 American Journal of Plant Sciences, 2011, 2, 781-789 doi :1 0.4236/ aj ps.2011 .26093 Publ i s hed Online December 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS 781 Phylogeography of Asparagus schoberioides Kunth (Asparagaceae) in Japan Tatsuya Fukuda1*, In-Ja Song2, Hokuto Nakayama3, Takuro Ito4, Akira Kanno5, Hiroshi Hayakawa6, Yukio Minamiya6, Jun Yokoyama7 1Faculty of Agriculture, Kochi University, Nankoku, Japan; 2Subtropical Horticulture Research Institute, Jeju National University, Jeju, Ko rea; 3Graduate School of Science, University of Tokyo, Tokyo, Japan; 4Institute for Advanced Biosciences, Keio Un iversity, Tsuruoka, Japan; 5Graduate School of Life Sciences, Tohoku University, Sendai, Japan; 6United Graduate School of Agricultural Sciences, Ehime University, Monobe, Japan; 7Department of Biology, Faculity of Science, Yamagata University, Yamagata, Ja- pan. Email: *tfukuda@kochi-u.ac.jp Received April 8th, 2011; revised May 3rd, 2011; accepted May 24th, 2011. ABSTRACT To describe the phylogeographic structures of Asparagus schoberioides Kunth (Asparagaceae) in Japan, we investi- gated its nucleotide sequence variations with respect to its geographic distribution pattern. Sequencing of the internal transcribed spacer (ITS) 1 region in 29 samples of A. schoberioides revealed 20 polymorphic nucleotide sites. As a re- sult, the 29 samples of A. schoberioides fell in to 15 distinct haplotypes and phylogenetic analyses revealed these haplo- types fell into two major clad es, Clade 1 and Clade 2. The haplotypes of Clade 1 were distributed chiefly along the Pa- cific Ocean side of Japan, while those of Clade 2 occurred mainly along the Japan Sea side. This result suggests that A. schoberioides has migrated via two routes in Japan. Keywords: Asparagus schoberioides, Internal Transcribed Spacer (ITS), Phyloge ny, Phylogeography 1. Introduction The genetic and geographic structures of natural plant populations are a consequence of ecological factors and historical events. Consequently, the amount of genetic variation within a species depends on its history and life strategy. Comparison of nucleotide sequences are the best way to analyze the history of plant populations, and phylogeographic studies employing nuclear DNA data have been used to test ever more sophisticated historical models [1-3]. In particular, nucleotide divergences of the nuclear DNA (nrDNA) showed relative higher values than those of chloroplast DNA (cpDNA) [4], although recombination of nrDNA is biased the concepts of ho- mology [5]. Asparagus schoberioides Kunth is a perennial herb bearing small flowers with yellowish green perianth in a raceme-like inflorescence and reddish colored berries. This species ranged from Far East Russia, northern China, Korea, Sakhalin, Japan and Taiwan [6,7]. In Japan, its distribution extends from the northern-most part of Hok- kaido down to Shikoku and Kyushu [6]. A. schoberioides is one of the most closely related species to the garden asparagus, A. officinalis L. [8], which is the most eco- nomically important species in this genus. While the gar- den asparagus is not nati ve to Jap an, it is cultivated t here and has escaped from the crop fields [6]. Recently, Ochiai et al. [9] and Ito et al. [10] reported successfully generating of interspecific hybrids between A. schobe- rioides and the garden asparagus. A natural hybrid be- tween A. schoberioides and the garden asparagus has not been found to date, but the possibility of genetic intro- gression from crop to wild relatives suggests the escape of the garden asparagus from the crop fields may pose a substantial ecological risk for the environment and its biodiversity [2,11-16]. In Japan, molecular approaches have been used to ana- lyze the intraspecific genetic variation of a number of different plant species (e.g., Abies mariesii [17]; Aucuba chinensis and A. japonica [18]; Fagus crenata [19-21]; Purimula cunefolia [22]; Quercus serrata and its allied species [23]; Stachyurus praecox [24], Alpinia