 American Journal of Plant Sciences, 2011, 2, 765-775 doi :1 0.4236/ aj ps.2011 .26091 Publ i s hed Online December 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS 765 Nucleotide Sequence Variations in a Medicinal Relative of Asparagus, Asparagus cochinchinensis (Lour.) Merrill (Asparagaceae) Tatsuya Fukuda1*, In-Ja Song2, Takuro Ito3, Hiroshi Hayakawa4, Yukio Minamiya4, Akira Kanno5, Hokuto Nakayama6, Jun Yokoyama7 1Faculty of Agriculture, Kochi University, Nankoku, Japan; 2Subtropical Horticulture Research Institute, Jeju National University, Jeju, Korea; 3Institute for Advanced Biosciences, Keio University, Tsuruoka, Japan; 4United Graduate School of Agricultural Sci- ences, Eh ime Universit y, Mon obe, Jap an ; 5Graduate School of Life Sciences, Tohoku University, Sendai, Japan; 6Graduate School of Science, University of Tokyo, Tokyo, Japan; 7Depertment of Biology, Faculity of Science, Yamagata University, Yamagata, Japan. Email: *tfukuda@kochi-u.ac.jp Received February 20th, 2011; revised March 15th, 2011; accepted April 6th, 2011. ABSTRACT To determine the evolutionary history of Asparagus cochinchinensis (Asparagaceae), we investigated the geographic pattern of its n ucleotide sequence variations in Japan, Taiwan, South Korea and China. We found 21 polym orphic nu - cleotide sites by sequencing the internal transcribed spacer (ITS) region, which gave rise to a total of 15 haplotypes, labeled A to O. The A-type was found only in inland China (Guizhou and Sichuan), and the other haplotypes in China extended to several lineages; therefore, A. cochinchinensis may have its origin in the interior of China. The I-type has large distribu tion area and it also experienced a quick expansion in relative recent yea rs. Haplotype differentia tion was observed between the eastern and western side of the Central Mountain Ridge in Taiwan. Two lineages were found in Japan, one in the Yaeyama Islands and the other in remaining areas of Japan, implying that A. cochinchinensis inde- pendently colonized in Japan at least twice. Keywords: Asparagus cochinchinensis, Internal Transcribed Spacer (ITS), Phylogeny, Phylogeography 1. Introduction Genetic variation within species is one o f the fundamen- tal underpinnings of biological diversity [1]. The distri- bution of genetic variation is shaped by processes that are extrinsic to the species, such as ecological events or selective regimes, as well as intrinsic factors, such as the type of mating system. When considered in a geographi- cal context, the structure of this variation is a product of current genetic exchange within a species as well as his- torical relationships between populations. By taking into account the genealogical relationships between haplo- types as well as their geographical distribution, phylo- geographical methods can potentially determine the his- torical and recurrent population-level processes that shape current patterns of variation. Moreover, genetic li- neages are often localized geographically and may share a common history, such as episodes of vicariance, routes of migration, colonization, and recent population expan- sions [2-4]. Such historical insights can be obtained by identifying the haplotype variations, which are then over- laid upon a geographical frame, reflecting the spatiotem- poral dynamics of the organi sm studied. The number of phylogenetic studies based on molecu- lar data has grown enormously in recent years, and most of the recent studies are concerned with closely related species or variation within species. In particular, the use of molecular markers has considerably improved our knowledge about how past events shape the genetic di- versity within a species [2-4]. In angiosperms, for exam- ple, the application of phylogenetic analysis is emerging as an important and practical tool for the study of eco- nomically important species such as rice, maize, sun- flower and cassava, and their relatives [5-13]. These ana- lyses also helped to unveil the domestication processes of these crops, and the ancestors and close relatives of a given species have repeatedly provided sources of useful traits to genetically impoverished crops. Moreover, con-  Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 766 (Lour.) Merril l (Asparagace ae) scious efforts to search for desirable traits in plants have been underway for the past century, and in recent de- cades species with desirable traits have come to be re- garded as important biological resources in need of con- servation [14]. The population structure and history of a given species is thus an important research focus, be- cause this knowledge is needed to design concerted ef- forts for conservation of the species and to understand the evolutionary processes leading to genetic diversity. Recently, various intergenic spacers have been widely analyzed to assess the genetic variability of wild plants [15-22]. These markers are known to be mostly neutral with relative high mutation rates, and, in associatio n with the geological history, provide information to estimate the extent of seed dispersal and track migration routes [23-27]. The genus Asparagus (Asparagaceae) is a large group distributed in arid and sub-arid regions of the Old World [28-35]. This