Paper Menu >>
Journal Menu >>
 J. Biomedical Science and Engineering, 2009, 2, 559-563 doi: 10.4236/jbise.2009.27081 Published Online November 2009 (http://www.SciRP.org/journal/jbise/ JBiSE ). Published Online November 2009 in SciRes. http://www.scirp.org/journal/jbise MRTF-A transactivates COMT gene and decreases the anti-tumor effects of tamoxifen Zhi-Peng Liu*, Xue-Gang Luo*, Shu Guo, Jian-Xin Wang, Xin Zhang, Nan Wang, Yong Jiang, Tong-Cun Zhang Key Laboratory of Industrial Microbiology, Ministry of Education, College of Biotechnology, Tianjin University of Science and Technology, Tianjin, China; *The authors contributed equally to this work; Correspondence to: Pro. Tong-Cun Zhang. Email: tony@tust.edu.cn Received 29 June 2009; revised 21 August 2009; accepted 1 September 2009. ABSTRACT Myocardin-related transcription factors A (MR TF-A) is a myocardin-related transcription factor that have been found strongly activated CarG box–containing genes through its direct binding to serum response factor (SRF). In the present study, the MRTF-A ex- pression vector was constructed. The MTT assay showed that transfection of MRTF-A could signifi- cantly decrease the anti-tumor effect of tamoxifen on MCF-7 breast cancer cells. The bioinformatics analy- sis found that the CarG element existed in the pro- moter region of COMT gene of many familiar verte- brates, including of human, rhesus macaque, chim- panzee, etc. The results of RT-PCR assay further showed that MRTF-A could enhance the transcrip- tion level of COMT. These results are the first to in- dicate that COMT might be a target gene which could be regulated by MRTF-A/SRF, and such transactivation event might be involved in the proc- ess of tamoxifen resistance. Keywords: MRTF-A; Tamoxifen; COMT; Breast Cancer 1. INTRODUCTION Breast cancer is the most common cancer diagnosed in women in the world. Seventy percent of diagnosed breast cancers express Estrogen Receptor alpha (ERa) and are likely to be hormone-responsive. The most common therapy for ERa–positive breast cancers has employed the use of selective estrogen receptor modula- tors (SERMs) such as tamoxifen. As an adjuvant therapy in breast cancer, tamoxifen improves overall survival, and its widespread use is thought to have made a sig- nificant contribution to the reduction in breast cancer mortality seen over the last decade. However, although many patients benefit from tamoxifen, the resistance is an important clinical problem [1,2]. Myocardin-related transcription factors (MRTFs); in- cluding myocardin, MRTF-A/MKL1/MAL, and MRTF- B/MKL2, comprise a family of related transcriptional coactivators. MRTFs drives transcription through inter- action with the ubiquitous transcription factor serum response factor (SRF), which acts on a responsive ele- ment CC(A/T)6GG (known as CarG box) that is com- monly found in many gene promoters. Myocardin is expressed specifically in cardiac and smooth muscle cells and activates muscle genes associated with the dif- ferentiation of these cell types. MRTF-A and MRTF-B are expressed in a broad range of cell types [3,4]. Previous publications have suggested that MRTF-A might plays a role in development of mammary myoepi- thelial cells and breast cancer [5,6]. In the present paper, we further found that MRTF-A could decrease the anti-tumor effect of tamoxifen on MCF-7 human breast cancer cells, and the transcriptional activation of cath- echol-O-methyltransferase (COMT), a phase II metabo- lising enzyme of tamoxifen, might be involved in this process. 