japonica, Arachnioides sporadosora, A. aristata, Daphne kiusiana, Elaeocarpus sylvestris var. ellip ticus, Prunus zippeliana [25]). Most of these studies were conducted based on the chloroplast DNA (cpDNA) sequence. Similarly, in our  Phylogeography of Asparagus schoberioides Kunth (Asparagaceae) in Japan 782 previous study, we used primers designed by Taberlet et al. [26] and Nishizawa and Watano [27] to sequence approximately 3000 bp of 16 regions in the cpDNA of some Asparagus species [8]. However, we detected little variation in these regions a nd concluded that the cpDN A is not suitable for phylogenetic analyses of this species. The internal transcribed spacer (ITS) region is a power- ful tool for resolving historical relationships among po- pulations of widespread plants [28]. In fact, many re- searches with various species using ITS region had been reported in recent years [29-32]. For example, Yoko- yama et al. [33] analyzed the ITS region of Mitchella undulata Sieblod et Zucc. in Yakushima Island, they detected intraspecific variations that suggested this popu- lation had undergone rapid morphological modification. Thus, analysis of this region may permit plant relation- ships to be evaluated down to the intraspecific level. The aims of this study were to describe the phylo- geographic structures of A. schoberioides especially in Japan on the basis of its ITS region and to analyze the distribution of t hese genetic variations relative to the di s- tribution of A. schoberioides. 2. Materials and Methods 2.1. Plant Materials Twenty-nine samples of Asparagus schoberioides were examined in this study (Table 1). A. kiusianus Makino and A. officina lis were selected as outgroups on the basis of phylogenetic analyses of the genus Asparagus [8]. Vouchers for all species sa mpled in this study have been deposited in the Herbarium, Graduate School of Science, Tohoku University (TUS) and the Herbarium of Tsumura Laboratory (THS). 2.2. DNA Extraction, Amplification, and Sequencing Total DNAs were isolated from 200 - 300 mg of phyllo- clades with a Plant Genomic DNA Mini Kit (VIOGENE, Sunnyvale, USA) used according to the manufacturers’ protocols. The isolated DNA was resuspended in TE and stored at –20˚C until use. For phylogenetic analysis, we amplified the ITS1 re- gion with primers designed by White et al. [34]. Dou- ble-stranded DNA was amplified by incubation at 94˚C for 2 min followed by 40 cycles of incubation at 94˚C for 1.5 min, 48˚C for 2 min, and 72˚C for 3 min, wit h a fi nal extension at 72˚C for 15 min. After the amplification, reaction mixtures were subjected to electrophoresis in 1% low-melting-temperature agarose gels to purify of the amplified products. We sequenced the purified PCR pro- ducts using a DYEnamic ET-terminator Cycle Sequenc- ing Kit (Amersham Pharmacia) and a Model 373A auto- mated sequencer (Applied BioSystems) according to the manufacturers’ instructions. For sequencing, we used the same primers as those used for amplification. 2.3. Data Analysis ITS region sequences were aligned with the CLUSTAL X program [35]. Phylogenetic analysis and a test of clade support were conducted using the PAUP* program (ver- sion 4.0b10; [36]). Maximum parsimony analyses were carried out via a heuristic search with TBR branch swap- ping and MULPERS optio n. Multiple islands of the most parsimonious trees [37] were identified using the heuris- tic option with 100 random sequence additions. To esti- mate confidence levels of monophyletic groups, the bootstrap method with 1000 replications were employed [38]. 