genus is well known by virtue of the fact that it includes commercially important species of vege- tables, most notably A. officinalis, as well as some orna- mentally or medicinally important species such as A. asparagoides, A. scandens, A. plumosus, and A. falcatus [36]. A. cochinchinensis (Lour.) Merrill is one of the medicinally important species in the genus, and has small flowers in raceme-like inflorescences, whitish-colored berries, and swollen roots [37]. It is mainly distributed along seashores in temperate regions from China to Ja- pan [31,37]. Here, to enhance our understanding of the evolutionary history of Asparagaceae, we describe the DNA polymorphisms of the internal transcribed spacer (ITS) region in A. cochinchinensis and discuss how these genetic variations spread within the present distribution with morphological differentiation. 2. Materials and Methods 2.1. Plant Materials Thirty-seven samples of Asparagus cochinchinensis were examined in this study (Table 1). A. lycopodineus Wall. ex Baker, A. schoberioides Kunth and A. officinalis L. were selected as outgroups on the basis of phylogenetic analyses of the genus Asparagus [38]. Vouchers for all species sampled in this study have been deposited in the Herb ar iu m, Gr ad uate Sc ho ol of Sc ie nc e, T o hoku Uni ver - sity (TUS), and the Herbarium of Tsumura Laboratory (THS). 2.2. Morphological Analysis Length and width of a phylloclade vary extensively in Asparagus cochinchinensis [37]. Therefore, a prelimi- nary morphological analysis of A. cochinchinensis and its allied species was conducted by measuring the con- tinuous macromorphological variables of length and wid- th of 5 phylloclades in each individual. Measurements were taken with a digital caliper. Phylloclade measure- ments we re take n fr om flo weri ng ind i vid ua l s. 2.3. DNA Extraction, Amplification, and Sequencing Total DNA was isolated from 200 - 300 mg of phyllo- clades with a Plant Genomic DNA Mini Kit (VIOGENE, Sunnyvale, USA), according to the manufacturer’s pro- tocol. Isolated DNA was resuspended in Tris-EDTA (TE) buffer and stored at –20˚C until use . Of Asparagus species, only A. cochinchinensis has two relatively large deletions, which are located between the ndhC and trnV genes in chloroplast DNA (cpDNA) and are 95 bp and 347 bp in length [39]. To identify A. cochinchinensis, therefore, two regions of cpDNA in- cluding indels were amplified using two pairs of primers (Table 2) [40]. For phylogenetic analysis in this species, we amplified the ITS1 region with primers designed by White et al. [41]. DNA was amplified by PCR in a 50 µl reaction volume containing approximately 50 ng total DNA, 10 mmol/l Tris-HCl buffer (pH 8.3) with 50 mmol/L KCl and 1.5 mmol/L MgCl2, 0.2 mmol/L of each dNTP, 1.25 units Taq DNA polymerase (TAKARA) and 0.5 µmol/L of each primer. We used the following thermal cycle profile for amplification: 1.5 min at 94˚C, 2 min at 48˚C, and 3 min at 72˚C for 40 cycles, followed by 15 min of final extension at 72˚C. After amplification, reaction mixtures were subjected to electrophoresis in 1% - 2% low-melting-temperature agarose gels for purification of amplified products. We sequenced the purified PCR pro- ducts using a DYEnamic ET-terminator Cycle Sequenc- ing Kit (Amersham Pharmacia) and a Model 373A auto- mated sequencer (Applied BioSystems) according to the manufacturer’s instructions. For sequencing, we used the same primers as those used for amplification. 2.4. Data Analysis Sequences for the ITS region were prealigned with the CLUSTAL X program [42]. Alignment for this region required the inclusion of an indel. This indel was coded as binary (0 or 1) and treated as the fifth character. Phylogenetic analysis and a test of clade support were conducted using the PAUP* program (version 4.0b10) [43]. Maximum parsimony analyses were carried out via a heuristic search with TBR branch swapping and the MULPERS option. Multiple islands of the most parsi- monious trees [44] were identified using the heuristic option with 100 random sequence additions. To estimate confidence levels of monophyletic groups, the bootstrap Copyright © 2011 SciRes. AJPS  Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis (Lour.) Merril l (Asparagaceae) Copyright © 2011 SciRes. AJPS 767 Table 1. List of taxa, sources and haplotypes of plant materials. Taxon OTU name Locality Haplotype Aspa ragus cochinchinens is (Lour.) Merril l var. cochinchinensis ACChinaSC1 CHINA: Sichuan, Gulin A ACChinaSC2 CHINA: Sichuan, Gulin A ACChinaGZ CHINA: Guizhou, Anshun A ACChinaHN CHINA: Hainan, Sanya B ACChinaAH CHINA: Anhui, Shitai J ACChinaHB CHINA: Hubei, Xianning K ACChinaHK CHINA: Hon gKong, M t.Victoria I ACTaiwanHL TAIWAN: Hualien, Chosi C ACTaiwanTC TAIWAN: Taichung, Hoping H ACTaiwanNT TAIWAN: Nantou, Mt. Nankao-shan F ACTaiwanTP TAIWAN: Taipei, Kungliao C ACTaiwanKS TAIWAN: Kaohsiung, Mt. Tsuyun-shan G ACTaiwanKL TAIWAN: Keelung, Hepingdao I ACSouthKorea SOUTH KOREA: Jeollanam-do, Yochon I ACIriomote JAPAN: Okinawa, Iriomote Isl. D ACYonaguni JAPAN: Okinawa, Yonaguni Isl. E ACMiyako JAPAN: Okinawa, Miyako Isl. I ACYakushima JAPAN: Kagoshima, Yakushima Isl. I ACKagoshima1 JAPAN: Kagoshima, Kimotsuki, Uchinoura I ACKagos hima2 JAP AN: