Advances in Bioscience and Biotechnology
Vol.5 No.2(2014), Article ID:42429,5 pages DOI:10.4236/abb.2014.52019
Juvenile myelo-monocytic leukemia (JMML): No effect of granulocyte monocyte-colony stimulating factor (GM-CSF) on Wilms Tumor gene (WT1) by nested Polymerase Chain Reaction (nPCR) and flow cytometry
![]()
1Stem Cell Transplant Chimerism Laboratory, Depts. of Hematology-Oncology Transplant; Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, USA
2Northwestern University Feinberg School of Medicine, Chicago, USA
Email: *mkletzel16@northwestern.edu
Copyright © 2014 Sana Khan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Sana Khan et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 27 November 2013; revised 15 December 2013; accepted 13 January 2014
KEYWORDS
Juvenile Myelo-Monocytic Leukemia; Wilms Tumor; Nested PCR; JMML; WT1; GM-CSF; nPCR
ABSTRACT
This study was to determine whether GM-CSF induced WT1 gene expression and to establish an association with markers of proliferation CD71+CD34+ using nPCR and flow cytometry respectively, in samples obtained from 5 newly diagnosed JMML patients. Overtime (day 0 to day 14) there was an insignificant difference in WT1 gene expression and CD71+CD34+ in JMML samples when compared to peripheral blood of normal volunteers (n = 3). Our study suggests that there is a correlation between WT1 gene expression and cellular proliferation and that GMCSF in vitro does not create a significant difference in JMML samples.
1. INTRODUCTION
Juvenile myelomonocytic leukemia (JMML) is a rare hematopoietic malignancy of early childhood with features exhibiting of both myeloproliferative neoplasm (MPN) and myelodysplastic syndrome (MDS) [1]. JMML affects very young children (median age at diagnosis is 2 years) with a significant male predominance (male:female = 2.5). This leukemia clinically presents with marked hepatosplenomegaly together with lymphadenopathy, pallor and skin rash [2]. It is a representative of a clonal disease of pluripotent cell origin and in vitro granulocyte-macrophage colony-stimulating factor (GMCSF) hypersensitivity is the hallmark of JMML [3-7].
WT1 gene was first identified in patients with Wilms’ tumor, codes for a zinc-finger transcription factor located on chromosome 11p13 [8]. Although expressed at low levels in bone marrow by normal progenitor cells, WT1 has been reported to be expressed at high levels in almost all hematological malignancies, including JMML [9,10]. WT1 plays an important role in regulating cellular growth and differentiation. Furthermore, WT1 gene expression has been utilized as a diagnostic tool in assessing minimal residual disease (MRD) in varieties of leukemias [9,11-15] and disease progression in MDS [11- 15].
In this study, we correlate the effect of GM-CSF stimulation and WT1 gene expression over a period of time, obtaining samples from JMML patients, to establish a direct association of its expression with markers of proliferation (CD71+CD34+) [16-20].
2. MATERIALS & METHODS
2.1. Patient Samples
Age at diagnosis ranged from 30 days to 9 years old. There were 3 males and 2 females. All five patients were diagnosed as JMML according to the World Health Organization (WHO) and met the laboratory criteria: Ph chromosome negative, no BCR/ABL rearrangement, peripheral blood monocyte count >1 × 109/L, and bone marrow blasts <20%. Additionally, all patients met at least two laboratory findings: white blood count >10 × 109/L and GM-CSF hypersensitivity of myeloid progenitors in vitro. Informed consent were obtained from the patients at Ann and Robert H. Lurie Children’s Hospital of Chicago.