3. Results We determined the ITS1 region sequence in 29 samples of Asparagus schoberioides and two outgroup species, namely A. kiusianus and A. officinalis. The ITS1 region of all A. schoberioides plants was 249 bp in length. Variable sites in this region are shown in Table 2. Fif- teen ITS1 haplotypes were obtained from A. schoberi- oides, and the sequence of each haplotype has been de- posited in the DDBJ/EMBL/GenBank international DNA data bank (Table 1). There are no indels among the ITS sequences of A. schoberioides and its allied species. When we used the ITS1 sequence data set in phyloge- netic analyses, we obtained 78 most parsimonious trees of the 28 steps with a consistency index (CI) of 0.71 and a retention index (RI) of 0.92. One of the most parsimo- nious trees is shown in Figure 1. In our phylogenetic tree, all A. schoberioides samples formed the monophyletic group and two clades (Clade 1 and Clade 2 hereafter) in A. schoberioides were recog- nized. The relationship between the clades and the lo- calities of the individuals is indicated in Table 1. The correspondence between haplotypes and samples is shown in Figure 2. The geographic distribution of the samples is shown in Figure 3. Clade 1 consists of the following fourteen samples: China, South Korea, Hok- kaido1, Hokkaido3, Hokkaido4, Hokkaido5, Hokkaido6, Miyagi1, Miyagi2, Saitama, Yamanashi, Nagano1, Na- gano2 and Shizuoka. Clade 2 contains the following fif- teen samples: Russia, Hokkaido2, Iwate, Niigata1, Nii gata2, Niigata3, Nagano3, Kyoto1, Kyoto2, Kyoto3, Hyogo, Shimane, Yamaguchi, Fukuoka and Nagasaki. Thus, in our phylogenetic analysis, the samples from Hokkaido and Nagano extend into the two lineages that form Clades 1 and 2 (Figures 1, 3). Apart from these samples, Clade 1 consists of samples from the Pacific Copyright © 2011 SciRes. AJPS  Phylogeography of Asparagus schoberioides Kunth (Asparagaceae ) in Japan Copyright © 2011 SciRes. AJPS 783 Table 1. List of taxa, sources, haplotypes and accession numbers of plant materials. Taxon Sample name Locality Collector HaplotypeAcc. no. Asparagus scho berioides Ku nth Russia RUSSIA: Sakhalin, Dovie-alexandrowsk Kudo 25406 K AB196766 China CHINA: Liaoning, Dalian, Lushun Suzuki s.n . D AB196739 SouthKorea SOUTH KOREA: Junranam, Gwangyang, OkryongIm 21864 E AB196767 Hokkaido1 JAPAN: Hokkaido, Kamikawa, Toma Deguchi 8326 A AB196744 Hokkaido2 JAPAN: Hokkaido, Oshima, Matsumae Kudo 25410 F AB196741 Hokkaido3 JAPAN: Hokkaido, Iburi, Tomakomai Kudo 25407 A AB196745 Hokkaido4 JAPAN: Hokkaido, Okushiri Isl. Mt. Kamui Kudo 25411 B AB196743 Hokkaido5 JAPAN: Hokkaido, Abashiri, Abashiri Kudo 25409 A AB196742 Hokkaido6 JAPAN: Hokkaido, Hidaka, Samani Yamaji s.n. A AB196760 Iwate JAPAN: Iwate, Mt. Hayachine Endo s.n. M AB196747 Miyagi 1 JAPAN: Miyagi, Tome, Towa Sugaya & Soma 4203 A AB196752 Miyagi2 JAPAN: Miyagi, Shiroishi, Mt. Ohagi Suzuki 366 A AB196751 Saitama JAPAN: Saitama, Chichibu Yamaji s.n. A AB196759 Yamanashi JAPAN: Yamanashi, Hokuto Yamaji s.n. A AB196765 Niigata1 JAPAN: Niigata , Nakakubiki, Kasu ga Iwano 1177 H AB19675 6 Niigata2 JAPAN: Niigata, Nishikubiki, Nou Iwano 5257 N AB196758 Niigata3 JAPAN: Niigata, Nishikanbara, Maze Takeuchi s.n. I AB196757 Nagano1 JAPAN: Nagano, Minamisaku, Minamimaki Takahashi 21353 A AB196755 Nagano2 JAPAN: Nagano, Chiisagata, Aoki Hisauchi s.n . C AB196754 Nagano3 JAPAN: Nagano, Kitasaku, Karuizawa Kimura s.n. L AB196753 Shizuoka JAPAN: Shizuoka, Fujinomiya, Nehara Konta et al. 771 A AB196 7 62 Kyoto1 JAPAN: Kyoto, Takeno, Amino Tsugaru & Miyah ara 26674 I AB196748 Kyoto2 JAPAN: Kyoto, Kuma no, Kumiha ma Tsugaru & Takahashi 27970 H AB19674 9 Kyoto3 JAPAN: Kyoto, Maizuru, Kan mu r i Isl. Tsugaru & Takahashi 30054 I AB19 6750 Hyogo JAPAN: Hyogo, Kobe, Fukiai Miyake 3869 G AB196746 Shimane JAPAN: Shimane, Yatsuk a, Shi mane Miki s.n. I AB196761 Yamaguchi JAPAN: Yamaguch i, Toyoura, Toyoura Imada 4414 I AB196764 Fukuoka JAPAN: Fukuoka, Munakata Watanabe s.n. J AB196740 Nagasaki JAPAN: Naga s aki, Tsushima Isl., Shimoa gata Ohash i & Soma 7454 I AB196763 Outgroup A. kiusianus Makino JAPAN: Fukuoka, Munakata Watanabe 93 AB196738 A. offic inalis L. JAPAN: Mi yagi, Furuka wa (cult. ) Komatsu 041118 AB195716  Phylogeography of Asparagus schoberioides Kunth (Asparagaceae) in Japan 784 Table 2. Apomorphic characters of the ITS1 sequences obtained from Asparagus schoberioides and two outgroups. 