Kagoshima, Kimotsuki, Nejime I ACMiya zaki JAP AN: Miyazaki, Koyu, Kawaminami L ACFukuoka1 JAPAN: Fukuoka, Munakata, Oshima I ACFukuoka2 JAPAN: Fukuoka, Munakata, Genkai I ACKoch i JAPAN: Kochi, Tos a, Kagami N ACKagawa JAPAN: Kagawa, Takamatsu M ACEhime JAPAN: Ehime, Hojo I ACTokushima1 JAPAN: Tokushima, Kaifu, Mugi N ACTokushima2 JAPAN: Tokushima, Tokushima M ACOkayama JAPAN: Okayama, Okayama N ACAwaji1 JAPAN: Hyogo, Awaji Isl. (Tsuna, Ichinomiya) N ACAwaji2 JAPAN: Hyogo, Awaji Isl. (Mihara, Nandan) N ACWakayama1 JAPAN: Wakayama, Higashimuro, Taiji I ACWakayama2 JAPAN: Wakayama, Nishimuro, Shirahama O ACShizuoka1 JAPAN: Shizuoka, Numazu I ACShizuoka2 JAPAN: Shizuoka, Ito I ACKanagawa JAPAN: Kanagawa, Kamakura I A. cochinchinensis (Lour.) Merrill var. pygmaeus Makino ACpygmarus JAPAN: cultivated in Tohoku Univ. I Outgroup A. officina lis L. JAPAN: cult ivat ed in Tohoku Univ. ***** A. schoberioides Knuth JAPAN: Shimane, Ya tsuka, Shimane ***** A. lycopodineus Wall. Ex Baker CHINA: Sichuan, Tianquan ***** 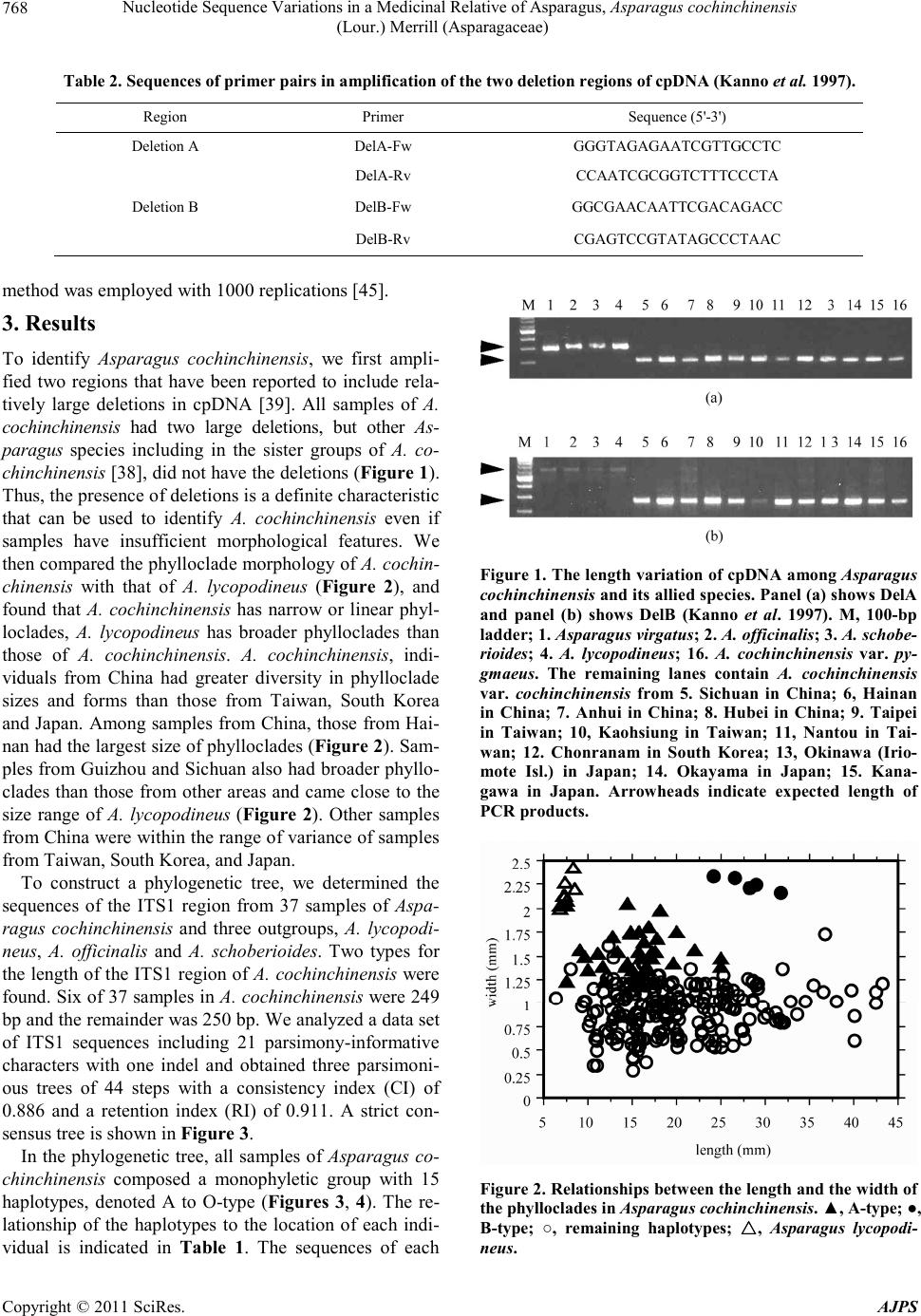 Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 768 (Lour.) Merril l (Asparagace ae) Table 2. Sequences of primer pairs in amplification of the two deletion regions of cpDNA (Kanno et al. 1997). Region Primer Sequence (5'-3') Deletion A DelA-Fw GGGTAGAGAATCGTTGCCTC DelA-Rv CCAATCGCGGTCTTTCCCTA Deletion B DelB -Fw GGCGAACAATTC GACAGACC DelB-Rv CGAGTCCGTATAGCCCTAAC method was employed with 1000 replications [45]. 3. Results To identify Asparagus cochinchinensis, we first ampli- fied two regions that have been reported to include rela- tively large deletions in cpDNA [39]. All samples of A. cochinchinensis had two large deletions, but other As- paragus species including in the sister groups of A. co- chinchinensis [38] , did no t have t he de leti ons ( Figure 1). Thus, the presence of deletions is a definite characteristic that can be used to identify A. cochinchinensis even if samples have insufficient morphological features. We then compared the phylloclade morphology of A. cochin- chinensis with that of A. lycopodineus (Figure 2), and found that A. cochinchinensis has narrow or linear phyl- loclades, A. lycopodineus has broader phylloclades than those of A. cochinchinensis. A. cochinchinensis, indi- viduals from China had greater diversity in phylloclade sizes and forms than those from Taiwan, South Korea and Japan. Among samples from China, those from Hai- nan had the largest size of phylloclades (Figure 2). Sam- ples from Guizhou and Sichuan also had broader phyllo- clades than those from other areas and came close to the size range of A. lycopodineus (Figure 2). Other samples from China were within the range of variance of samples from Taiwan, South Korea, and Japan. To construct a phylogenetic tree, we determined the sequences of the ITS1 region from 37 samples of Aspa- ragus cochinchinensis and three outgroups, A. lycopodi- neus, A. officinalis and A. schoberioides. Two types for the length of the ITS1 region of A. cochinchinensis were found. Six of 37 samples in A. cochinchinensis were 249 bp and the remainder was 250 bp. We analyzed a data set of ITS1 sequences including 21 parsimony-informative characters with one indel and obtained three parsimoni- ous trees of 44 steps with a consistency index (CI) of 0.886 and a retention index (RI) of 0.911. A strict con- sensus tree is shown in Figure 3. In the phylogenetic tree, all samples of Asparagus co- chinchinensis composed a monophyletic group with 15 haplotypes, denoted A to O-type (Figures 3, 4). The re- lationship of the haplotypes to the location of each indi- vidual is indicated in Table 1. The sequences of each Figure 1. The length variation of cpDNA among Asparagus coch inchinensis and its allied species. Panel (a) shows DelA and panel (b) shows DelB (Kanno et al. 1997). M, 100-bp ladder; 1. Asparagus virgatus; 2. A. officinalis; 3. A. schobe- rioid es ; 4. A. lycopodineus; 16. A. cochinchinensis var. py- gmaeus. The remaining lanes contain A. cochinchinensis var. coch i nchinensis from 5. Sichuan in China; 6, Hainan in China; 7. Anhui in China; 8. Hubei in China; 9. Taipei in Taiwan; 10, Kaohsiung in Taiwan; 11, Nantou in Tai- wan; 12. Chonranam in South Korea; 13, Okinawa (Irio- mote Isl.) in Japan; 14. Okayama in Japan; 15. Kana- gawa in Japan. Arrowheads indicate expected length of PCR products. Figure 2. Relationships between the length and the width of the phylloclades in Asparagus cochinchinensis. ▲, A-type; ●, B-type; ○, remaining haplotypes; , △Asparagus lycopodi- neus. Copyright © 2011 SciRes. AJPS 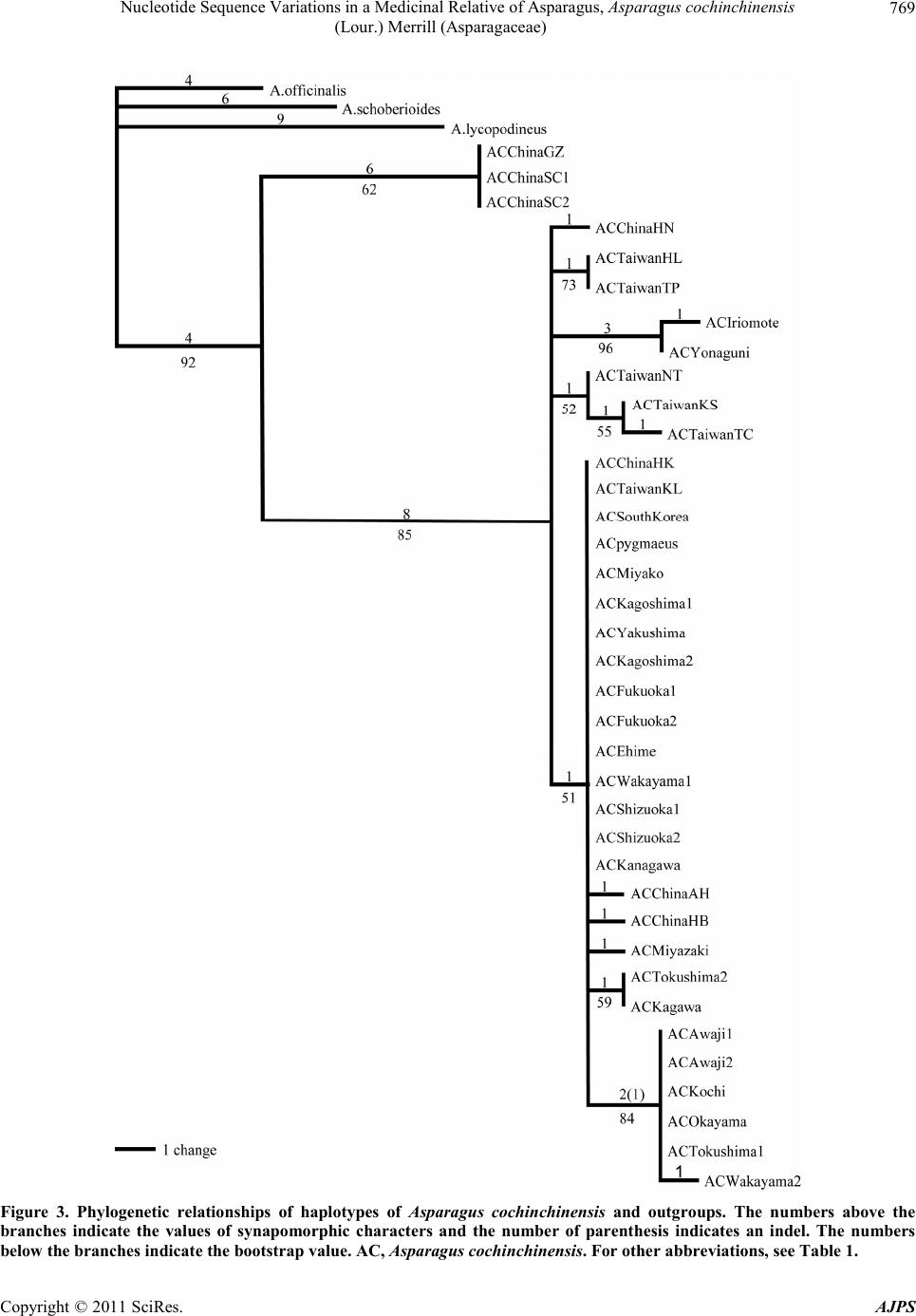 Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 769 (Lour.) Merril l (Asparagaceae) Figure 3. Phylogenetic relationships of haplotypes of Asparagus cochinchinensis and outgroups. The numbers above the branches indicate the values of synapomorphic characters and the number of parenthesis indicates an indel. The numbers below the branches indicate the bootstrap value. AC, Asparagus co chinchinensis. For other abbreviations, see Table 1. Copyright © 2011 SciRes. AJPS 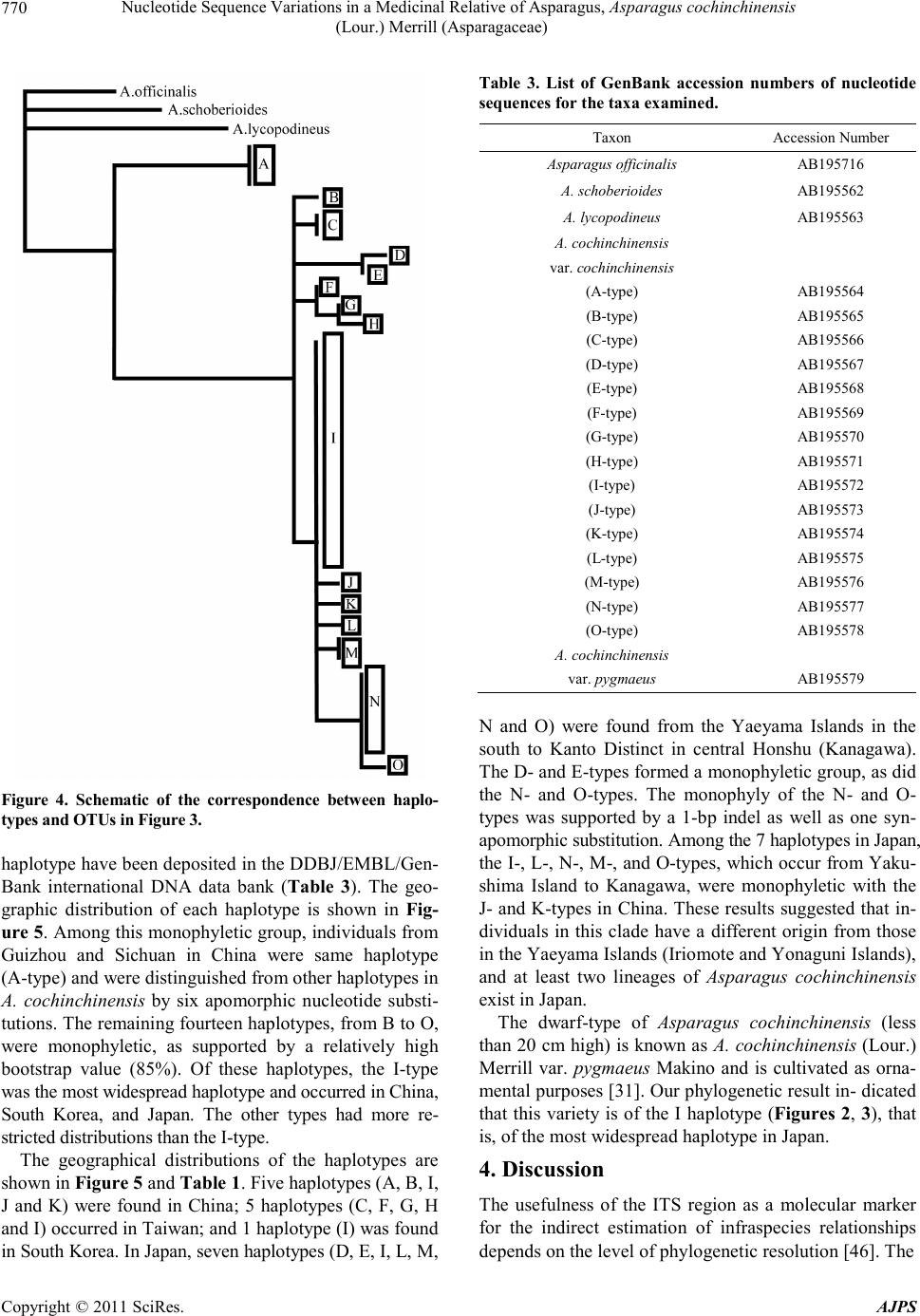 Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 770 (Lour.) Merril l (Asparagace ae) Figure 4. Schematic of the