 American Journal of Plant Sciences, 2011, 2, 521-526 doi:10.4236/ajps.2011.24061 Published Online October 2011 (http://www.SciRP.org/journal/ajps) Copyright © 2011 SciRes. AJPS 521 Interspecific Hybridization between Arisaema sikokianum and A. serratum (Araceae) Confirmed through Nuclear and Chloroplast DNA Comparisons Hiroshi Hayakawa1,2, Hidenori Hamachi3, Kanako Matsuyama3, Yuko Muramatsu3, Yukio Minamiya1, Katsura Ito1, Jun Yokoyama4, Tatsuya Fukuda1* 1Faculty of Agriculture, Kochi University, Monobe, Nankoku 783-8502, Japan; 2The United Graduate School of Agricultural Sci- ences, Ehime University, Monobe, Nankoku 783-8502, Japan; 3The Graduate School of Integrated Arts and Sciences, Kochi Univer- sity, Monobe, Nankoku 783-8502, Japan; 4Faculty of Science, Yamagata University, 1-4-12 Kojirakawa-machi, Yamagata 990-8560, Japan. Email: *tfukuda@kochi-u.ac.jp Received January 22nd, 2011; revised March 24th, 2011; accepted April 1st, 2011. ABSTRACT A morphologically intermediate plant between Arisaema sikokianum Franch. et Sav. and A. serratum (Thunb.) Schott has been newly found in Kochi Prefecture, Shikoku, Japan. The putative hybrid has the intermediate morphological characteristics of the parental species. Molecular analysis using PCR-RFLP of internal transcribed spacer (ITS) in nuclear DNA (nrDNA) indicates that the putative hybrid has a combined pattern of the two putative parent species. Moreover, the sequence result of chloroplast DNA (cpDNA) of the putative hybrid was identical to that of A. si- kokianum. These results suggest that the putative hybrid is a hybrid between A. sikokianum and A. serratum and that it was formed by interactive gene excha nging via pollens from A. serratum to A. sikokianum. It is the first record o f a hy- brid between A. sikokianum and A. serratum. Keywords: Araceae, Arisaema, A. serratum, A. sikokianum, Chloroplast DNA, Interspecific Hybrid, Molecular Analysis, Nuclear DNA 1. Introduction The genus Arisaema Martius (Araceae), which has a large, often colored and conspicuous bract (spathe), and subtending and enveloping bisexual or unisexual spadix with numerous small flowers, comprises approximately 40 - 85 species in Japan [1,2]. Species of Arisaema in the section Pedatisecta Schott ex Engler have a slender ap- pendage at the base and are mostly distributed in Japan [2]. Section Pedatisecta is included in 35 - 80 species in Japan [2], and presents many taxonomic difficulties caused by the concentration of closely related species with few morphological differences [3]. Sixteen patterns of putative natural hybrids have been reported in Arisaema sect. Pedatisecta (e.g., [4]). Of them, A. sikokianum Franch. et Sav. and A. tosaense Ma- kino make the hybrids [5,6]. Moreover, A. ehimense J. Murata et Ohno is of hybrid origin between A. serratum (Thunb.) Schott and A. tosaense based on allozyme analysis [7]. Therefore, it is possible that hybrid speci- ation or reticulate evolution or both has occurred among A. tosaense, A. sikokianum and A. serratum. However, the hybridization between A. sikokianum and A. serratum was unknown until now. Arisaema sikokianum has a purple upward spathe, a white capitate appendage and leaves with 3 - 5 leaflets (Figure 1(a), Table 1), while Arisaema serratum has a green cylindrical appendage and leaves with 7 - 17 leaflets (Figure 1(b), Table 1). Although the spathe of A. serra- tum varies