2.2. Sample Preparations
Peripheral blood samples were collected in EDTA tube and prepared by density gradient preparation to isolate mononuclear cells using Ficoll-Paque Plus (Pharmacia Amersham Bioscience AB, Uppsala, Sweden). A cell concentration of 2 × 106/mL/plate mononuclear cells was used as a seeding density on a 6-well culture plate. Samples were conducted in RPMI-1640 containing 100 U/ mL penicillin, 100 ug/mL streptomycin, 10% FBS and 10 ng/mL GM-CSF added at day 0. Cells were cultured with and without the addition of GM-CSF to assessWT1 gene expression. Cultures were incubated at 37˚C in a humidified atmosphere with 5% CO2 for a period of 14 days. At days 0 and 14 culture cells were harvested for total RNA extraction using a QIAamp RNA Blood Mini kit (QIAGEN Inc., Valencia, CA, USA) to determine WT1 gene expression. Cells were additionally stained for CD 34+ early stem cell markers and CD 71+ a transferrin receptor, all indicators of cellular proliferation [16-20] were executed by flow cytometry using a FACSort flow cytometer (FacSort Becton-Dickinson, San Jose, CA, USA) and was analyzed with CELLQuest software (Becton Dickinson).
2.3. Nested PCR (nPCR)
The composition of the reaction buffer was 2 µL 10× PCR buffer II, 4 µL of 25 nm MgCl2, 8 µL dNTP, 1 µL Oligo d (T), 1 µL MuLV reverse transcriptase, and 1 µL RNAase inhibitor (Applied Biosystem, Foster City, CA, USA). Primer pairs were devised to locate precise nucleotide fragments for mRNA of the WT1 gene illustrated in Table 1 [21].
RT-PCR reaction was performed in two rounds in a 20-µL reaction volume containing 1 ug/µL total RNA from each sample. In preparation of the first round, RT was incubated at 42˚C in a waterbath for 60 min, followed by denaturation at 96˚C for 10 min. The first step included amplification of 10 µL of cDNA which was then subjugated to 30 cycles of denaturation for 5 min, amplification for 5 seconds at 95˚C, annulment at 55˚C for 5 seconds, and extension at 72˚C for 10 seconds. Step 2 was then conducted by taking 1 µL of the amplified product from the first step and which was further re-amplified with inner primers using the LightCycler System with SYBR-Green I RNA master mix reagent, acquired from Roche Diagnostics, Biochemica (Indianapolis, IN, USA) [21]. After the termination of PCR the

Table 1. WT1 Primer Sequences [21].
LightCycler software through computation from the external curve figured the titration of target molecules. The housekeeping gene GAPDH was admitted as an internal control to establish the integrity of the RNA in question. A tenfold serial dilution of the K562 leukemia cell line yielded a standard curve wherein sample results were interpolated. Values ranging from 1 × 10 ng/µL and higher were considered positive for WT1 mRNA expression (Table 2) [21]. The level of WT1 gene expression in K562 was arbitrarily defined as 1.0 [14-15,21-26]. Samples were analyzed in triplicate. Any PCR results illustrating a discrepancy greater than one cycle threshold were expelled and reprised.
2.4. Statistical Analysis
Statistical analysis was performed using SPSS software (IBM Corporation, Armonk, New York, USA) to obtain a mean, median, ± SEM and range along with 95% confidence interval (CI) by column statistics. To compare and contrast sample groups and cultured groups an F-test and paired t-test were analyzed respectively for the determination of WT1 expression. A p value of <0.05 was considered statistically significant.