correspondence between haplo- types and OTUs in Figure 3. haplotype have been deposited in the DDBJ/EMBL/Gen- Bank international DNA data bank (Table 3). The geo- graphic distribution of each haplotype is shown in Fig- ure 5. Among this monophyletic group, individuals from Guizhou and Sichuan in China were same haplotype (A-type) and were distinguished from other haplotypes in A. cochinchinensis by six apomorphic nucleotide substi- tutions. The remaining fourteen haplotypes, from B to O, were monophyletic, as supported by a relatively high bootstrap value (85%). Of these haplotypes, the I-type was the most widespread haplotype and occurred in China, South Korea, and Japan. The other types had more re- st r i cted d i stributi ons th an the I- typ e . The geographical distributions of the haplotypes are shown in Figure 5 and Table 1. Five haplotypes (A, B, I, J and K) were found in China; 5 haplotypes (C, F, G, H and I) occurred in Taiwan; and 1 haplotype (I) was found in Sout h K o re a. I n J a pan, se ven ha p lot ype s ( D , E , I , L, M , Table 3. List of GenBank accession numbers of nucleotide sequences for the taxa examined. Taxon Accession Number Asparagus officinalis AB195716 A. schoberioides AB195562 A. lycopodineus AB195563 A. cochinchinensis var. cochinchinensis (A-type) AB195564 (B-type) AB195565 (C-type) AB195566 (D-type) AB195567 (E-type) AB195568 (F-type) AB195569 (G-type) AB195570 (H-type) AB195571 (I-type) AB195572 (J-type) AB195573 (K-type) AB195574 (L-type) AB195575 (M-type) AB195576 (N-type) AB195577 (O-type) AB195578 A. cochinchinensis var. pygmaeus AB195579 N and O) were found from the Yaeyama Islands in the south to Kanto Distinct in central Honshu (Kanagawa). The D- and E-types formed a monophyletic group, as did the N- and O-types. The monophyly of the N- and O- types was supported by a 1-bp indel as well as one syn- apomorphic substitution. Among the 7 haplotypes in Japan, the I-, L-, N-, M-, and O-types, which occur from Yaku- shima Island to Kanagawa, were monophyletic with the J- and K-types in China. These results suggested that in- dividuals in this clade have a different origin from those in the Yaeyama Islands (Iriomote and Yonaguni Islands), and at least two lineages of Asparagus cochinchinensis exist in Japan. The dwarf-type of Asparagus cochinchinensis (less than 20 cm high) is known a s A. cochinchinensis (Lour.) Merrill var. pygmaeus Makino and is cultivated as orna- mental purposes [31]. Our phylogenetic result in- dicated that this variety is of the I haplotype (Figures 2, 3), that is, of the most widespread haplotype in Japan. 4. Discussion The usefulness of the ITS region as a molecular marker for the indirect estimation of infraspecies relationships depends on the level of phylogenetic resolution [46]. The Copyright © 2011 SciRes. AJPS 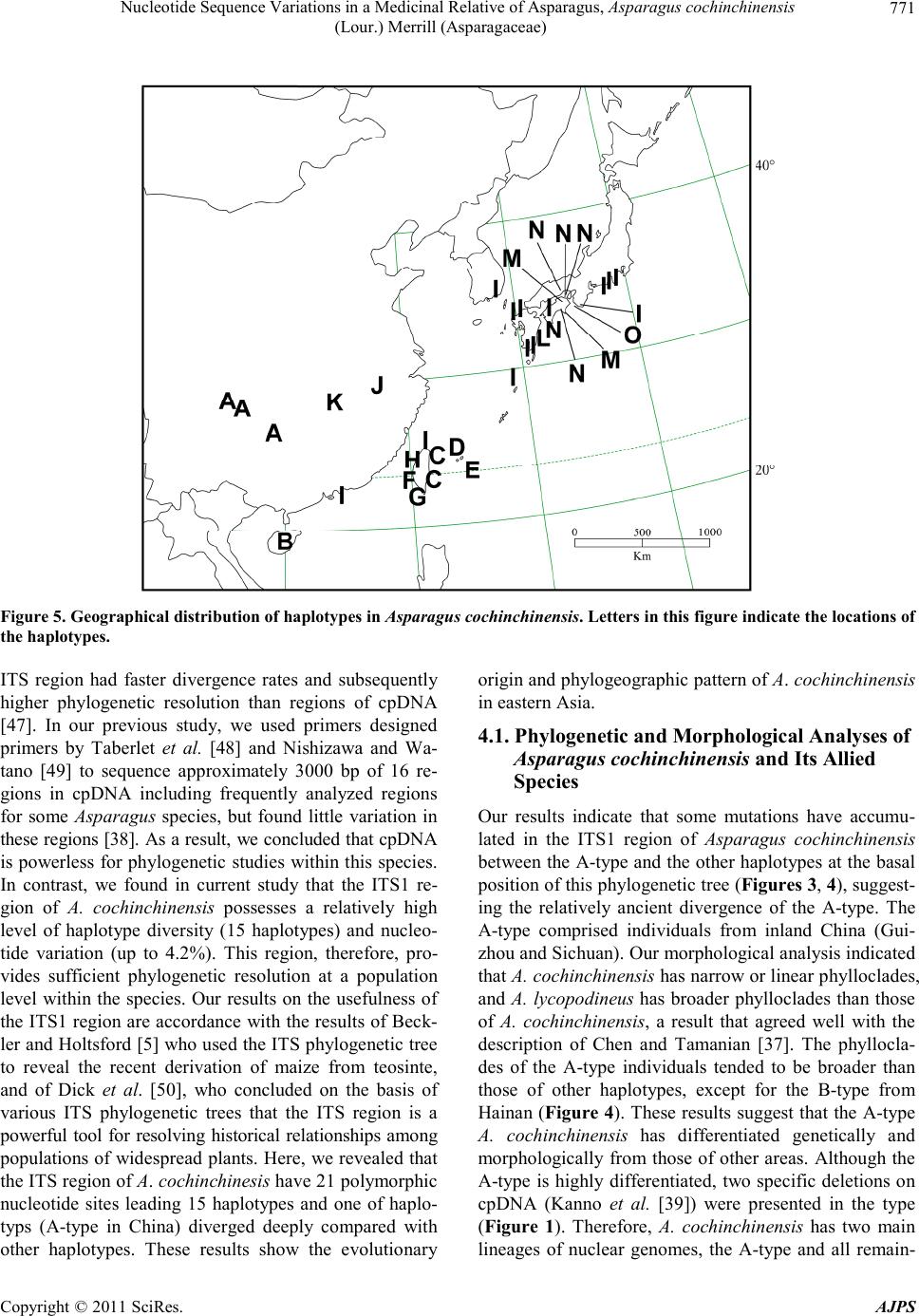 Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 771 (Lour.) Merril l (Asparagaceae) Figure 5. Geographical distribution of haplotypes in Asp ar ag us cochi nch inen sis. Letters in this figure indicate the locations of the haplotypes. ITS region had faster divergence rates and subsequently higher phylogenetic resolution than regions of cpDNA [47]. In our previous study, we used primers designed primers by Taberlet et al. [48] and Nishizawa and Wa- tano [49] to sequence approximately 3000 bp of 16 re- gions in cpDNA including frequently analyzed regions for some Asparagus species, but found little variation in these regions [38]. As a result, we concluded that cpDN A is powerless for phylogenetic studies within t his species. In contrast, we found in current study that the ITS1 re- gion of A. cochinchinensis possesses a relatively high level of haplotype diversity (15 haplotypes) and nucleo- tide variation (up to 4.2%). This region, therefore, pro- vides sufficient phylogenetic resolution at a population level within the species. Our results on the usefulness of the ITS1 region are accordance with the results of Beck- ler and Holtsford [5] who used the ITS phylogenetic tree to reveal the recent derivation of maize from teosinte, and of Dick et al. [50], who concluded on the basis of various ITS phylogenetic trees that the ITS region is a powerful tool for resolving historical relationships among populations of widespread plants. Here, we revealed that the ITS region of A. cochinchinesis have 21 polymorphic nucleotide sites leading 15 haplotypes and one of haplo- typs (A-type in China) diverged deeply compared with other haplotypes. These results show the evolutionary origin and phylogeographic pattern of A. cochinchinensis in eastern Asia. 4.1. Phylogenetic and Morphological Analyses of Asparagus cochinchinensis and Its Allied Species Our results indicate that some mutations have accumu- lated in the ITS1 region of Asparagus cochinchinensis between the A-type and the other haplotypes at the basal position of this phylogenetic t ree (Figures 3, 4), suggest- ing the relatively ancient divergence of the A-type. The A-type comprised individuals from inland China (Gui- zhou and Sichuan). Our morphological analysis indicated that A. cochinchinensis has narrow or linear phylloclades, and A. lycopodineus has broader phylloclades than those of A. cochinchinensis, a