widely in various areas of Japan [2,3], our ob- servation is that almost all A. serratum in Kochi Prefecture show a green spathe and appendage (Figure 1(b)). In Ko- chi Prefecture of Shikoku, A. sikokianum and A. serratum are found in sympatry. The flowering phenology of the two species overlaps (Table 1). The chromosome num-  Interspecific Hybridization between Arisaema sikokianum and A. serratum (Araceae) Confirmed through Nuclear 522 and Chloroplast DNA Comparisons Figure 1. Arisaema species examined in this study. (a) A. sikokianum; (b) A. serratum; (c) putative hybrid in 2007; (d) putative hybrid in 2006; (e) putative hybrid in 2010; (f) newly formed putative hybrids in 2010. ber of A. sikokianu m and A. serratum is 2 n = 28 in both species [8]. Therefore, it is possible to generate a hybrid between A. sikokianum and A. serratum. Molecular approaches can reveal the processes of past event such as hybridization and reticulate evolution [9]. Nuclear markers provide information to estimate the pu- tative parents in hybrids [10,11]. The polymorphisms of the internal transcribed spacer (ITS) region in nuclear DNA (nrDNA) are good tools for clarifying the relation- ship of closely related taxa in many plant groups [12-14], and can provide evidence of hybridization. In fact, inter- specific hybrids are most commonly identified by the heterogeneity of nrDNA [5,15,16]. Moreover, chloroplast DNA (cpDNA) data can indicate maternal and paternal parents of hybrids, because cpDNA normally inherit from maternal parent [17]. In this study, we found a pu- tative natural hybrid of A. sikokianum and A. serratum in Kochi Prefecture from the viewpoint of its morphological characteristics (Figure 1(d), Figure 2). To clarify the maternal and paternal parents of the putative hybrid, we conducted molecular analysis using nrDNA and cpDNA sequences. Our finding suggests that hybridization may occur between A. sikokianum and A. serratum. 2. Materials and Methods A morphologically intermediate plant was found at one locality in Kochi Prefecture (Figure 2, Table 2). The intermediate hybrid was growing in situ with Arisaema sikokianum, A. serratum and A. tosaense. A voucher specimen is deposited in the Herbarium of the Makino Botanical Garden, Kochi (MBK). For the molecular analysis, total DNA was isolated from 200 - 300 mg of leaves with a Plant Genomic DNA Mini Kit (VIOGENE, Sunnyvale, CA, USA), according to the manufacturer’s protocol. We amplified the internal transcribed spacer (ITS) region from nrDNA and the trnL intron from cpDNA with primers designed by Taberlet et al. [18] and White et al. [19], respectively. Isolated DNA was amplified by PCR in a 50 µl reaction solution con- taining approximately 50 ng total DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM of each dNTP, 1.25 units Taq DNA polymerase (Ta Ka Ra) and 0.5 µM of each primer. We used the following thermal cycle profile for amplification: 1 min at 94˚C, 2 min at 48˚C, and 2 min at 72˚C for 45 cycles, followed by 15 min of final extension at 72˚C. A preliminary sequence of the ITS region was taken from the parent species to confirm whether PCR-RFLP can effectively demonstrate Table 1. Morphological characteristics of samples used in this study. Trait A. sikokianum Putative Hybrid A. serratum Pseudostem Pseudostem Length Short Short-Long Long Pseudostem Color Green Purplish Dark Brown Purplish