3. RESULTS
JMML (n = 5) and negative control (n = 3) PB samples were tested on days 0 and 14 for WT1 gene expression (Figure 1). Differentiation of cultures, un-stimulated vs. stimulated by GM-CSF was measured on day 14. There was a difference found on day 14 in JMML samples for WT1 transcription in assessment to the initial day. The detection of WT1 transcription in JMML cultured groups on day 14 represented an insignificant p-value (p < 0.738, 95% CI −63.9 to 49.3) signifying that despite of the GMCSF stimulation WT1 transcripts remained persistently positive. In contrast, normal PB measured low to undetectable levels of WT1 transcripts from day 0 to day 14 in both culture groups (p < 0.0182, 95% CI −0.259 to −0.0669). Furthermore, this level of expression was significantly lower in comparison to the positive control, K562 cell line. A significant difference was noted when comparing WT1 transcriptions of patient samples to the negative control (Table 3) throughout the study on day 0 (P < 0.0001), day 14 un-stimulated (p < 0.0001) and sti-
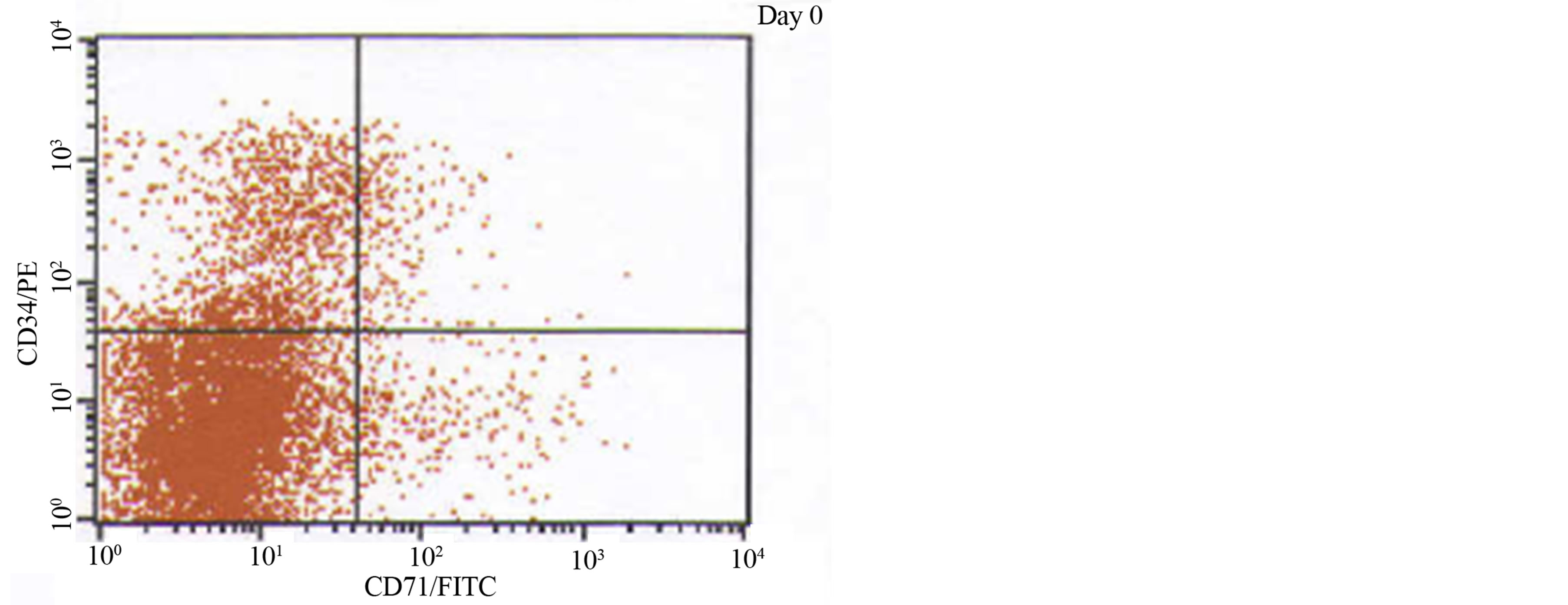
Figure 1. Comparison of WT1 gene expression in JMML patients vs. negative control (CTL) samples with a significant (p < 0.0001) difference to be found at days 0 and 14: un-stimulated vs. stimulated by GM-CSF. The bars indicate standard errors.

Table 2. Value ranges for WT1 gene expression analysis.
