Advances in Bioscience and Biotechnology
Vol.5 No.1(2014), Article ID:41984,8 pages DOI:10.4236/abb.2014.51008
Cost effective filamentous phage based immunization nanoparticles displaying a full-length hepatitis B virus surface antigen
![]()
1Immunogenetics Laboratory, TUBITAK Marmara Research Center, Genetic Engineering and Biotechnology Institute, Gebze, Turkey
2Istanbul Technical University, Molecular Biology and Genetics, Istanbul, Turkey
3Department of Mechanical Engineering, University of Kansas, Lawrence, USA
4Bioengineering Research Center, University of Kansas, Lawrence, USA
Email: berrin.erdag@tubitak.gov.tr, koray.balcioglu@tubitak.gov.tr
Copyright © 2014 Bertan Koray Balcioglu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Bertan Koray Balcioglu et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 5 November 2013; revised 16 December 2013; accepted 3 January 2014
KEYWORDS
Phage Display; Hepatitis B Virus Surface Antigen; Protein Expression; Phage Immunization; Nano Vector System
ABSTRACT
Hepatitis B virus (HBV) is one of the major causes of chronic hepatitis, cirrhosis and liver cancer. In combating HBV infections, HBV diagnosis and vaccination are therefore critical. The hepatitis B virus surface antigen (HBsAg) is a key target molecule in developing vaccines and diagnostic systems. To date, although HBsAg has been expressed in bacteria, yeasts and mammalian cells, there are still limitations in the existing ones, which leave the necessity for searching new HBsAg production methods. In this study, a simple phage display-based method was developed to produce the purified full-length HBsAg molecules for further immunization studies. For this purpose, the HBsAg coding gene was cloned into a pCANTAB5E phagemid vector and expressed on the surface of M13 filamentous phages. The HBsAg-expressing phage nanosystem was then used as immunization agent in BALB/cJ mice. The ELISA results for sera obtained from mice immunized with HBsAg-displaying phage particles revealed an immune response against HBsAg. These results demonstrate the potential use of a full-length antigen to be displayed on phages as cost effective adjuvant-free immunization agents as an alternative to the highly purified and more expensive antigens conjugated with carrier molecules.
1. INTRODUCTION
Hepatitis B virus (HBV) is one of the major causes of chronic hepatitis, cirrhosis and liver cancer [1]. According to the World Health Organization (WHO), the number of infected people in 2009 was greater than 2 billion, 350 million of whom were HBV carriers. Every year, approximately 500,000 to 700,000 people die from HBVrelated diseases. Although the incidence of HBV infection has decreased in developed countries due to the implementation of vaccination programs [2], HBV still remains an important health problem in developing countries.
HBV belongs to the Hepadnaviridae virus family, which infects human hepatocytes. HBV infection results in the release of infectious virions and excessive hepatitis B surface antigens (HBsAg) from host cells to the bloodstream. Therefore, the presence of HBsAg in circulating blood is an indicator of HBV infection. ELISA kits are widely used throughout the world to detect HBsAg and anti-HBsAg antibodies, and thus, HBsAg production is important for the manufacture of ELISA-based HBV diagnostic kits. However, the primary need for HBsAg production is to develop vaccines against HBV. Thus, the development of a cost-effective method of producing HBsAg for vaccine advancement is of great importance.
HBsAg was first purified from human plasma in 1980 [3]. It was then produced in a recombinant yeast system [4], followed by expression in mammalian cells [5]. However, the production and purification processes for HBsAg are expensive and time consuming. Several years ago, a lower-cost method using phage display for the expression of HBsAg for immunization purposes was demonstrated [6]. Tan et al. fused the 111 - 156 amino acid-coding DNA region of HBsAg, including the dominant neutralizing epitope “a”, with T7 phage capsid proteins. T7 phages expressing the 45 amino acid-length partial HBsAg on their surfaces were used as immunization agents in rabbits.