Dark Brown Leaf Characteristics Leaflets 3 to 5 9 7 to 17 Width of Leaflets Wide Narrow-Wide Narrow Petiole Length Long Short-Long Short Reproductive Characteristics Spathe Tip Direction Upward Middle Downward Spathe Color Purple Purple Green Appendage Shape Capitate Capitate-Cylindrical Cylindrical Appendage Color White White Green Flowering Phenology April to May Early April Late April to June Copyright © 2011 SciRes. AJPS 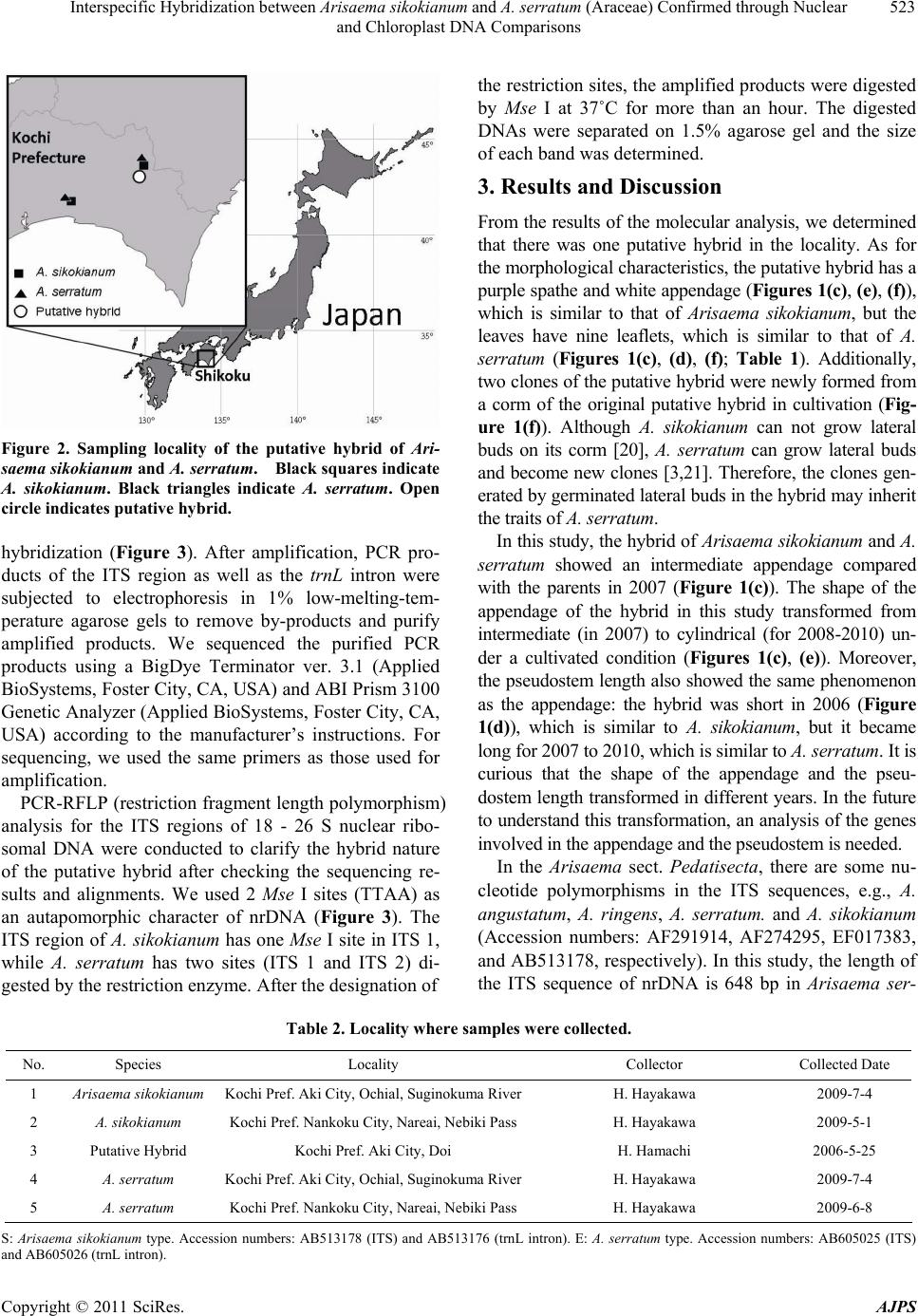 Interspecific Hybridization between Arisaema sikokianum and A. serratum (Araceae) Confirmed through Nuclear 523 and Chloroplast DNA Comparisons Figure 2. Sampling locality of the putative hybrid of Ari- saema sikokianum and A. serratum. Black squar es indicate A. sikokianum. Black triangles indicate A. serratum. Open circle indicates putative hybr id. hybridization (Figure 3). After amplification, PCR pro- ducts of the ITS region as well as the trnL intron were subjected to electrophoresis in 1% low-melting-tem- perature agarose gels to remove by-products and purify amplified products. We sequenced the purified PCR products using a BigDye Terminator ver. 3.1 (Applied BioSystems, Foster City, CA, USA) and ABI Prism 3100 Genetic Analyzer (Applied BioSystems, Foster City, CA, USA) according to the manufacturer’s instructions. For sequencing, we used the same primers as those used for amplification. PCR-RFLP (restriction fragment length polymorphism) analysis for the ITS regions of 18 - 26 S nuclear ribo- somal DNA were conducted to clarify the hybrid nature of the putative hybrid after checking the sequencing re- sults and alignments. We used 2 Mse I sites (TTAA) as an autapomorphic character of nrDNA (Figure 3). The ITS region of A. sikokianum has one Mse I site in ITS 1, while A. serratum has two sites (ITS 1 and ITS 2) di- gested by the restriction enzyme. After the designation of the restriction sites, the amplified products were digested by Mse I at 37˚C for more than an hour. The digested DNAs were separated on 1.5% agarose gel and the size of each band was determined. 3. Results and Discussion From the results of the molecular analysis, we determined that there was one putative hybrid in the locality. As for the morphological characteristics, the putative hybrid has a purple spathe and white appendage (Figures 1(c), (e), (f)), which is similar to that of Arisaema sikokianum, but the leaves have nine leaflets, which is similar to that of A. serratum (Figures 1(c), (d), (f); Table 1). Additionally, two clones of the putative hybrid were newly formed from a corm of the original putative hybrid in cultivation (Fig- ure 1(f)). Although A. sikokianum can not grow lateral buds on its corm [20], A. serratum can grow lateral buds and become new clones [3,21]. Therefore, the clones gen- erated by germinated lateral buds in the hybrid may inherit the traits of A. serratum. In this study, the hybrid of Arisaema sikokianum and A. serratum showed an intermediate appendage compared with the parents in 2007 (Figure 1(c)). The shape of the appendage of the hybrid in this study transformed from intermediate (in 2007) to cylindrical (for 2008-2010) un- der a cultivated condition (Figures 1(c), (e)). Moreover, the pseudostem length also showed the same phenomenon as the appendage: the hybrid was short in 2006 (Figure 1(d)), which is similar to A. sikokianum, but it became long for 2007 to 2010, which is