result that agreed well with the description of Chen and Tamanian [37]. The phyllocla- des of the A-type individuals tended to be broader than those of other haplotypes, except for the B-type from Hainan (Figure 4). These results suggest that the A-type A. cochinchinensis has differentiated genetically and morphologically from those of other areas. Although the A-type is highly differentiated, two specific deletions on cpDNA (Kanno et al. [39]) were presented in the type (Figure 1). Therefore, A. cochinchinensis has two main lineages of nuclear genomes, the A-type and all remain- Copyright © 2011 SciRes. AJPS  Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 772 (Lour.) Merril l (Asparagace ae) ing hapl o t ypes. What is the history of the evolutio n of the A-haplotype? The morphological characteristics of the A-type are in- termediate bet ween other Asparagus cochinchinensis hap- lotypes and its most closely related species, A. lycopodi- neus on the basis of the ITS phylogeny of Asparagus (Fukuda et al. unpublished data). Two possible expla- nations would be consistent with the establishme nt of the A-type. One is that the A-type of A. cochinchinensis ap- peared as an intermediate form in the course of gradual differentiation from A. lycopodineus to A. cochinchinen- sis. The other is that A-type of this species may have ex- perienced ancient hybridization events with related spe- cies and subsequent allelic recombination and concerted evolution. A previous phylogenetic study using cpDNA sequences suggested that genetic differentiation among species of the genus Asparagus is poor [38]. Therefore, it is possible that interspecific hybrids are generated in various combinations of species. In fact, successful pro- duction of interspecific hybrids between A. officinalis and A. schoberioides, and A. officinalis and A. kiusianus are known [51,52]. Moreover, interlocus-concerted evo- lution of the ITS region is well known [53-55]. Interlo- cus-concerted evolution has affected the heterogeneity of nucleotide substitution and is generally regarded as a mechanism that homogenizes copies of a gene in a ge- nome. Once an interspecific hybrid occurs and its pro- portion by chance increases interlocus-concerted evolu- tion may operate to fix a new haplotype in this species. From the view of the hybridization explanatio n, however, the ITS1 sequences of A-type must have experienced quite complicated recombination events with repeated crossing over throughout its range. Thus, the simpler ex- planation of the former view that A. cochinchinensis dif- ferentiated gradually from A. lycopodineus may be more plausible based on the present data. Extensive compari- sons of genetic data between A. lycopodineus and A. cochinchinensis should be made to confirm these expla- nations for the origin of the A-type. 4.2. Phylogeographic Pattern of Asparagus cochinchinensis In this study, we investigated the geographical structure of haplotype distribution in Asparagus cochinchinensis. Although the A-type varied from the other haplotypes, a relatively low level of genetic variation was detected in the ITS1 region among the haplotypes from B to O. This may reflect geographical patterns of haplotype distribu- tion associated with more recent events. China had the greatest diversity of haplotypes. Our results indicated that haplotypes in China (A, B, I, J, and K) were spread throughout the phylogenetic tree and included the most deeply divergent lineage (the A-type). This suggests that A. cochinchinensis had its origin in China. A recent phy- logenetic analysis of the genus Asparagus indicated that A. cochinchinensis is most closely related to A. lycopo- dineus (Fukuda et al. unpublished data), which occurs fro m inla nd China to India [37]. Ta ken together with t he fact, A. cochinchinensis appears to have originated in the interior of China. Then, how did the haplotypes expand their distribution beyond inland China? The large monophyletic group of haplotypes except A-type is divided into five lineages: one in Hainan as mentioned above, two in Taiwan, one in the Yaeyama Islands, and one involved mixed localit y from China, Tai- wan, South Korea, and Japan. Except haplotypes found i n inland China (J- and K-type), all of these haplotypes are distributed in the archipelago along eastern coast of the Eurasian continent. These facts indicate that Asparagus cochinchinensis experienced relative rapid expansion of its distribution range to the east and islands in the archi- pelago. The monophyletic group consisting of I- to O- type includes samples from wid e range of localit y: J apan, South Korea, Taiwan and China. Especially, I-type is found in the coastal area from Japan to south China. These facts suggest that the I-type of A. cochinchinensis might have expanded its range to these areas in more recent years than the expansion event with haplotype diversification mentioned above. Therefore, from our analysis, A. cochinchinensis experienced relative quick expansions of distribution range at least twice. In T aiwan, a loft y backb one ra nge, the Central Moun - tain Ridge, basically runs the axis of the island and has numerous peaks above 3000 m in elevation. Therefore, some local topographical barriers in the