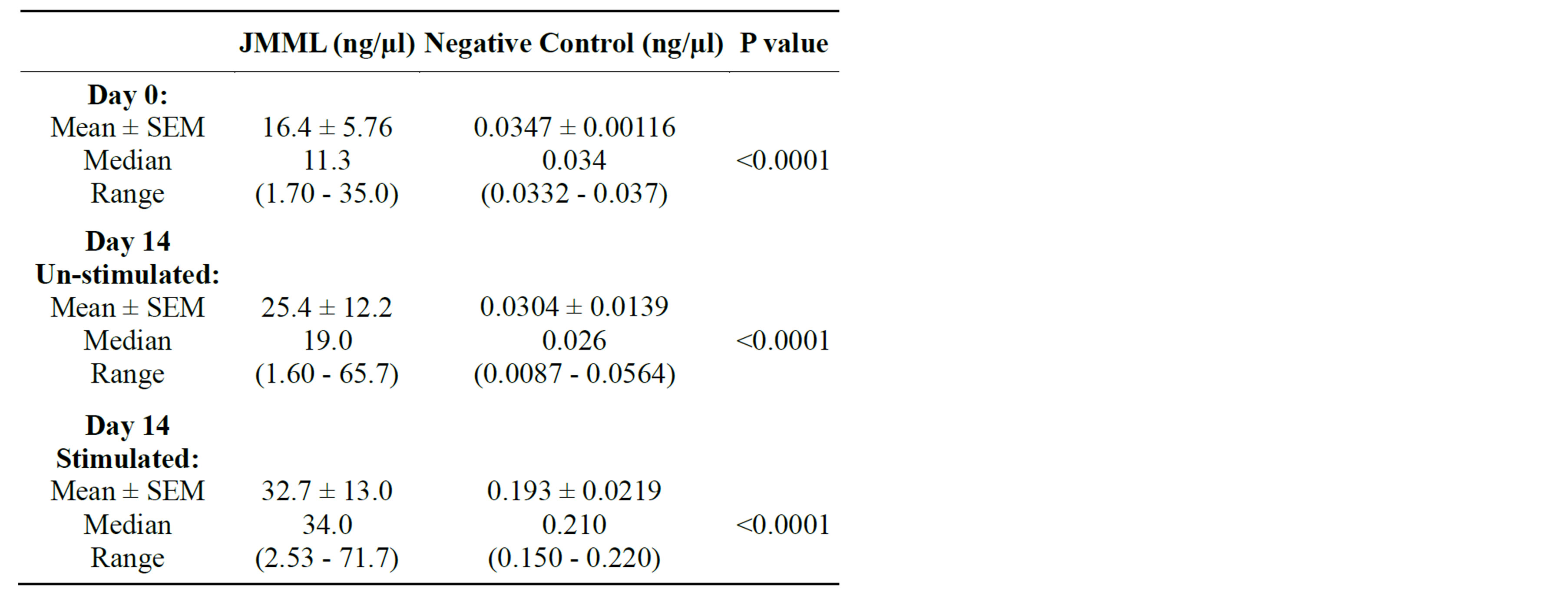
Table 3. Measurement of WT1 transcription by nPCR.
mulated (p < 0.0001).