similar to A. serratum. It is curious that the shape of the appendage and the pseu- dostem length transformed in different years. In the future to understand this transformation, an analysis of the genes involved in the appendage and the pseudostem is needed. In the Arisaema sect. Pedatisecta, there are some nu- cleotide polymorphisms in the ITS sequences, e.g., A. angustatum, A. ringens, A. serratum. and A. sikokianum (Accession numbers: AF291914, AF274295, EF017383, and AB513178, respectively). In this study, the length of the ITS sequence of nrDNA is 648 bp in Arisaema ser- Table 2. Locality where samples were collected. No. Species Locality Collector Collected Date 1 Arisaema sikokianum Kochi Pref. Aki City, Ochial, Suginokuma RiverH. Hayakawa 2009-7-4 2 A. sikokianum Kochi Pref. Nankoku City, Nareai, Nebiki PassH. Hayakawa 2009-5-1 3 Putative Hybrid Kochi Pref. Aki City, Doi H. Hamachi 2006-5-25 4 A. serratum Kochi Pref. Aki City, Ochial, Suginokuma RiverH. Hayakawa 2009-7-4 5 A. serratum Kochi Pref. Nankoku City, Nareai, Nebiki PassH. Hayakawa 2009-6-8 S: Arisaema sikokianum type. Accession numbers: AB513178 (ITS) and AB513176 (trnL intron). E: A. serratum type. Accession numbers: AB605025 (ITS) and AB605026 (trnL intron). Copyright © 2011 SciRes. AJPS  Interspecific Hybridization between Arisaema sikokianum and A. serratum (Araceae) Confirmed through Nuclear 524 and Chloroplast DNA Comparisons M M M M A. sikokian um A.serratum A. sikokian um A. serratum A. sikokian um A. serratum A. sikokian um A. serratum A. sikokian um A. serratum A. sikokian um A. serratum A. sikokian um A. serratum A. sikokian um A. serratum A. sikokian um A. serrarum A. sikokian um A. serratum A. sikokian um A. serratum Figure 3. Expected restriction sites of Mse I for molecular characteristics of ITS regions by PCR-RFLP. M: restriction site. ratum (Accession number: AB605025). This sequence is the same as A. tosaense (Accession number: AB513179). Therefore, we conducted PCR-RFLP in the nrDNA to obtain further evidence of the hybrid nature, because the ITS region of A. sikokianum has one Mse I site, while A. serratum and A. tosaense have two sites digested by this restriction enzyme (Figure 3) [5]. The digestion patterns of all samples of A. sikokianum and A. serratum showed expected patterns and the putative hybrid showed the combined pattern of A. sikokianum and A. serratum (Figure 4). Thus, from the evidence of morphological and molecular analyses, we confirmed that the putative hybrid was truly a hybrid of A. sikokianum and A. serra- tum. The length of the trnL intron of cpDNA is 467 bp in Arisaema serratum (Accession number: AB605026). This region is a good molecular marker for distinguish- ing Arisaema sikokianum, because the trnL intron has a 17 bp-insertion or deletion (indel) in the sequence of A. sikokianum (450 bp) (Figure 5) [5]. We therefore deter- mined the sequences of the trnL intron. In the hybrid, the sequence result from the cpDNA analysis was identical to that of A. sikokianum (Table 1). Therefore, the hybrid Figure 4. PCR-RFLP profile of Arisaema sikokianum, A. serratum and putative hybrid. Arrowheads indicate ex- pected fragments of both A. sikokianum and A. serratum. M: size marker. The cpDNA types correspond to the types in Table 2 and Figure 5. transferred pollen from A. serratum to A. sikokianum. In Shikoku, Arisaema ehimense J. Murata et Ohno is