2. METERIALS AND METHODS 2.1. Regents and Cell Culcure Tamoxifen (Wako Pure Chemicals, Osaka, Japan) was dissolved in Phosphate Buffered Saline (PBS). 1kb DNA ladder marker was purchased from Solarbio Science & Technology Co., Ltd (Beijing, China). Human liver cell lines L02 and breast cancer cell line MCF-7 was cul- tured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Paisley, UK) containing 10% fetal bovine serum (FBS; Gibco). 2.2. Construction of MRTF-A Expression Vector Total RNA of the LO2 cells was isolated using the me- thod described in molecular cloning. First-strand cDNA was synthesized from the total RNA using M-MLV re- verse transcriptase (Promega, Madison, USA) and oligo (dT)18 (Sangon, Shanghai, China). The full-ength MR-  Z. P. Liu et al. / J. Biomedical Science and Engineering 2 (2009) 559-563 SciRes Copyright © 2009 JBiSE 560 TF-A gene was amplified by polymerase chain reaction (PCR) using the cDNA and the following primer pair: 5'- CAAGGTACCATGCCGCCTTTGAAAAG -3' (forward) and 5'- CCCGAATTCAGCCAGAGAGCTA- CAAGC -3' (reverse). PCR was performed at 94°C for 5 min, then 28 cycles at 94°C for 45 s, at 60°C for 45 s, and at 72°C for 3 min; extension was carried out at 72°C for 10 min. The PCR product (2824 bp) was double- digested with kpnI and EcoRI (Takara, Kyoto) and inserted into the pcDNA3.1 (+) mammalian expression vector (Invi- trogen, Carlsbad, Canada). The recombinant construc- tion was analyzed by restriction-enzyme digestion and sequencing to determine reading frame orientation and confirm sequence fidelity, and the positive recombinant plasmid was named pcDNA-MRTF-A. 2.3. Cell Transfection Transient transfection of pcDNA-MRTF-A plasmid into the MCF-7 cells using LipofectaminTM 2000 transfection reagents (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. As a negative control, the empty vector pcDNA3.1 (+) (mock) was transfected in parallel, and to assess the transfection efficiency, the pEGFP-C3 plasmid was also transfected as a positive control simultaneously. 2.4. Evaluation of the Inhibition Rate of Tamoxifen on MCF-7 Cells The MCF-7 human breast cancer cells were plated in 96-well plates at a density of 8×103 cells/well and trans- fected with the pcDNA-MRTF-A plasmid or pcDNA3.1 (+). After 24h, the medium was removed and replaced with fresh medium containing tamoxifen at concentra- tions of 5, 10 and 20 μM. Forty-eight hours later, MTT assay was performed. In brief, MTT (5 mg/ml) was added to the wells (20 µl/well). The plate was incubated in a cell incubator for 4 h, then the supernatant was re- moved and 150 µl of dimethyl sulfoxide was added into each well. After incubation for 10 min, the absorbance of each well was measured using a microplate reader (Bio-Rad) with a wavelength of 570 nm, with the refer- ence wavelength set at 630 nm. Absorbance (A) was directly proportional to the number of viable cells. All assays were performed using