In vitro patient samples were examined for the expression of WT1 and CD71+CD34+ simultaneously throughout the study. Phenotypic analysis was completed to establish the level of cellular proliferation using markers CD71+CD34+ (Figure 2). Although cellular proliferation increased as time progressed from day 0 (mean 9.00 ± SEM 3.81, median 6.20, range 3.30% - 24.0%) to day 14, under the influence of GM-CSF un-stimulated (mean 51.5 ± SEM 6.59, median 46.9, range 39.2% - 77.0%) vs. stimulated (mean 56.4 ± SEM 2.20, median 55.7, range
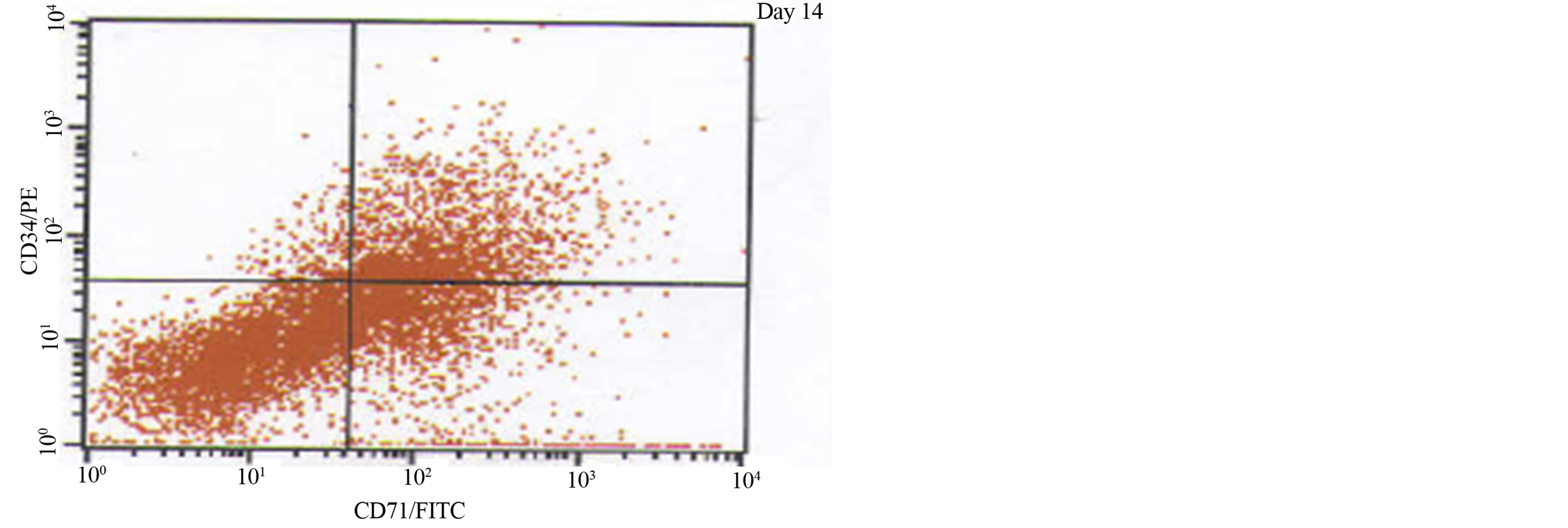
Figure 2. A phenotypic analysis of one JMML patient using cellular proliferative markers CD71+CD34+ measured on day 0 and day 14, stimulated by GM-CSF.
49.7% - 63.1%) there was no significant difference (p < 0.4689, 95% CI −22.1 to 12.2). CD71+CD34+ levels were further compared with WT1 transcripts to understand the relationship between cellular proliferation and gene expression in JMML. The insignificance found between cultured groups on day 14 concurs with the results of WT1 expression.
4. DISCUSSION
WT1 transcription is associated with presence, persistence or reappearance of leukemic hematopoiesis [12]. Tamaki et al. [22] as well as Cilloni et al. [12] reported a strong correlation between leukemic progression and WT1 gene expression. It is over expressed in all leukemic cell lines regardless of lineage and is also the most reliable risk index of disease progression available for adult MDS and JMML patients [23-26].
In order to establish WT1 transcription as a proliferative marker, phenotypic analysis was performed using CD71+CD34+ [16-20]. WT1 expression was measured with a standard K562 cell line, arbitrarily defined as 1.0 as a positive expression [14-15,21-26], which was demonstrated in all 5 JMML patients. Measuring CD71+CD34+ coherently with WT1 using K562 clearly shows the consistency of proliferation throughout the study.WT1 is an unalterable gene that is highly expressed irrespective of the presence of GM-CSF, allowing us to assume that it is a proliferative marker not only in the genotypic level as well as in the phenotypic level. This observation concurs with the nature of JMML, being that is a myeloproliferative neoplasm (MPN) [1,3,4]. The fact that there is no statistical significance between the cultured groups defines the characteristic of random proliferation in JMML [3,4]. The inability of WT1 expression as a quantitative assay using nPCR does not show the degree of proliferation but proliferation by itself.