of hybrid origin between A. serratum and A. tosaense [7,22]. Additionally, the hybrids between A. sikokianum and A. tosaense are known [5,6]. Moreover, this study eveals a hybrid between A. sikokianum and A. serratum. r Copyright © 2011 SciRes. AJPS  Interspecific Hybridization between Arisaema sikokianum and A. serratum (Araceae) Confirmed through Nuclear 525 and Chloroplast DNA Comparisons A. sikokia nu m A. serratum A. sikokia nu m A. serratum A. sikokia nu m A. serratum A. sikokia nu m A. serratum A. sikokia nu m A. serratum A. sikokia nu m A. serratum A. sikokia nu m A. serratum Figure 5. The result of alignment of trnL intron sequences of Arisaema sikokianum and A. serratum. To our knowledge, it is first report of a hybrid between this Arisaema pair. These facts indicate that all the com- bination patterns of hybridization among A. tosaense, A. sikokianum and A. serratum have been in Shikoku. Therefore, these facts suggest that hybrid speciation or reticulated evolution or both might have occurred among the three species in Shikoku. 4. Acknowledgements We wish to thank N. Tanaka, the curator of the MBK herbarium, for allowing us to examine specimens of Ari- saema, and R. Arakawa, M. Saito, K. Ohga and N. Yo- koyama for providing much help. I would also like to thank Dennis Murphy from the United Graduate School of Agricultural Sciences, Ehime University, for checking the English in this manuscript. This study was partly sup- ported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan (to TF and JY). 5. Conclusions In Figure 3 is reported the shifting of the three switching frequencies and also in this case it is evident. REFERENCES [1] H. Ohashi, “Araceae,” In: Y. Satake, J. Ohwi, S. Kita- mura, S. Watari and T. Tominari, eds., Wild Flowers of Japan, Vol. 1, 1982, pp. 127-139. [2] J. Murata, “Should Arisaema serratum Group Be Classified into One or 30 Species? With Reference to the Geographic Structure of Aucuba japonica Widely Distributed in Ja- pan,” Seibutu Kagaku, Vol. 55, No. 2, 2004, pp. 87-94. [3] J. Murata, “Diversity in the Arisaema serratum Group,” Acta Phytotaxonomica et Geobotanica, Vol. 46, No. 2, 1995, pp. 185-208. [4] T. Kobayashi, J. Murata and K. Watanabe, “Two New Putative Natural Hybrids in Japanese Arisaema (Arac eae),” Acta Phytotaxonomica et Geobotanica, Vol. 56, No. 1, 2005, pp. 105-110. [5] H. Hayakawa, H. Hamachi, Y. Muramatsu, A. Hirata, Y. Minamiya, K. Matsuyama, K. Ito, J. Yokoyama and T. Fukuda, “Interspesific Hybridization between Arisaema sikokianum and A. tosaense (Araceae) Confirmed through Nuclear and Chloroplast DNA Comparisons,” Acta Phy- totaxonomica et Geobotanica, Vol. 61, No. 2, 2010, pp. 57-63. [6] G. Murata, “Taxonomical Note 7,” Acta Phytotaxonomica et Geobotanica, Vol. 19, No. 2-3, 1962, pp. 67-72. [7] M. Maki and J. Murata, “Allozyme Analysis of the Hy- brid Origin of Arisaema ehimense (Araceae),” Heredity, Vol. 86, No. 1, 2001, pp. 87-93. doi:10.1046/j.1365-2540.2001.00813.x [8] J. Murata and M. Iijima, “New or Noteworthy Chromo- some Records in Arisaema,” Journal of Japanese Botany, Vol. 58, No. 9, 1983, pp. 270-280. [9] H. Yamaji, T. Fukuda, J. Yokoyama, J. -H. Pak, C. -Z. Zhou, C. -S. Yang, K. Kondo, T. Morota, S. Yakeda, H. Sasaki and M. Maki, “Reticulate Evolution and Phy- logeograhy in Asarum sect. Asiasarum (Aristolochiaceae) Documented in Internal Transcribed Spacer Sequences (ITS) of