Phage display is a powerful technology in which fusion proteins are displayed on phage surfaces using coat proteins [7-9]. This technology has been widely used for the selection of recombinant antibodies, such as Fab, single chain variable fragments or peptides that are able to bind various target molecules such as growth factor receptors, growth factors, viral antigens and even inorganic materials for diagnosis, tumor targeting and drug discovery [10-14]. Phage display has recently been used in new applications for vaccine development by displaying peptides derived from virus antigens on the phage surface [15-17]. Until now, the full-length HBsAg has not been displayed on the phage surface for use in immunization applications.
Here, we describe the expression of the entire small HBsAg on phage surfaces as a pIII fusion protein. We further use these recombinant phage nanoparticles as immunization agents in immunocompetant BALB/cJ mouse strain.
2. MATERIAL AND METHODS
2.1. Materials
The plasmid construct containing the HBsAg coding gene was kindly provided by Assoc. Prof. Mehmet Yapar of the Virology Department from Gulhane Military Medical Faculty, Gulhane Military Medical Faculty (GATA) [18]. Escherichia coli strain TG1 (Δ(lac-pro), supE, thi, hsdD5/Fʹ,traD36, proAB, LacIq, lac ZΔM15) was used as a bacterial host. The phagemid vector pCANTAB5E (Pharmacia, Stockholm, Sweden) was used for the phage display (Figure 1). BALB/cJ mice were obtained from the Animal Genetics and Reproduction Biology Laboratory of the TUBITAK Marmara Research Center, Genetic Engineering and Biotechnology Institute, Turkey. Animal studies were approved by the Ethics Committee of the Genetic Engineering and Biotechnology Institute, TUBITAK Marmara Research Center.
2.2. HBsAg-Encoding Gene Cloning into a Phagemid Vector
The HBsAg-encoding gene was amplified by PCR using a sense strand primer (5’-ACTCGCGGCCCAGCCGGC
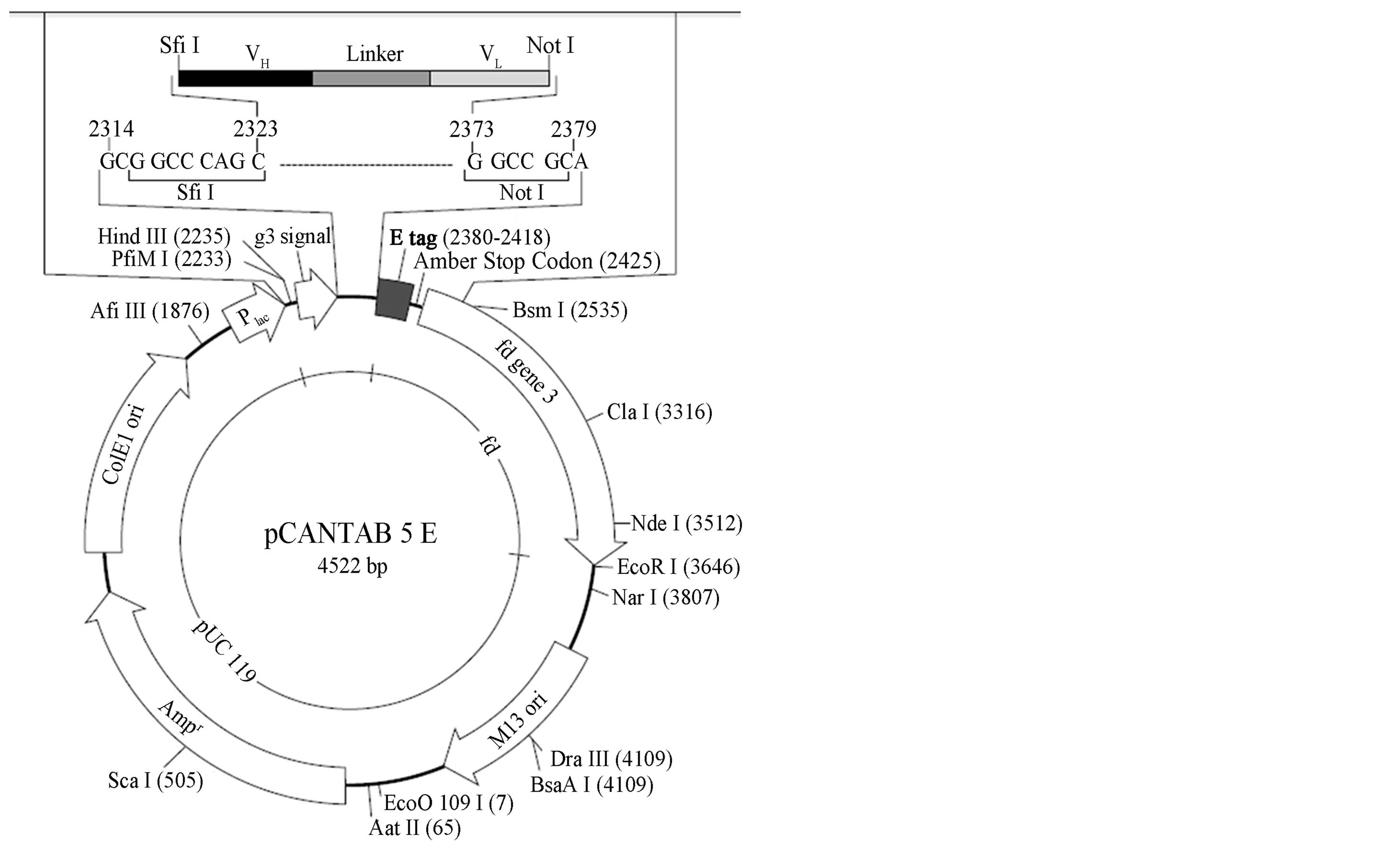