Although the WT1 gene expression was positively amplified regardless of GM-CSF stimulation, due to insignificant p-values our observations are invalid. Minimal response to GM-CSF can be attributed to the fact that our study included a small number of patients. Thus, we cannot conclude that WT1 gene expression has a relationship with GM-CSF.
5. CONCLUSION
Our findings suggest that over time there is a trend of WT1 gene expression in JMML cells, and that GM-CSF does not induce higher transcription in liquid culture. Phenotypic analysis concurs with these results. Thus our study supports the hypothesis that WT1 gene expression is a marker of proliferation and may not have any role in the pathogenesis of JMML.
DECLARATION OF INTEREST
The authors report no conflicts of interest.
REFERENCES
- Grillo, Niemeyer, C.M., Arico, M. and Basso, G. (1997) Chronic myelomonocytic leukemia in childhood: A retrospective analysis of 110 cases. Blood, 89, 3534-3543.
- Emanuel, P.D. (2008) Juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia. Leukemia, 22, 1335-1342. http://dx.doi.org/10.1038/leu.2008.162
- Emanuel, P.D., Bates, L.J., Castleberry, R.P., Gualtieri, R.J. and Zuckerman, K.S. (1991) Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile myeloid leukemia hematopoietic progenitors. Blood, 77, 925-929.
- de Vries, A.C., Zwaan, C.M. and van den Heuvel-Eibrink, M.M. (2010) Molecular basis of juvenile myelomonocytic leukemia. Hematologica, 95, 179-181. http://dx.doi.org/10.3324/haematol.2009.016865
- Kenichi, K. and Kazuyuki, M. (2008) Recent advances in the pathogenesis and management ofjuvenile myelomonocyticleukaemia. British Journal of Haematology, 141, 567-575. http://dx.doi.org/10.1111/j.1365-2141.2008.07104.x
- Yoshida, N., Yagasaki, H., Xu, Y.Y., et al. (2009) Correlation of clinical features with the mutational status of GMCSF signaling pathway-related genes in juvenile myelomonocytic leukemia. Pediatric Research, 65, 334-340. http://dx.doi.org/10.1203/PDR.0b013e3181961d2a
- Gualtieri, R.J., Emanuel, P.D., Zuckerman, K.S., et al. (1989) Granulocyte-macrophage colony-stimulating factor is an endogenous regulator of cell proliferation in juvenile chronic myelogenous leukemia. Blood, 74, 2360- 2367.
- Scharnhorst, V., van der Eb, A.J. and Jochenmsen, A.G. (2001) WT1 proteins: Functions in growth and differentiation. Gene, 273, 141-161. http://dx.doi.org/10.1016/S0378-1119(01)00593-5
- Inoue, K., Sugiyama, H., Ogawa, H., et al. (1994) WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood, 84, 3071-3079.
- Chan, R.J., Cooper, T., Kratz, C.P., Weiss, B. and Loh, M.L. (2009) Juvenile myelomonocytic leukemia: A report from the 2nd International JMML symposium. Leukemia Research, 33, 355-362. http://dx.doi.org/10.1016/j.leukres.2008.08.022
- Jacobsohn, D.A., Tse, W.T., Cheleff, S., et al. (2009) High WT1 gene expression before hematopoietic stem cell transplant in children with acute myeloid leukemia predicts poor event-free survival. British Journal of Haematology, 146, 669-674. http://dx.doi.org/10.1111/j.1365-2141.2009.07770.x
- Cilloni, D., Gottardi, E., Messa, F., et al. (2003) Significant correlation between the degree if WT1 expression and the international prognostic scoring system score in patients with myelodysplastic syndromes. Journal of Clinical Oncology, 21, 1988-1995. http://dx.doi.org/10.1200/JCO.2003.10.503
- Kletzel, M., Olzewski, M., Huang, W. and Chou, P.M. (2002) Utility of WT1 as a reliable tool for the detection of minimal residual disease in children with leukemia. Pediatric and Developmental Pathology, 5, 269-275. http://dx.doi.org/10.1007/s10024-001-0208-x
- Sugiyama, H. (1998) Wilms’ tumor gene (WT1) as a new marker for the detection of minimal residual disease in leukemia. Leuk Lymphoma, 30, 55-61.