six replicates. The inhibi- tion rate was calculated as follows: 2.5. Bioinformatics Screen of the CarG Box in the Promoter of COMT Gene The genomic sequence of COMT gene was extracted from the Database of GenBank. Their transcription start sites were then located by using the University of Cali- fornia at Santa Cruz (UCSC) genome brower, and the Table 1. Primers used in RT-PCR analysis. (a. F: forward primer. R: reverse primer) Gene Primersa Product (bp) F: 5’-ATTCAACGGCACAGTCAAGG-3’ GAPDH R: 5’-GCAGAAGGGGCGGAGATGA-3’ 213 F: 5’-ACCGTGACCAATAAGAATGC-3’ MRTFA R: 5’-CCGCTCTGAATGAGAATGTC-3’ 269 F: 5’-GGATTTCGCTGGCGTGAAG-3’ COMT R: 5’-TCCACCACCTCCCTGTATTCC-3’ 306 sequence from 2000 bp upstream of the transcription sites to 200 bp downstream of the transcription start sites (–2000~+200) was extracted by using the UCSC data- base assemblage, and then the potential CarG box was analyzed using the primer premier 5.0 software [7]. 2.6. RT-PCR Analysis Total RNA of the transfected cells was extracted, and then the potential residual genomic DNA was eliminated with RNase-free Dnase I (Bio Basic Inc, Ontario, Can- ada). First-strand cDNA was synthesized as described above. For PCR amplification, primers specific for the cDNA of the MRTF-A, COMT gene and the constitutive glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene were used (Ta b l e 1 ). PCR was performed at 94°C for 5 min, then 25 cycles at 94°C for 30 s, at 54°C for 30 s, and at 72°C for 45 s; extension was carried out at 72°C for 10 min. PCR products were electrophoretically separated in 1.5% agarose gels and visualized by ethidium bromide staining. The densities (D) of the bands were analyzed with Quantity One software and relative mRNA levels were deduced from the ratio of the mean values of MRTF-A or COMT to that of GAPDH. Cells transfected with the empty plasmid pcDNA3.1 (+) was used as blank control. The assay was performed using three replicates. The relative mRNA level was defined as: GAPDH genetarget D D levelmRNA Relative= 2.7. Statistical Analysis The data from the above mentioned experiments were expressed as mean ± SD. The statistical significance of differences was determined using Student’s t test. The minimal level of significance was P < 0.05. 3. RESULTS 3.1. Construction of MRTF-A Expression Vector The cDNA encoding human MRTF-A was obtained by RT-PCR from Human liver cell lines L02 and cloned into the pcDNA3.1 (+) vector. By the methods of restric- tion digestion and sequence analyses, we confirmed that the length, position and orientation of the inserted MRTF-A gene were all correct, suggesting that the re- 100 ) A ( control Aexperimental 1(%) rateInhibition =  Z. P. Liu et al. / J. Biomedical Science and Engineering 2 (2009) 559-563 SciRes Copyright © 2009 561 3.5. Effect of MRTF-A on the Transcri ption Level of COMT combinant pcDNA-MRTF-A plasmid was constructed successfully (Figure 1). 3.2. Cell Transfection To further detect whether the transcription of COMT could be regulated by MRTF-A, the RT-PCR analysis was performed to detect the effect of MRTF-A on the The endotoxin-free plasmids were extracted and trans- fected into the MCF-7 cells using LipofectaminTM 2000 transfection reagents. To assess the transfection effi- ciency, 0.5μg pEGFP-C3 plasmid was also transfected as a positive control simultaneously. As shown in Figure 2, the MCF-7 cell transfected with pEGFP-C3 exhibited bright fluorescence, indicating that the transfection effi- ciency in the experiment was high enough to perform the following experiments. 