111111 1 1 1 111 3 3 3 4 4 9 023 556 6 7 7 789 sample 5 9 2 5 8 6 8 1 195 454 6 1 2 330 Russia C A A T T GACCCCATC T C T GAG China . G C C C . . G. .. . . T . T A .G . South Korea . G C C C . . G. .. . . . . T A .G . Hokkaido1 . G C C C . . G. .. . . . . . A .G . Hokkaido2 . G . . C A. G. .. C. . . . A .. . Hokkaido3 . G C C C . . G. .. . . . . . A .G . Hokkaido4 . G C C . . . G. .. . . T . . A .G . Hokkaido5 . G C C C . . G. .. . . Y . . A .G . Hokkaido6 . G C C C . . G. .. . . . . . A .G . Iwate . . . . . . . G. .. C. . . . A .. . Miyagi1 . G C C C . . G. .. . . . . . A .G . Miyagi2 . G C C C . . G. .. . . . . . A .G . Saitama . G C C C . . G. .. . . . . . A .G . Yamanashi . G C C C . . G. .. . . . . . A .G . Niigata1 . . C . . . . G. .. C. . . . . .. . Niigata2 . . . . . . . G. .. C. . . . . .. . Niigata3 . . . . . . . G. .. C. . . . A .. T Nagano1 . G C C C . . G. .. . . . . . A .G . Nagano2 . G C C C . . G. T. . . . . . A . G. Nagano3 . . . . . . . G. .. . . . . . . .. . Shizuoka . G C C C . . G. .. . . . . . A .G . Kyoto1 . . . . . . . G. .. C. . . . . .. . Kyoto2 . . C . . . . G. .. C. . . . . .. . Kyoto3 . . . . . . . G. .. C. . . . . .. . Hyogo . G C . . . . G. .. . . . . . . .. . Shimane . . . . . . . G. .. C. . . . . .. . Yamaguchi . . . . . . . G. .. C. . . . . .. . Fukuoka . . . . . A. G. .. C. . . . . .. . Nagasaki . . . . . . . G. .. C. . . . . .. . A. kiusianus . G C . C ACG. .ACC. C . A TG. A. officina lis T G C . C . C GT .. CC . . . A TG. Numbers indicat e the position from the first nucleotide of 5’ region of ITS sequence rem oving the primer region. Copyright © 2011 SciRes. AJPS 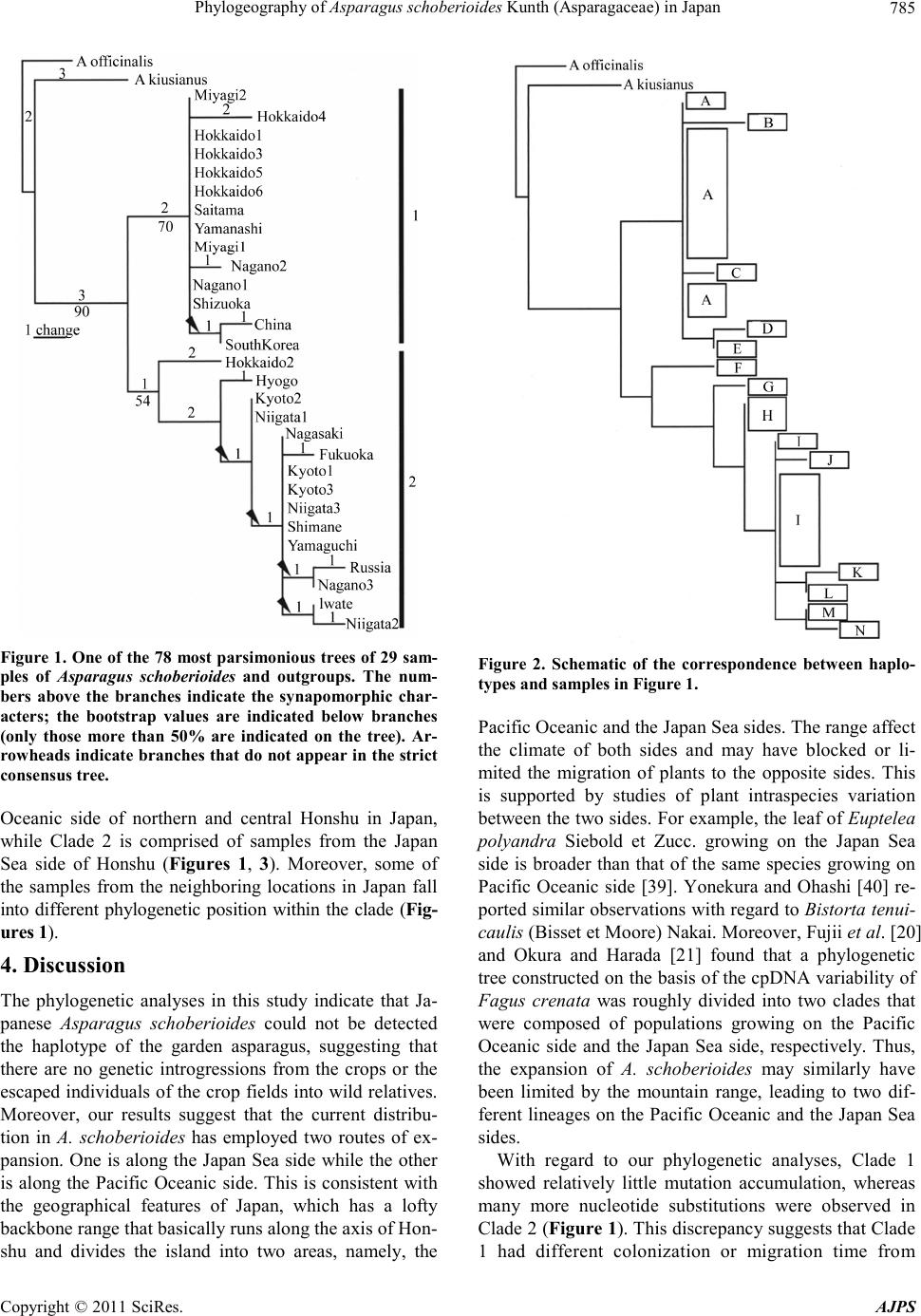 Phylogeography of Asparagus schoberioides Kunth (Asparagaceae ) in Japan785 Figure 1. One of the 78 most parsimonious trees of 29 sam- ples of Asparagus schoberioides and outgroups. The num- bers above the branches indicate the synapomorphic char- acters; the bootstrap values are indicated below branches (only those more than 50% are indicated on the tree). Ar- rowheads indicate branches that do not appear in the strict consensus tree. Oceanic side of northern and central Honshu in Japan, while Clade 2 is comprised of samples from the Japan Sea side of Honshu (Figures 1, 3). Moreover, some of the samples from the neighboring locations in Japan fall into different phylogenetic position within the clade (Fig- ures 1). 4. Discussion The phylogenetic analyses in this study indicate that Ja- panese Asparagus schoberioides could not be detected the haplotype of the garden asparagus, suggesting that there are no genetic introgressions from the crops or the escaped individuals of the crop fields into wild relatives. Moreover, our results suggest that the current distribu- tion in A. schoberioides has employed two routes of ex- pansion. One is along the Japan Sea side while t he other is along the Pacific Oceanic side. This is consistent with the geographical features of Japan, which has a lofty backbone range that basically runs along the axis of Hon- shu and divides the island into two areas, namely, the Figure 2. Schematic of the correspondence between haplo- types and samples in Figure 1. Pacific Oceanic and the Japan Sea sides. The range affect the climate of both sides and may have blocked or li- mited the migration of plants to the opposite sides. This is supported by studies of plant intraspecies variation between the two sides. For example, the leaf of Euptelea polyandra Siebold et Zucc. growing on the Japan Sea side is broader than that of the same species growing on Pacific Oceanic side [39]. Yonekura and Ohashi [40] re- ported similar observations with regard to Bistorta tenui- caulis (Bisset et Moore) Nakai. Moreover, Fujii et al. [20] and Okura and Harada [21] found that a phylogenetic tree constructed on the basis of the cpDNA variability of Fagus crenata was roughly divided into two clades that were composed of populations growing on the Pacific Oceanic side and the Japan Sea side, respectively. Thus, the expansion of A. schoberioides may similarly have been limited by the mountain range, leading to two