central zone may have blocked or limited the migration to the opposite coasts, with considerable effects on the genetic structure of plant species. In our study, significant genetic diffe- rences were detected between Asparagus cochinchinen- sis individuals on the island of Taiwan. Except for the presence of haplotype I in Keelung, which is the northern most area in Taiwan, the samples from the eastern region in Taiwan (Huelien and Taipei) had the same haplotype (C-type), and the haplotypes from Nantou, Kaohsiung and Taichung in the region of coastal to western Taiwan formed a single monophyletic group (F, G, and H). A further analysis using more samples from Taiwan is needed to confirm hypotheses of the geographical struc- ture of haplotype distribution in this area. The D- and E-types are only found in the Yaeyama Is- lands (Iriomote and Yonaguni Islands), may have been involved in a different colonization event from the latter group. The islands are geographically located in the south- western-most part of Japan and close to Taiwan. This Copyright © 2011 SciRes. AJPS  Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 773 (Lour.) Merril l (Asparagaceae) area is the transition zone from subtropical to temperate climate, and vascular plants from both temperate and tro- pical floras are observed [56]. Although our phylogenetic analysis could not resolve whether Asparagus cochin- chinensis in the Yaeyama Islands is phylogenetically grouped with Japan or Taiwan. The second lineage in Japan (the I- to O-types) formed a monophyletic group with haplotypes from Anhui and Hubei in China, Kee- lung in Taiwan and Jeollanam-do in South Korea (Fig- ures 2, 3). Moreover, few apomorphic characters have accumulated within this monophyletic group. These facts suggest that A. cochinchinensis might have expanded its range to these areas in recent years. Considering these results, the ITS variation we see today may have been generated after Japan became separated from the Eura- sian continent. The interest in trying to link geographic distribution and evolution of the ITS region with organism evolution rests on the wide use of such markers for phylogeny as well as on the increasing number of cases documenting complicated evolution in plants. The ITS region analyzed in this work provides a robust phylogeographic hypothe- sis for the evolution of Asparagus cochinchinensis. This hypothesis, based on molecular information, is a timely contribution that allows unbiased interpretation of evolu- tionary history. Additional molecular markers may lend support to this scenario, and multiple markers should be surveyed within this species to find any variation. This work will help to reinterpret existing ITS data sets and facilitate the interpretation of new ones. 5. Acknowledgements We wish to thank Drs. T. Nakamura, H. Tsukaya, H. Yamaji, M. Kawata, M. Maki, T. Yamashiro, T. Ochiai, P.-Y. Yun, H. Ashizawa, Y. Mashiko, M. Nakada, R. Shinohara, M. Komatsu, S.-Y. Kim, M. Hirai, M. Shiba, K. Kondo, E. Miki, and H. Tokairin for providing much help and advice. This study was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan, by the Sa- sakawa Scientific Research Grant from The Japan Sci- ence Society, and Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education Science and Technology (2010-0029630). REFERENCES [1] M. Nei and S. Kumar, “Molecular EVolution and Phy- logenetics,” Oxford University Press, New York, 2000, pp. 3-16. [2] J. C. Avise, J. Arnold, R. M. Ball, E. Bermingham, T. Lamb, J. E. Neigel, C. A. Reeb and N. C. Saunders, “In- traspecific Phylogeography: The Mitochondrial DNA Bridge between Population Genetics and Systematics,” Annual Review of Ecology, Evolution, and Systematics, Vol. 18, 1987, pp. 489-552. [3] J. C. Avise, “Molecular Markers, Natural History, and Evolution,” Chapman & Hall, New York, 1994. doi:10.1007/978-1-4615-2381-9 [4] J. C. Avise, “Phylogeography: The History and Forma- tion of Species,” Harvard University Press, Cambridge, 2000. [5] E. Beckler and T. Holtsford, “Zea Systematics: Ribo- somal ITS Evidence,” Molecular Biology and Evolution, Vol. 13, No. 4, 1996, pp. 612-622. [6] B. A. Schaal and K. M. Olsen, “Gene Genealogies and Population Variation in Plants,” Proceedings of the Na- tional Academic of Sciences of the United States of Ame- rica, Vol. 97, No. 13, 1999, pp. 7024-7029. [7] K. M. Olsen, “Population History of Manihot esculenta (Euphorbiaceae) Inferred from Nuclear DNA Sequences,” Molecular Ec ology, Vol. 11, N o. 5, 2002 , pp. 901-911. doi:10.1046/j.1365-294X.2002.01493.x [8] K. M. Olsen and M. D. Purugganan, “Molecular Evi- dence on the Origin and Evolution of Glutinous Rice,” Genetics, Vol. 162, No. 2, 2002, pp. 941-950. [9] K. Liu, M. M. Goodman, S. Muse, J. S. C. Smith, E. S. Buckler and J. Doebley, “Genetic St ructure and Diversit y among Maize Inbred Lines as Inferred from DNA Mi- crosatellites,” Genetics, Vo l. 165, No. 4, 2003 , pp. 2117- 2128. [10] Y. Vigouroux, Y. Matsuoka and J. Doebley, “Directional Evolution for Microsatellite Size in Maize,” Molecular Biology and Evolution, Vol. 20, No. 9, 2003, pp. 1480- 1483. doi:10.1093/molbev/msg156 [11] A. V. Harter, K. A. Gardner, D. Falush, D. L. Lentz, R. A. Bye and L. H. Ri eseberg, “ Ori gin of Extant Domest icated Sunflowers in E astern North America,” Nature, V o l. 43 0, 2004, pp . 