3.3. Effect of MRTF-A on the Anti-Tumor Activity of Tamoxifen To investigate the effect of MRTF-A on the anti-tumor activity of tamoxifen, the MCF-7 breast cancer cells were transfected with the MRTF-A or the mock plasmid, and then treated with tamoxifen at different doses for 48 h. The inhibition rate was determined by MTT assay. As inhibition rates of cells transfected with MRTF-A were shown in Figure 2, at 5, 10 and 20 μM tamoxifen, the all significantly lower than the cells transfected with mock plasmid (p<0.05). Figure 1. Identification of recombinant pcDNA- MRTF-A plasmid by restriction endonuclease. Lane 1: pcDNA3.1(+) plasmid double-digested with kpnI and EcoRI; Lane 2: pcDNA-MRTF-A double- di- gested with kpnI and EcoRI; Lane M: 1kb DNA ladder marker. 3.4. Bioinformatics Screen of the Potential Carg Box in the Promoter of COMT Gene To investigate whether COMT was a potential target gene of MRTF-A, the sequence from 2000 bp upstream of the transcription sites to 200 bp downstream of the transcription start sites (–2000~+200) of COMT gene of familiar vertebrate was analyzed. As shown in Ta b l e 2 , the CarG-like element existed in many vertebrates, in- cluding of human, rhesus macaque, chimpanzee, etc. This result indicated that COMT might be a target gene which could be regulated by MRTF-A/SRF. Figure 2. Cell image of MCF-7 transfected with PEGFP-C3- C3 plasmid. The left one was the normal micrograph and the right one was the fluorescence micrograph in the same visual field. Table 2. The carg box in the promoter region of comt gene. Organism Sciencific name Common name GenBank ID CarG seq Position Homo sapiens Human NT_011519 CCTTTTATGG -1105 Macaca mulatta Rhesus macaque NW_001095157 CCTTTATTGG -933 Pan troglodytes Chimpanzee NW_001230944 CCTTTTATGG -1187 Mus musculus Laboratory mouse NT_039624 CCTATTGGGG -834 Rattus norvegicus Rat NW_047358 CCTGTTTTGG -616 Bos taurus Cattle NW_001493554 CCAATACTGG -9 CCTAAAGTGG -1175 Gallus gallus Chicken NW_001471459 CCTTATCAGG -21 CCTAAAAGGG -1582 CCCTAAAAGG -1583 Monodelphis domestica (Opossum), Ornithorhynchus anatinus (duck-billed platypus), Bubalus bubalis (river buffalo), Canis lupus familiaris (Dog), Felis catus (Cat), Ovis aries (Sheep), Sus scrofa (Pig), Danio rerio (Zebrafish) The typical CarG box was not found in the promoter of COMT gene. Alternatively, the COMT gene still remained unclear JBiSE 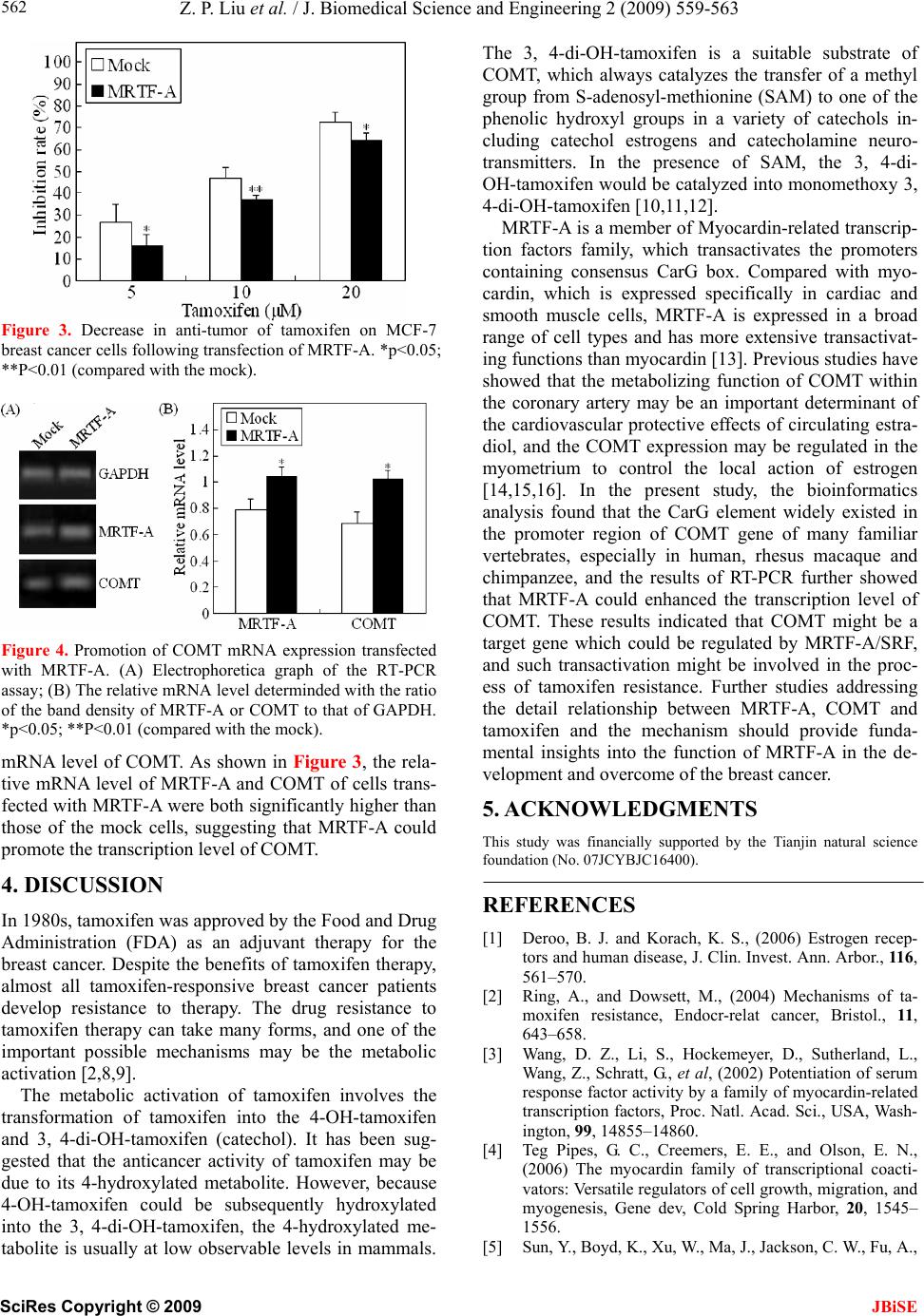 Z. P. Liu et al. / J. Biomedical Science and Engineering 2 (2009) 559-563 SciRes Copyright © 2009 JBiSE 562 Figure 3. Decrease in anti-tumor of tamoxifen on MCF-7 breast cancer cells following transfection of MRTF-A. *p<0.05; **P<0.01 (compared with the mock). Figure 4. Promotion of COMT mRNA expression transfected with MRTF-A. (A) Electrophoretica graph of the RT-PCR assay; (B) The relative mRNA level determinded with the ratio of the band density of MRTF-A or COMT to that of