- Ostergaard, M., Olesen, L.H., Hasle, H., Kjeldsen, E. and Hokland, P. (2004) WT1 gene expression: An excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukemia patients-results from a singlecenter study. British Journal of Haematology, 125, 590- 600. http://dx.doi.org/10.1111/j.1365-2141.2004.04952.x
- Bender, J.G., Unverzagt, K., Walker, D.E., et al. (1994) Phenotypic analysis and characterization of CD34+ cells from normal human bone marrow, cord blood, peripheral blood, and mobilized peripheral blood from patients undergoing autologous stem cell transplantation. Clinical Immunology and Immunopathology, 70, 10-18. http://dx.doi.org/10.1006/clin.1994.1003
- D’Arena, G., Cascavilla, N., Musto, P., et al. (1996) Flow cytometric characterization of CD34+ hematopoietic progenitor cells in mobilized peripheral blood and bone marrow of cancer patients. Haematologica, 81, 216-223.
- Hoffman, R., Tong, J., Brandt, J., et al. (1993) The in vitro and in vivo effects of stem cell factor on human hematopoiesis. Stem Cells, 2, 76-82.
- Hosen, N., Sonoda, Y., Oji, Y., et al. (2002) Very low frequencies of human normal CD34+ haematopoietic progenitor cells express the wilms’ tumour gene WT1 at levels similar to those in leukaemia cells. British Journal of Haematology, 116, 409-420. http://dx.doi.org/10.1046/j.1365-2141.2002.03261.x
- Campana, D. and Coustan-Smith, E. (2004) Minimal residual disease studies by flow cytometry in acute leukemia. Acta Haematologica, 112, 8-15. http://dx.doi.org/10.1159/000077554
- Olszewski, M., Huang, W., Chou, P.M., Duerst, R. and Kletzel, M. (2005) Wilms’ tumor 1 (WT1) gene in hematopoiesis: A surrogate marker of cell proliferation as a possible mechanism of action? Cytotherapy, 7, 57-61.
- Tamaki, H., Ogawa, H., Ohyashiki, J.H., et al. (2005) The Wilms’ tumor is a good marker for diagnosis of disease progression of myelodysplastic syndrome. Leukemia, 29, 343-345.
- Bader, P., Niemeyer, C., Weber, G., et al. (2004) WT1 gene expression: Useful marker for minimal residual disease in childhood myelodysplastic syndromes and juvenile myelomonocytic leukemia? European Journal of Haematology, 73, 25-28. http://dx.doi.org/10.1111/j.1600-0609.2004.00260.x
- Ogawa, H., Tamaki, H., Ikegame, K., et al. (2003) The usefulness of monitoring WT1 gene transcripts for the prediction and management of relapse following allogeneic stem cell transplantation in acute type leukemia. Blood, 101, 5. http://dx.doi.org/10.1182/blood-2002-06-1831
- Qin, Y.Z., Zhu, H.H., Jiang, B. et al. (2009) Expression patterns of WT1 and PRAME in acute myeloid leukemia patients and their usefulness for monitoring minimal residual disease. Leukemia Research, 33, 384-390. http://dx.doi.org/10.1016/j.leukres.2008.08.026
- Phelan, S.A., Lindberg, C. and Call, K.M. (1994) Wilms’ tumor gene, WT1, mRNA, is down-regulated during induction of erythroid and megakaryocytic differentiation of K562 cells. Cell Growth & Differentiation, 5, 677-686.
ABBREVIATIONS
JMML WT1 GM-CSF nPCR
NOTES
*Corresponding author.