dif- ferent lineages on the Pacific Oceanic and the Japan Sea sides. With regard to our phylogenetic analyses, Clade 1 showed relatively little mutation accumulation, whereas many more nucleotide substitutions were observed in Clade 2 (Figure 1). This discrepancy suggests that Clade 1 had different colonization or migration time from Copyright © 2011 SciRes. AJPS 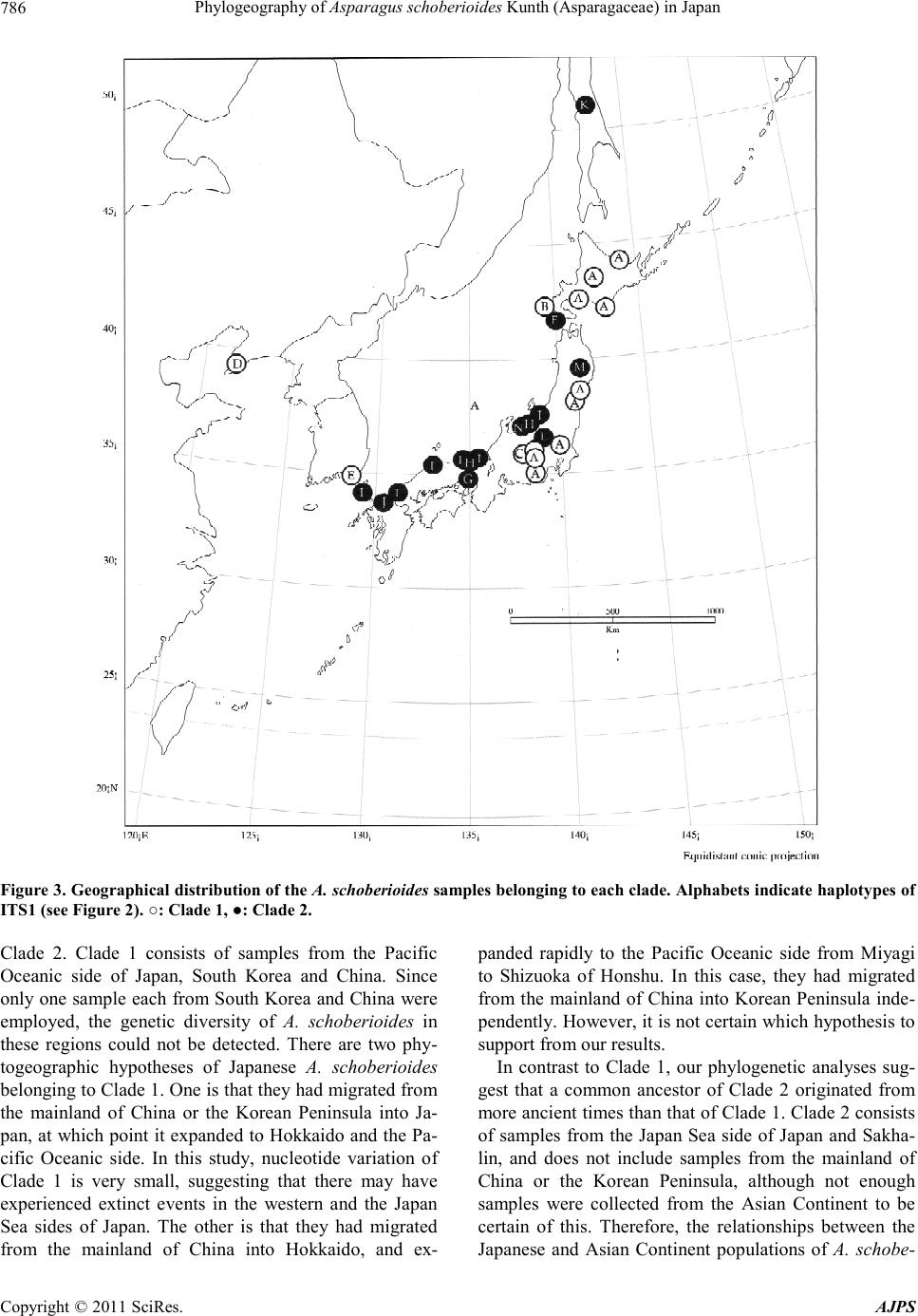 Phylogeography of Asparagus schoberioides Kunth (Asparagaceae) in Japan 786 Figure 3. Geographical distribution of the A. schoberioides samples belonging to each clade. Alphabets indicate haplotypes of ITS1 (see Figure 2). ○: Clade 1, ●: Clade 2. Clade 2. Clade 1 consists of samples from the Pacific Oceanic side of Japan, South Korea and China. Since only one sample each from South Korea and China were employed, the genetic diversity of A. schoberioides in these regions could not be detected. There are two phy- togeographic hypotheses of Japanese A. schoberioides belonging to Clade 1. One is that they had migrated from the mainland of China or the Korean Peninsula into Ja- pan, at which point it expanded to Hokkaido and the Pa- cific Oceanic side. In this study, nucleotide variation of Clade 1 is very small, suggesting that there may have experienced extinct events in the western and the Japan Sea sides of Japan. The other is that they had migrated from the mainland of China into Hokkaido, and ex- panded rapidly to the Pacific Oceanic side from Miyagi to Shizuoka of Honshu. In this case, they had migrated from the mainland of China into Korean Peninsula inde- pendently. However, it is not certain which hypothesis to support from our results. In contrast to Clade 1, our phylogenetic analyses sug- gest that a common ancestor of Clade 2 originated from more ancient times than that of Clade 1. Clade 2 consists of samples from the Japan Sea side of Japan and Sakha- lin, and does not include samples from the mainland of China or the Korean Peninsula, although not enough samples were collected from the Asian Continent to be certain of this. Therefore, the relationships between the Japanese and Asian Continent populations of A. schobe- Copyright © 2011 SciRes. AJPS  Phylogeography of Asparagus schoberioides Kunth (Asparagaceae ) in Japan787 rioides remain unclear at present. Our phylogenetic analyses also indicate that within each clade, the phylogenetic relationship and geographic distribution do not correlate. For example, some neigh- boring locations in Japan belong to different phyloge- netic position within the clade. Examples of this are the three samples from Niigata and the three Kyoto samples in Clade 2 (Figure 1). This suggests that A. schobe- rioides populations in Niigata and Kyoto consist of indi- viduals that have experienced different histories. Fur- thermore, northern samples such as those from Sakhalin, Hokkaido and Iwate also appeared in various phyloge- netic positions in Clade 2 (Figure 1). These results indi- cate that the migration of A. schoberioides along the Ja- pan Sea side of Japan has occurred repeatedly. Thus, our phylogeographic analysis has outlined the different histories of A. schoberioides between the Pa- cific Oceanic and the Japan Sea sides in Japan. What has aided the rapid migration of samples in Clade 1 and the frequent colonization of samples in Clade? One answer may lie in the fact that A. schoberioides bears reddish berries that may be bird-dispersed [41-44]. This may aid the rapid dispersal of seeds of A. schoberioides through- out the two routes of Japan. In general, a morphologically recognized species may be regarded as a composite of subtly defined cryptic spe- cies each of which has equal status [45]. In fact, such a definition takes the systematic approach to an extreme which would appear to be unworkable in plants. For example, Yatabe et al. [46] concluded that there are some cryptic species of Asplenium nidus L. (Asplenia- ceae) in West Jawa on the basis of sequence variations using rbcL of cpDNA. In this study, A. schoberioides had nucleotide differentiation between Clade 1 and Clade 2, suggesting that there are cryptic species with the geo- graphical features of Japan. 