Nuclear Ribosomal DNA,” Molecular Phyloge- netics and Evolution, Vol. 44, No. 2, 2007, pp. 863-884. doi:10.1016/j.ympev.2007.01.011 [10] L. H. Rieseberg, J. Whitton and C. R. Linder, “Molecular Marker Incongruence in Plant Hybrid Zones and Phy- logenetic Trees,” Acta Botanica Neerlandica, Vol. 45, No. 3, 1996, pp. 243-262. [11] J. F. Wendel and J. J. Doyle, “Phylogenetic Incongruence: Copyright © 2011 SciRes. AJPS  Interspecific Hybridization between Arisaema sikokianum and A. serratum (Araceae) Confirmed through Nuclear 526 and Chloroplast DNA Comparisons Window into Genome History and Molecular Evolution,” In: P. S. Soltis, D. E. Soltis and J. J. Doyle, eds., Molecu- lar Systematics of Plants II, Dordrecht, Kluwer, 1998, pp. 265-296. doi:10.1007/978-1-4615-5419-6_10 [12] S. Markos and B. G. Baldwin, “Higher-Level Relation- ships and Major Lineages of Lessingia (Compositae, As- tereae) Based on Nuclear rDNA Internal and External Transcribed Spacers (ITS and ETS) Sequences,” System- atic Botany, Vol. 26, No. 1, 2001, pp. 168-183. [13] L. E. Urbatsch, R. P. Roberts and V. Karaman, “Phy- logenetic Evaluation of Xylothamia, Gundlachia, and Re- lated Genera (Asteraceae, Astereae) Based on ETS and ITS nrDNA Sequence Data,” American Journal of Botany, Vol. 90, No. 4, 2003, pp. 634-649. doi:10.3732/ajb.90.4.634 [14] O. Hidalgo, N. Garcia-Jacas, T. Garnatje and A. Susanna, “Phylogeny of Rhaponticum (Asteraceae, Cardue- ae―Centaureinae) and Related Genera Inferred from Nu- clear and Chloroplast DNA Sequence Data: Taxonomic and Biogeographic Implications,” Annals of Botany, Vol. 97, No. 5, 2006, pp. 704-714. doi:10.1093/aob/mcl029 [15] J. Yokoyama, T. Fukuda, A. Yokoyama and M. Maki, “The Intersectional Hybrid between Weigela hortensis and W. maximowiczii (Caprifoliaceae),” Botanical Jour- nal of the Linnean Society, Vol. 138, No. 3, 2002, pp. 369-380. doi:10.1046/j.1095-8339.2002.00033.x [16] H. Hayakawa, M. Yukio, K. Ito, Y. Yamamoto and T. Fukuda, “Difference of Curcumin Content in Curcuma longa L. (Zingiberaceae) Caused by Hybridization with Other Curcuma species,” American Journal of Plant Sci- ences, Vol. 2, 2011, pp. 111-119. doi:10.4236/ajps.2011.22013 [17] Y. Ukai, “Breeding Utility of Tissue Culture,” Plant Breeding―Crossing to Geneticaly Modification, Univer- sity of Tokyo Press, Tokyo, 2003, p. 334. [18] P. Taberlet, L. Gielly, G. Pautou and J. Bouvet, “Univer- sal Primers for Amplification of Three Non-Coding Re- gions of Chloroplast DNA,” Plant Molecular Biology, Vol. 17, No. 5, 1991, pp. 1105-1109. doi:10.1007/BF00037152 [19] T. J. White, T. Bruns, S. Lee and J. Taylor, “Amplifica- tion and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics,” In: M. Innis, D. Gelfand, J. Sninsky and T. J. White, eds., PCR Protocols: A Guide to Methods and Application, Academic Press, San Diego, 1990, pp. 315-322. [20] S. Fukai, “A Morphological Study on Arisaema si- kokianum (Araceae),” Technical Bulletin of Faculty of Agriculture, Kagawa University, Vol. 59, 2007, pp. 15-25. [21] T. Maekawa, “On the Phenomena of Sex Transition in Arisaema japonica BL,” Journal of the College of Agri- culture, Vol. 13, No. 3, 1924, pp. 217-305. [22] J. Murata and J. Ohno, “Arisaema ehimense J. Murata et Ohno (Araceae), a New Species from Shikoku, Japan, of Putative Hybrid Origin,” Journal of Japanese Botany, Vol. 64, No. 11, 1989, pp. 341-351. Copyright © 2011 SciRes. AJPS
|