costa, G. and Martin, F. (1991) Ectomycorrhizin synthesis and polypeptide changes during the early stage of eucalypt mycorrhiza development. Plant Physiology, 97, 977-984.
- Burgess, T., Laurent, P., Dell, B., Malajczuk, N. and Martin, F. (1995) Effect of fungal-isolate aggressivity on the biosynthesis of symbiosis-related polypeptides in differentiating eucalypt ectomycorrhizas. Planta, 195, 408- 417. doi:10.1007/BF00202599
- Martin, F., Duplessis, S., Ditengou, F., Lagrange, H., Voiblet, C. and Lapeyrie, F. (2001) Developmental cross talking in the ectomycorrhizal symbiosis: signal and communication genes. New Phytologist, 151, 145-154. doi:10.1046/j.1469-8137.2001.00169.x
- Hilbert, J.L. and Martin, F. (1988) Regulation of gene expression in ectomycorrhizas. I. Protein changes and the presence of the ectomycorrhiza-specific polypeptides in the Pisolithus-Eucalyptus symbiosis. New Phytologist, 110, 339-346. doi:10.1111/j.1469-8137.1988.tb00270.x
- Voiblet, C., Duplessis, S., Encelot, N. and Martin, F. (2001) Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant Journal, 25, 181-191. doi:10.1046/j.1365-313x.2001.00953.x
- Duplessis, S., Courty, P.E., Tagu, D. and Martin, F. (2005) Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytologist, 165, 599-611. doi:10.1111/j.1469-8137.2004.01248.x
- Martin, F., Aerts, A., Ahren, D., Brun, A., Danchin, E.G., Duchaussoy, F., Gibon, J., Kohler, A., Lindquist, E. and Pereda, V., et al. (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature, 452, 88-92. doi:10.1038/nature06556
- Acioli-Santos, B., Malosso, E., Calzavara-Silva, C.E., Lima, C.E.P., Figueiredo, A., Sebastiana, M. and Pais, M.S. (2009) PtSRR1, a putative Pisolithus tinctorius symbiosis related receptor gene is expressed during the first hours of mycorrhizal interaction with Castanea sativa roots. Brazilian Journal of Microbiology, 40, 292- 295. doi:10.1590/S1517-83822009000200015
- Tagu, D., Nasse, B. and Martin, F. (1996) Cloning and and characterization of hydrophobins-encoding cDNAs from the ectomycorrhizal basdiomycete Pisolithus tinctorius. Gene, 168, 93-97. doi:10.1016/0378-1119(95)00725-3
- Laurent, P., Voiblet, C., Tagu, D., de Carvalho, D., Nehls, U., De Bellis, R., Balestrini, R., Bauw, G., Bonfante, P. and Martin, F. (1999) A novel class of ectomycorrhizaregulated cell wall polypeptides in Pisolithus tinctorius. Molecular Plant-Microbe Interactions, 12, 862-871. doi:10.1094/MPMI.1999.12.10.862
- Peter, M., Courty, P., Kohler, A., Delaruelle, C., Martin, D., Tagu, D., Frey-klett, P., Duplessis, S., Chalot, M., Podila, G. et al. (2003) Analysis of expressed sequence tags from the ectomycorrhizal basiodiomycetes Laccaria bicolor and Pisolithus microcarpus. New Phytologist, 159, 117-129. doi:10.1046/j.1469-8137.2003.00796.x
- De Angioletti, M., Lacerra, G., Sabato, V., and Carestia, C. (2004) Beta + 45 G → C: a novel silent beta-thalassaemia mutation, the first in the Kozak sequence. British Journal of Haematology, 124, 224-231. doi:10.1046/j.1365-2141.2003.04754.x
- Kozak, M. (1984) Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature, 308, 241-246. doi:10.1038/308241a0
- Kozak, M. (1987) An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Research, 15, 8125-8148. doi:10.1093/nar/15.20.8125
- Cavener, D.R. (1987) Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Research, 15, 1353-1361. doi:10.1093/nar/15.4.1353
- Hamilton, R., Watanabe, C.K. and de Boer, H.A. (1987) Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Research, 15, 3581-3593. doi:10.1093/nar/15.8.3581
- Lutcke, H.A., Chow, K.C., Mickel, F.S., Moss, K.A., Kern, H.F. and Scheele, G.A. (1987) Selection of AUG initiation codons differs in plants and animals. EMBO Journal, 6, 43-48.
- Ruoslahti, E. (1996) RGD and other recognition sequences for integrins. Annual Review of Cell and Developmental Biology, 12, 697-715. doi:10.1146/annurev.cellbio.12.1.697
- Nehls, U., Mikolajewski, S., Ecke, M. and Hampp, R. (1999) Identification and expression analysis of two fungal cDNA regulated by ectomycorrhiza and fruit body formation. New Phytologist, 144, 195-202. doi:10.1046/j.1469-8137.1999.00488.x
- Sundaram, S., Kim, S.J., Suzuki, H., McQuattie, C.J., Hiremah, S.T. and Podila, G.K. (2001) Isolation and characterization of a symbiosis-regulated ras from the ectomycorrhizal fungus Laccaria bicolor. Molecular Plant-Microbe Interactions, 14, 618-628. doi:10.1094/MPMI.2001.14.5.618
- Baptista, P., Martins, A., Pais, M.S., Tavares, R.M. and Lino-Neto, T. (2007) Involvement of reactive oxygen species during early stages of ectomycorrhiza establishment between Castanea sativa and Pisolithus tinctorius. Mycorrhiza, 17, 185-193. doi:10.1007/s00572-006-0091-4
- Marx, D.H. (1969) The influence of ectotrophic mycorrhizal fungi on the resistance of pine roots to pathogenic infections. II. Production, identification, and biological activity of antibiotics produced by Leucopaxillus cerealis var. piceina. Phytopathology, 59, 411-417.
- McGinnis, S., Madden, T.L. (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research, 32, 20-25.
- Rost, B. and Liu, J. (2003) The Predict Protein server. Nucleic Acids Research, 31, 3300-3304. doi:10.1093/nar/gkg508
- Hunter, S., Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Binns, D., Bork, P., Das, U., Daugherty, L., Duquenne, L. et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Research, 37, 211- 215. doi:10.1093/nar/gkn785
- Emanuelsson, O., Nielsen, H., Brunak, S. and von Heijne, G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology, 300, 1005-1016. doi:10.1006/jmbi.2000.3903
- Nielsen, H., Engelbrecht, J., Brunak, S. and von Heijne, G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering, 10, 1-6. doi:10.1093/protein/10.1.1
- Emanuelsson, O., Nielsen, H. and von Heijne, G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science, 8, 978-984. doi:10.1110/ps.8.5.978
- Bendtsen, J.D., Nielsen, H., von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology, 340, 783-795. doi:10.1016/j.jmb.2004.05.028
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N. and Bourne, P.E. (2000) The protein data bank. Nucleic Acids Research, 28, 235-242. doi:10.1093/nar/28.1.235
NOTES
*Competing interests: The authors declare that they have no competing interests.
#Corresponding author.