5. Conclusions There are no genetic introgression between A. schobe- rioides and A. officinalis (garden asparagus) and two migr ation r outes o f A. schoberioides exist in Japan. Such research, when applied to A. schoberioides and to other Japanese plant taxa, will provide new insights into the phytogeography of Japanese plants. 6. Acknowledgements We wish to thank H. Yamaji, M. Kawata, M. Maki, T. Yamashiro, P.-Y. Yun, T. Ito, H. Ashizawa, Y. Mashiko, M. Nakada, R. Shinohara, S.-Y. Kim, M. Hirai and H. To kairin for providing muc h help and a dvice. T his stud y was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan and Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education Science and Technology (2010-0029630) and in part by the Sasakawa Scientific Research Grant from The Japan Science Soci- ety. REFERENCES [1] J. C. Avise, J. Arnold, R. M. Ball, E. Bermingham, T. Lamb, J. E. Neigel, C. A. Reeb and N. C. Saunders, “In- traspecific Phylogeography: The Mitochondrial DNA Bri- dge between Population Genetics and Systematics,” An- nual Review of Ecology, Evolution, and Systematics, Vol. 18, 1987, pp. 4 89- 5 52. [2] J. C. Avise, “Molecular Markers, Natural History, and Evolution,” Chapman & Hall, New York, 1994. doi:10.1007/978-1-4615-2381-9 [3] J. C. Avise, “Phylogeography: The History and Forma- tion of Spe ci e s,” H a rvard U niv er s ity Pres s , L ondon, 200 0. [4] D. E. Soltis and P. S. Soltis, “Choosing an Approach and an Appropriate Gene for Phylogenetic Analysis,” In: D. E. Soltis, P. S. Soltis and J. J. Doyle, Eds., Molecular Sys- tematics of Plant II: DNA Sequencin g, Kluwer Academic Publishe r s , Mass a c hus e tts , 19 98, pp. 1- 42. [5] J. J. Doyle and J. I. Davis, “Homology in Molecular Phylogenetics: A Parsimony Perspective,” In: D. E. Soltis, P. S. Soltis and J. J. Doyle, Eds., Molecular Systematics of Plant II: DNA Sequencing, Kluwer Academic Publish- ers, Massachusetts, 1998, pp. 101-131. do i:10.1007/978-1-4615-5419-6_4 [6] J. Ohwi, “Asparagus L.,” In: F. G. Meyer and E. H. Walker, Eds., Flora of Japan, Smithonian Institution, Washington , DC, 1965, p. 300. [7] X. Chen and K. G. Tamanian, “Asparagus L.,” In: Z. Y. Wu and P. H. Raven, Eds., Flora of China, Vol. 24, (Flagellariaceae through Marantaceae), Missouri Botani- cal Garden Press, St. Louis, 2000, pp. 139-146. [8] T. Fukuda, H. Ashizawa, T. Nakamura, T. Ochiai, A. Kanno, T. Kameya and J. Yokoyama, “Molecular Phy- logeny of the Genus Asparagus (Asparagaceae) Inferred from Plastid petB Intron and petD—rpoA Intergenic Spacer Seq uen ces,” P lan t Sp ecies B io log y, Vol. 20, No. 2, 2005, pp . 123-134 . [9] T. Ochiai, T. Sonoda, A. Kanno and T. Kameya, “Inter- specific Hybri ds between Asparagus schoberioides Kunth and A. officinalis L.,” Acta Horticulturae, Vol. 589, 2002 , pp. 225-229. [10] T. Ito, T. Ochiai, T. Fukuda, H. Ashizawa, T. Sonoda, T. Kameya and A. Kanno, “Potential of Interspecific Hy- brids in Asparagaceae,” Acta Horticulturae, Vol. 776, 2008, pp. 279-28 4. [11] M. L. Arnold, “Natural Hybridization and Evolution,” Oxford University Press, 1997, New York. [12] D. Bartsch, M. Lehnen, J. Clegg, M. Pohi-Orf, I. Schu- phan and N. C. Ellstrand, “Impact of Gene Flow from Cultivated Beet on Genetic Diversity of Wild Sea Beet Populations,” Molecular Ecology, Vol. 8, No. 10, 1999, Copyright © 2011 SciRes. AJPS  Phylogeography of Asparagus schoberioides Kunth (Asparagaceae) in Japan 788 pp. 1733 -1741. doi:10.1046/j.1365-294x.1999.00769.x [13] P. Gepts and R. Papa, “Possible Effects of (trans) Gene Flow from Crops on the Genetic Diversity from Land Races and Wild Relatives,” Environmental Biosafety Re- search, Vol. 2, No. 2, 2003, pp. 89-103. do i:10.1051/ebr:2003009 [14] M. A. Chapman and J. M. Burke, “Letting the Gene Out of the Bottle: The Population Genetics of Genetically Mod ified Crop s,” New Ph ytologist, Vol. 170, No. 3, 2006, pp. 429- 443. doi:10.1111/j.1469-8137.2006.01710.x [15] E. Bitocchi, L. Nanni, M. Rossi, D. Rau, E. Bellucci, G. G. Vendramin and R. Papa, “Introgression from Modern Hybrid Varieties into Landrace Poplation of Maize (Zea mays ssp. Mays L.) in Central Italy,” Molecular Ecology, Vol. 18, No. 4, 2009, pp. 603-621. doi:10.1111/j.1365-294X.2008.04064.x [16] J.