201-205. doi:10.1038/nature02710 [12] I. M. Ehrenreich an d M. D. Purugganan, “ The Mo lecular Genetic Basis of Plant Adaptation,” American Journal of Botany, V o l. 93, No. 7, 2008, pp. 953-962. doi:10.3732/ajb.93.7.953 [13] B. L. Gross and K. M. Olsen, “Genetic Perspectives on Crop Domestication,” Trends in Plant Science, Vol. 15, No. 9, 2010, pp. 529-537. doi:10.1016/j.tplants.2010.05.008 [14] S. D. Tanksley and S. R. McCouch, “Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild,” Science, Vol. 277, No. 53 2 9, 199 7, pp. 1063-1 066. doi:10.1126/science.277.5329.1063 [15] K. M. Olsen and B. A. Schaal, “Evidence on the Origin of Cassava: Phylogeography of Maniho t esculenta,” Pro- ceedings of the Nat ional Academic of Sciences of the Un i- ted States of America, Vol. 96, No. 10, 1999, pp. 5586- 5591. doi:10.1073/pnas.96.10.5586 Copyright © 2011 SciRes. AJPS  Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis 774 (Lour.) Merril l (Asparagace ae) [16] J. Yokoyama, T. Fukuda, A. Yokoyama and M. Maki, “The Intersectional Hybrid between Weigela hortensis and W. maximowiczii (Caprifoliaceae),” Botanical Jour- nal of the Linnean Society, Vol. 138, No. 3, 2002, pp. 369-380. doi:10.1046/j.1095-8339.2002.00033.x [17] T. Fukuda, J. Yokoyama and H. Tsukaya, “The Evolu- tionary Origin of Indeterminate Leaves in Meliaceae: Phylogenetic Relationships among Species in the Genera Chisocheton and Guarea, as Inferred from Sequences of Chloroplast DNA,” International Journal of Plant Sci- ences, Vol. 164, 2003, pp. 13-24. doi:10.1086/344741 [18] H. Tsukaya, T. Fukuda and J. Yokoyama, “Hybridization and Introgression between Callicarpa japonica and C. mollis (Verbenaceae) in Central Japan, as Inferred from Nuclear and Chloroplast DNA Sequences,” Molecular Ecology, Vol. 12, No. 11, 2003, pp. 3003-3011. doi:10.1046/j.1365-294X.2003.01961.x [19] J. Yokoyama, T. Fukuda and H. Tsukaya, “Morphological and Molecular Variation of Mitchella undulata Sieblod et Zucc., with Special Reference to Systematic Treatment of the Dwarf Form from Yakushima Island,” Journal of Plant Research, Vol. 11 6, No . 4, 200 3, pp . 309-316. do i:10.1007/s10265-003-0105-7 [20] T. Yamashiro, T. Fukuda, J. Yokoyama and M. Maki, “Molecular Phylogeny of Vincetoxicum (Apocynaceae- Asclepiadoideae) Based on the Nucleotide Sequences of cpDNA and nrDNA,” Molecular Phylogenetics and Evo- lution, Vol. 31, No. 2, 2004, pp. 689-700. doi:10.1016/j.ympev.2003.08.016 [21] H. Tsukaya, “Gen e Fl ow between Impatiens radicaus and I. javensis (Balsaminaceae) in Gunung Pangoran go, Cen- tral Java, Indonesia,” American Journal of Botany, Vol. 91, No. 12, 2005, pp. 2119-2123. [22] T. Feng, S. R. Downie, Y. Yu, X. Zhang, W. Chen, X. He and S. Liu, “Molecular Systematics of Angelica and Al- lied Genera (Apiaceae) from the Hengduan Mountains of China Based on nrDNA ITS Sequences: Phylogenetic Affinities and Biogeographic Implications,” Journal of Plant Research, Vol. 12 2, No . 4, 200 9, pp . 403-414. do i:10.1007/s10265-009-0238-4 [23] C. Ferris, R. P. Oliver, A. J. Davy and G. M. Hewitt, “Native Oak Chloroplasts Reveal an Ancient Divide across Europe,” Molecular Ecology, Vol. 2, No. 6, 1993, pp. 337- 344. doi:10.1111/j.1365-294X.1993.tb00026.x [24] S. Dumolin-Lapegue, M. H. Pemonge, L. Gielly, P. Ta- berlet and R. J. Petit, “Amplification of Oak DNA from Ancient and Modern Wood,” Molecula r Ecology, Vol. 8, No. 12, 1997, pp. 2137-2140. doi:10.1046/j.1365-294x.1999.00788.x [25] S. Dumolin-Lapegue, B. Demesure, S. Fineschi, V. L. Corre and R. J. Petit, “Phylogeographic Structure of White Oaks Throughout the European Continent,” Ge- netics, Vol. 146, No. 4, 1999, pp. 1475-1487. [26] T. Fukuda, J. Yokoyama and H. Ohashi, “Phylogeny and Biogeography in Lycium (Solanaceae): Inferences from Chroloplast DNA Sequences,” Molecular Phylogenetics and Evolution, Vol. 19, No. 2, 2001, pp. 246-258. doi:10.1006/mpev.2001.0921 [27] N. Fujii, N. Tomaru, K. Okuyama, K. Koike, T. Mikami and K. Ueda, “Chloroplast DNA Phylogeography of Fagus crenata (Fagaceae) in Japan,” Plant Systematics and Evo- lution, Vol. 232, No . 1-2, 2002, pp. 21-33. doi:10.1007/s006060200024 [28] L. H. Bailey, “Asparagus L.,” In: L. H. Bailey, Ed., The Standard Cyclopedia of Horticulture, The Macmillan Company, New York, 1944, pp. 406-411. [29] F. J. Chittenden, “Asparagus L.,” In; F. J. Chittenden, Ed., Dictionary of Gardening, Clarendon Press, Oxford, 1956, pp. 193-196. [30] B. Valdes, “Asparagus L.,” In: T. G. Tutin, V. H. Hey- wood, N. A. Burges, D. M. Moore, D. H. Valentine, S. M. Walters and D. A. Webb, Eds., Flora Europaeana, Cam- bridge University Press, Cambridge, 1964, pp. 71-73. [31] J. Ohwi, “ Asparagus L.,” In: F. G. Meyer and E. H. Walker , Eds., Flora of Japan, Smithonian Institution, Washington DC, 1965. [32] V. L. Komarov, “Asparagus L.,” In: V. L. Komarov, Ed., Flora of the USSR, Vol. IV., Israel Program for Scientific Translations, Jerusalem, 1968, pp. 325-339. [33] R. M. T. Dahlgren, H. T. Clifford and P. F. Yeo, “As- paragus L.,” In: R. M. T. Dahlgren, H. T. Clifford and P. F. Yeo, Eds., The Families of the Monocotyledons, Springer- Verlag Berlin, Heidelberg , 1985, pp. 