GAPDH. *p<0.05; **P<0.01 (compared with the mock). mRNA level of COMT. As shown in Figure 3, the rela- tive mRNA level of MRTF-A and COMT of cells trans- fected with MRTF-A were both significantly higher than those of the mock cells, suggesting that MRTF-A could promote the transcription level of COMT. 4. DISCUSSION In 1980s, tamoxifen was approved by the Food and Drug Administration (FDA) as an adjuvant therapy for the breast cancer. Despite the benefits of tamoxifen therapy, almost all tamoxifen-responsive breast cancer patients develop resistance to therapy. The drug resistance to tamoxifen therapy can take many forms, and one of the important possible mechanisms may be the metabolic activation [2,8,9]. The metabolic activation of tamoxifen involves the transformation of tamoxifen into the 4-OH-tamoxifen and 3, 4-di-OH-tamoxifen (catechol). It has been sug- gested that the anticancer activity of tamoxifen may be due to its 4-hydroxylated metabolite. However, because 4-OH-tamoxifen could be subsequently hydroxylated into the 3, 4-di-OH-tamoxifen, the 4-hydroxylated me- tabolite is usually at low observable levels in mammals. The 3, 4-di-OH-tamoxifen is a suitable substrate of COMT, which always catalyzes the transfer of a methyl group from S-adenosyl-methionine (SAM) to one of the phenolic hydroxyl groups in a variety of catechols in- cluding catechol estrogens and catecholamine neuro- transmitters. In the presence of SAM, the 3, 4-di- OH-tamoxifen would be catalyzed into monomethoxy 3, 4-di-OH-tamoxifen [10,11,12]. MRTF-A is a member of Myocardin-related transcrip- tion factors family, which transactivates the promoters containing consensus CarG box. Compared with myo- cardin, which is expressed specifically in cardiac and smooth muscle cells, MRTF-A is expressed in a broad range of cell types and has more extensive transactivat- ing functions than myocardin [13]. Previous studies have showed that the metabolizing function of COMT within the coronary artery may be an important determinant of the cardiovascular protective effects of circulating estra- diol, and the COMT expression may be regulated in the myometrium to control the local action of estrogen [14,15,16]. In the present study, the bioinformatics analysis found that the CarG element widely existed in the promoter region of COMT gene of many familiar vertebrates, especially in human, rhesus macaque and chimpanzee, and the results of RT-PCR further showed that MRTF-A could enhanced the transcription level of COMT. These results indicated that COMT might be a target gene which could be regulated by MRTF-A/SRF, and such transactivation might be involved in the proc- ess of tamoxifen resistance. Further studies addressing the detail relationship between MRTF-A, COMT and tamoxifen and the mechanism should provide funda- mental insights into the function of MRTF-A in the de- velopment and overcome of the breast cancer. 