-F. Arnaud, S. Fénart, C. Godé, S. Deledicque, P. Touzet and J. Cu guen, “Fine-S cale Geogr aphical Struct ur e of Ge- netic Diversity in Inland Wild Beet Populations,” Mo- lecular Ecology, Vol. 18, No. 15, 2009, pp. 3201-3215. doi:10.1111/j.1365-294X.2009.04279.x [17] Y. Tsumura, H. Taguchi, Y. Suyama and K. Ohba, “Geo- graphical Cline of Chloroplast DNA Variation in Abies mariesii,” Theoretical and Applied Genetics, Vol. 89, No. 7-8, 1994, pp. 922-925. doi:10.1007/BF00224518 [18] T. Ohi, T. Kajita and J. Murata, “Distinct Geographic Structure as Evidenced by Chloroplast DNA Haplotypes and Ploidy Level in Japanese Aucuba (Aucubaceae),” American Journal of Botany, Vol. 90, No. 11, 2003, pp. 1645-1652. do i:10. 3732/ajb.90.11.1645 [19] N. To maru , M. Taka h ash i , Y. Tsu mur a, Y. Taka h ash i an d K. Ohba, “Intraspecific Variation and Phylogeographic Patterns of Fagus crena ta (Fagaceae) Mitochondri al DNA,” American Journal of Botany, Vol. 85, No. 5, 1998, pp. 629-636. doi:10.2307/2446531 [20] N. Fujii, N. Tomaru, K. Okuyama, K. Koike, T. Mikami and K. Ueda, “Chloroplast DNA Phylogeography of Fagus crenata (Fagaceae) in Japan,” Plant Systematics and Evo- lution, Vol. 23 2, N o. 1-2 , 200 2, p p. 21- 33. doi:10.1007/s006060200024 [21] T. Okura and K. Harada, “Phylogeographical Structure Revealed by Chloroplast DNA Variation in Japanese Beech (Fagus crenata Blime),” Heredity, Vol. 88 , No. 4 , 20 02, pp. 322-329 . doi :10.103 8/sj. hd y.680004 8 [22] N. Fujii, K. Ueda, Y. Watano and T. Shimizu, “Further Analysis of Intraspecific Sequence Variation of Chloro- plast DNA in Primula cunefolia Ledeb. (Primulaceae): Implications for Biogeography of the Japanese Alpine flora,” Journal of Plant Research, Vol. 112, 1999, pp. 87-95. doi:10.1007/PL0 0013866 [23] M. Kanno , J. Yokoyama, Y. Suya ma, M. Oh yama, T. Ito and M. Suzuki, “Geographic Distribution of two Haplo- types of Chloroplast DNA in F our Oak Speci es (Quercus) in Japan,” Journal of Plant Research, Vol. 117, No. 4, 2004, pp . 311-317. doi:10.1007/s10265-004-0160-8 [24] T. Ohi, M. Wakabayashi, S. Wu and J. Murata, “Phy- logeography of Stachyurus praecox (Stachyuraceae) in the Japanese Archipelago Based on Chloroplast DNA Haplotypes,” Journal of Japanese Botany, Vol. 78, No. 1, 2003, pp . 1- 14. [25] K. Aoki, T. Suzuki, T.-W. Hsu and N. Murakami, “Phy- logeography of the Component Species of Broad-Leaved Evergreen Forests in Japan, Based on Chloroplast DNA Variation,” Journal of Plant Research, Vol. 117, No. 1, 2004, pp. 77- 9 4. doi:10.1007/s10265-003-0132-4 [26] P. Taberlet, L. Fumagalli, A. G. Wust-Saucy and J. F Coson, “Comparative Phylogeography and Postglacial Colonization Routes in Europe,” Molecular Ecology, Vol. 7, No. 4, 1998, pp. 453-464. doi:10.1046/j.1365-294x.1998.00289.x [27] T. Nishizawa and Y. Watano, “Primer Pairs Suitable for PCR-SSCP Analysis of Chloroplast DNA in Angio- sperms,” Journal of Phytogeography and Taxonomy, Vol. 48, No. 1, 2000 , pp. 67-70. [28] C. W. Dick, K. Abdul-Salim and E. Bermingham, “Mo- lecular Systematic Analysis Reveals Cryptic Tertiary Di- versificatio n of a Widesp read Tropical Rai n Forest Tree,” American Naturalist, Vol. 162, No. 6, 2003, pp. 691-703. doi:10.1086/379795 [29] J. Yokoyama, T. Fukuda, A. Yokoyama and M. Maki, “The intersectional hybrid between Weigela hortensis and W. maximowiczii (Caprifoliaceae),” Botanical Journal of the Linnean Society, Vol. 138, No. 3, 2002, pp. 369-380. doi:10.1046/j.1095-8339.2002.00033.x [30] T. Yamashiro, T. Fukuda, J. Yokoyama and M. Maki, “Molecular Phylogeny of Vincetoxicum (Apocynaceae- Asclepiadoideae) Based on the Nucleotide Sequences of cpDNA and nrDNA,” Molecular Phylogenetics