140-142. doi:10.1007/978-3-642-61663-1 [34] H. T. Clifford and J. G. Conran, “Asparagus L.,” In: A. S. George, Ed., Flora of Australia, Australian Government Publishing Service, Canberra, 1987, pp. 159-164. [35] K. Kubituki and P. J. Rudall, “Asparagus L.,” In: K. Ku- bituki, Ed., The Families and Genera of Vascular Plants, Vol. 3. Springer-Verlag Berlin, Heidenberg, 1998, pp. 125-128. [36] D. K. Kar and S. Sen, “Chromosome Characteristics of Asparagus-Sapogenin Yielding Plant,” Cytologia, Vol. 50, 1985, pp. 14 7- 1 55 . doi:10.1508/cytologia.50.147 [37] X. Chen and K. G. Tamanian , “Asparagus L.,” In: Z. Wu and P. H. Raven, Eds., Flora of China, Vol. 24, Missouri Botanical Garden Press, St. Louis, 2000, pp. 139-146. [38] T. Fukuda, H. Ashizawa, T. Nakamura, T. Ochiai, A. Kanno, T. Kameya and J. Yokoyama, “Molecular Phylo- geny of the Genus Asparagus (Asparagaceae),” Plant Species Biology, Vol. 20 , No. 2, 2005, pp. 121 -132. doi:10.1111/j.1442-1984.2005.00131.x [39] A. Kanno, Y.-O. Lee and T. Kameya, “The Structure of the Chloroplast Genome in Members of the Genus As- paragus,” Theoretical and Applied Genetics, Vol. 95, No. 8, 1997, pp. 1196 -1202. doi:10.1007/s001220050681 [40] T. Fukuda, I.-J. Song, T. Nakamura, M. Nakada, A. Kanno, T. Kameya, H. Yamaji, S. Terabayashi, S. Takeda, M. Aburada and J. Yokoyama, “Molecular Identification of Tiandong Derived from Asparagus cochinchinensis (Lour.) Merrill by Two Typical Deletions in cpDNA,” Copyright © 2011 SciRes. AJPS  Nucleot ide Sequen ce V ari at ions in a Medi cinal Relative of Asparagu s, Asparagus cochinchinensis (Lour.) Merril l (Asparagaceae) Copyright © 2011 SciRes. AJPS 775 Natural Medicines, Vol. 59, 2005, pp. 91-94. [41] T. J. White, T. Bruns, S. Lee and J. Taylor, “Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics,” In: M. Innis, D. Gelfand, J. Sninsky and T. J. White, Eds., PCR Protocols: A Guide to Methods and Application, Academic Press, San Diego, 1990, pp. 315-322. [42] J. D. Thompson, T. J. Gibson, F. Plewniak, F. Jeanmou- gin and D. G. Higgins, “The Clustal_X Windows Interface: Flexible Strategies f or Multiple Sequenc e Alig nment Ai ded by Quality Analysis Tools,” Nucleic Acids Research, Vol. 25, No. 24, 19 97, pp. 4 876-4882. doi:10.1093/nar/25.24.4876 [43] D. L. Swofford, “PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods) Version 4.0b10,” Sinauer Assoc iate s , Sunde rl and, 2002, [44] W. P. Maddison, “The Discovery and Importance of Mul- tiple Islands of Most-Parsimonious Trees,” Systematic Zoology, Vol. 40, No. 3, 1991, pp. 315-328. doi:10.2307/2992325 [45] J. Felsenstein, “Confidence Limits on Phylogenies: An Approach Using the Bootstrap,” Evolution, Vol. 39 , 1985, pp. 783- 791. doi:10.2307/2408678 [46] B. G. Baldwin, M. J. Sanderson, J. M. Porter, M. F. Wo- jciechowski, C. S. Campbell and M. J. Donoghue, “The ITS Region of Nuclear Ribosomal DNA: A Valuable Source of Evidence on Angiosperm Phylogeny,” Annals of the Missouri Botanica Garden, Vol. 82, No. 2, 1995, pp. 247- 277. doi:10.2307/2399880 [47] T. Sang, D. J. Crawford and T. F. Stuessy, “ITS Sequ- ences and the Phylogeny of the Genus Robinsonia (As- teraceae),” Systematic Zoology, Vol. 20, No. 1, 1995, pp. 55-64. [48] P. Taberlet, L. Gielly, G. P autou and J. Boub et, “Univer- sal Primers for Amplication of Three Non-Coding Re- gions of Chloroplast DNA,” Plant Molecular Biology, Vol. 17, No. 5, 1991, pp. 1105-1109. doi:10.1007/BF00037152 [49] T. Nishizawa and Y. Watano, “Primer Pairs Suitable for PCR-SSCP Analysis of Chloroplast DNA in Angiospe- rm s,” Journal of Phytogeography and Taxonomy, Vol. 48, 2000, pp. 67-70. [50] C. W. Dick, K. Abdul-Salim and E. Bermingham, “Mo- lecular Systematic Analysis Reveals Cryptic Tertiary Di- versificatio n of a Widesp read Tropical Rai n Forest Tree,” The American Naturalist, Vol. 160, No. 12, 2003, pp. 691- 703. doi:10.1086/379795 [51] T. Ochiai, T. Sonoda, A. Kanno and T. Kameya, “Inter- specific Hybri ds between Asparagus schoberioides Kunth and A. officinalis L.,” Acta Horticulturae, Vol. 589, 2002 , pp. 225-229. [52] T. Ito, T. Ochiai, T. Fukuda, H. Ashizawa, T. Sonoda, T. Kameya and A. Kanno, “Potential of Interspecific Hy- brids in Asparagaceae,” Acta Horticulturae, Vol. 776, 2008, pp . 279-284 . [53] D. M. Hillis, C. Moritz, C. A. Porter and R. J. Baker, “Evidence for Biased Gene Conversion in Concerted Evolution of Ribosomal DNA,” Science, Vol. 251, No. 4991, 1991 , pp. 30 8- 3 10 . doi:10.1126/science.1987647 [54] T. Sang, D. J. Crawford and T. F. Stuessy, “Documenta- tion of Reticulate Evolution in Peonies (Paeonia) Using Internal Transcribed Spacer Sequences of Nuclear Ribo- somal DNA: Implications for Biogeography and Con- certed Evolution,” Proceed ings of th e Nationa l Academic of Sciences of the United States of America, Vol. 92, No. 15, 1995 , pp. 6 8 13-6817. doi:10.1073/pnas.92.15.6813 [55] J. Fuertes Aguilar, J. A. Rosselo and G. Nieto Feliner, “Nuclear Ribosomal DNA (nrDNA) Concerted Evolution in Natural and Artificial Hybrid of Armeria (Plumbagi- naceae),” Molecular Ecology, Vol. 8, No. 8, 1999, pp. 1341-1346. doi:10.1046/j.1365-294X.1999.00690.x [56] E. H. Walker, “Flora of Okinawa and the Southern Ryu- kyu Islands,” Smithonian Institution Press, Washington DC, 1976.
|