5. ACKNOWLEDGMENTS This study was financially supported by the Tianjin natural science foundation (No. 07JCYBJC16400). REFERENCES [1] Deroo, B. J. and Korach, K. S., (2006) Estrogen recep- tors and human disease, J. Clin. Invest. Ann. Arbor., 116, 561–570. [2] Ring, A., and Dowsett, M., (2004) Mechanisms of ta- moxifen resistance, Endocr-relat cancer, Bristol., 11 , 643–658. [3] Wang, D. Z., Li, S., Hockemeyer, D., Sutherland, L., Wang, Z., Schratt, G., et al, (2002) Potentiation of serum response factor activity by a family of myocardin-related transcription factors, Proc. Natl. Acad. Sci., USA, Wash- ington, 99, 14855–14860. [4] Teg Pipes, G. C., Creemers, E. E., and Olson, E. N., (2006) The myocardin family of transcriptional coacti- vators: Versatile regulators of cell growth, migration, and myogenesis, Gene dev, Cold Spring Harbor, 20, 1545– 1556. [5] Sun, Y., Boyd, K., Xu, W., Ma, J., Jackson, C. W., Fu, A.,  Z. P. Liu et al. / J. Biomedical Science and Engineering 2 (2009) 559-563 SciRes Copyright © 2009 563 JBiSE et al, (2006) Acute myeloid leukemia-associated mkl1 (mrtf-a) is a key regulator of mammary gland function, Mol. Cell Biol., Washington, 26, 5809–5826. [6] Li, S., Chang, S., Qi, X., Richardson, J. A., and Olson, E. N., (2006) Requirement of a myocardin-ralated transcrip- tion factor for development of mammary myoepithelial cells, Mol. Cell Biol., Washington, 26, 5797–5808. [7] Sun, Q., Chen, G., Streb, J. W., Long, X., Yang, Y., Stoeckert, C. J., et al, (2006) Defining the mammalian CarGome, Genome. Res., Cold Spring Harbor, 16, 197– 207. [8] Jordan, V. C., (2003) Is tamoxifen the rosetta stone for breast cancer, J. Natl. Cancer. Inst. Bethesda, 95, 338– 340. [9] Hilsenbeck, S. G., Friedrichs, W. E., Schiff, R., O’Con- nell, P., Hansen, R. K., Osborne, C. K., et al, (1999) Sta- tistical analysis of array expression data as applied to the problem of tamoxifen resistance, J. Natl. Cancer. Inst. Bethesda, 91, 453–459. [10] Mannisto, P. T. and Kaakkola, S., (2008) Catechol-O- methyltransferase (COMT): Biochemistry, molecular bi- ology, pharmacology, and clinical efficacy of the new se- lective COMT inhibitors, Pharmacol. rev. Bethesda, 51, 593–628. [11] Singh, M. N., Stringfellow, H. E., Walsh, M. J., Ashton, K. M., Paraskevaidis, E., Abdo, K. R., et al, (2008) Quantifiable mRNA transcripts for tamoxifen-metabolising enzymes in human endometrium, Toxicology, Limerick, 249, 85–90. [12] Dehal, S. S. and Kupfer, D., (1995) Evidence that the catechol 3, 4-dihydroxytamosifen is a proximate inter- mediate to the reactive species binding covalently to proteins, Cancer Res. Baltimore, 56, 1283–1290. [13] Cen, B., Selvaraj, A., Burgess, R. C., Hitzler, J. K., Ma, Z., Morris, S. W., et al, (2003) Megakaryoblastic leuke- mia 1, a potent transcriptional coactivator for serum re- sponse factor (SRF), is required for serum induction of SRF target genes, Mol Cell Biol., Washington, 23, 6597– 6606. [14] Dubey, R. K., Gillespie, D. G., Zacharia, L. C., Barchiesi, F., Imthurn, B., and Jackson, E. K., (2003) CYP450- and COMT-derived estradiol metabolites inhibit activity of human coronary artery SMCs, Hypertension, Dallas, 41, 807–813. [15] Dubey, R. K., Tofovic, S. P., and Jackson, E. K., (2004) Cardiovascular pharmacology of estradiol metabolites, J. Pharmacol. Exp. Ther., Bethesda, 308, 403–409. [16] Wentz, M. J., Jamaluddin, M., Garfield, R. E., Al-Hendy, A., (2006) Regulation of catechol-O-methyltransferase expression in human myometrial cells, Obstet. Gynecol., Hagerstown, 108, 1439–1447. |