and Evo- lution, Vol. 31, No. 2, 2004, pp. 689-700. doi:10.1016/j.ympev.2003.08.016 [31] H. Tsukaya, “Gene Flow between Inpatiens radicans and I. javensis (Balsaminaceae) in Gunung Pangrango, Cen- tral Java, Indonesia,” American Journal of Botany, Vol. 91, No. 12, 2004, pp. 2119-2123. do i:10.3732/ajb.91.12.2119 [32] H. Tsukaya, Y. Lola wa, M. Kondo and H. Ohba, “Large- Scale General Collection of Wild-Plant DNA in Mustang, Nepal,” Journal of Plant Research, Vol. 118, No. 1, 2005, pp. 57-6 0. doi:10.1007/s10265-005-0196-4 [33] J. Yokoyama, T. Fukuda and H. Tsukaya, “Morphologi- cal and Molecular Variation of Mitchella undulata Sie- blod et Zucc., with S pecial Refer ence to S ystemati c Treat- ment o f the D warf Fo rm fro m Ya kushi ma Island ,” Journal of Plant Resear c h, Vol. 116, N o. 4, 2003, pp. 309-316. doi:10.1007/s10265-003-0105-7 [34] T. J. White, T. Bruns, S. Lee and J. Taylor, “Amplifica- tion and Direct Sequencing of Fungal Ribosomal RNA genes for Phylogenetics,” In: M. Innis, D. Gelfan, J. Sninsky and T. J. White, Eds., PCR Protocols: A Guide to Methods and Application, Academic Press, San Diego, 1990, pp . 315-322 . [35] J. D. Thompson, T. J. Gibson, F. Plewniak, F. Jean- mougin and D. G. Higgins, “The Clustal_X Windows In- terface: Flexible Strategies for Multiple Sequence Align- ment Aided by Quality Analysis Tools,” Nucleic Acids Copyright © 2011 SciRes. AJPS  Phylogeography of Asparagus schoberioides Kunth (Asparagaceae ) in Japan Copyright © 2011 SciRes. AJPS 789 Research, Vol. 25, No. 24, 19 97, pp. 4876-48 82 . doi:10.1093/nar/25.24.4876 [36] D. L. Swofford, “PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods). Version 4.0b10.,” Sinauer Assoc iate s , Sunde rl and, 2002. [37] W. P. Maddison, “The Discovery and Importance of Mul- tiple Islands of Most-Parsimonious Trees,” Systematic Zo- ology, Vol. 4 0, No. 3, 199 1, pp. 315-328. doi:10.2307/2992325 [38] J. Felsenstein, “Confidence Limits on Phylogenies: An Approach Using the Bootstrap,” Evolution, Vol. 39, No. 4, 1985, pp . 783-791. doi:10.2307/2408678 [39] H. Ikenoue and S. Okitsu, “Comparison of Leaf Area and Leaf Shape of Euptelea polyandra Sieb. Et Zucc. be- tween th e Pacific Ocean Side and t he Sea of Japan Side, ” Journal of Phytogeography and Taxonomy, Vol. 42, No. 2, 1995, pp. 125-131. [40] K. Yonekura and H. Ohashi, “Geographical Distribution and Variation in Bistorta tenuicaulis and Its New Variety from Japan, with Special Reference to Gynodioecy of B. tenuicaulis and B. abukumensis (Polygonaceae), ” Journal of Japanese Botany, Vol . 73, No. 1, 1998, pp. 1-1 1 . [41] T. J. Givnish, K. J. Sytsma, J. F. Smith and W. J. Hann, “Thorn-Like Prickles and Heterophylly in Cyanea: Ad- aptations to Extinct Avian Browsers on Hawaii?” Pro- ceedings of National Academy of Sciences of the USA, Vol. 91, No. 7, 1994, pp. 2810-2814. do i:10.1073/pnas.91.7.2810 [42] D. G. Howarth, D. E. Gardner and C. W. Morgan, “Phy- logeny of Rubus Subgenus Idaeobatus (Rosaceae) and It s Implications toward Colonization of the Hawaii Islands,” Systematic Botany, Vol. 22, No. 3, 1997, pp. 433-442. doi:10.2307/2419819 [43] T. Fukuda, J. Yokoyama and H. Ohashi, “Phylogeny and Biogeography of the Genus Lycium (Solanaceae): Infer- ences from Chloroplast DNA Sequences,” Molecular Phylogenetics and Evolution, Vol. 19, No. 2, 2001, pp. 246-258. doi:10.1006/mpev.2001.0921 [44] J. R. Worth, G. J. Jordan, J. R. Marthick, G. E. Mckinnon and R. E. Vaillancourt, “Chloroplast Evidence for Geo- graphic Stasis of the Australian Bird-Dispersed Shrub Tasmannia lanceolata (Winteraceae),” Molecular Ecol- ogy, Vol. 19, No. 14, 2010, pp. 2949-2963. doi:10.1111/j.1365-294X.2010.04725.x [45] M. King, “Species Evolution: The Role of Chromosome Change,” Cambridge University Press, Melbourne, 1993. [46] Y. Yatabe, D. Darnaedi and N. Murakami, “Allozyme Analysis of Cr yptic Species i n the Asplenium nidus Com- plex from West Java, Indonesia,” Journal of Plant Re- search, Vol. 115, No. 6, 2002, pp. 483-490. do i:10.1007/s10265-002-0060-8
|