Figure 1. Map of pCANTAB5E phagemid vector. “The Recombinant Phage Antibody System (RPAS) Expression Module/Recombinant Phage Antibody System protocol handbook (Pharmacia, Stockholm, Sweden).
CATGGAGAACATCACATCAGG-3’) including a SfiI restriction site (underlined) and an anti-sense strand primer (5’-CATTCTGCGGCCGCTTTGTTTTGTTAGGGT TTAA-3’) including a NotI restriction site (underlined). The PCR reaction was performed as follows: 1 min at 94˚C, 2 min at 55˚C and 2 min at 72˚C for a total of 30 cycles. The PCR-amplified fragment and the pCANTAB5E vector were first digested with SfiI and then with NotI. Agarose gel-purified SfiI/NotI digestion products were ligated to each other and then transformed into E. coli TG1 (Amersham Pharmacia Biotech, Buckinghamshire, England) by calcium chloride transformation [19]. The transformed cells were heated at 95˚C for 5 min to release the plasmids into the medium. The HBsAg gene insert in the plasmids was controlled by colony PCR using a heat-treated bacterial suspension as a template. Plasmids from the positive clones were purified and then sequenced with the GenomeLab DTCS Quick Start Kit (Beckman Coulter, Brea, USA) using a forward primer (TATGACCATGATTACGCCAAG) and a reverse primer (TTTTGTCGTCTTTCCAGACGTT). The raw sequencing data were analyzed using a CEQ 8800 Genetic Analysis System (Beckmann Coulter), and the DNA sequences were then uploaded to the SDCD Biology Workbench Internet website (http://workbench.sdsc.edu), where multiple alignments, database searches and protein translations were conducted.
2.3. HBsAg Expression on the Phage Surface
A single positive transformed colony was selected and grown overnight at 37˚C in 2XTY medium (10 g BactoTryptone, 5 g yeast extract and 10 g NaCl per liter) containing 100 μg/ml ampicillin and 2% glucose for phage rescue. The next day, the overnight culture was inoculated into fresh medium containing 100 μg/ml ampicillin and 2% glucose. When the absorbance at 600 nm reached 0.5, 1010 pfu of M13KO7 helper phage (New England Biolabs, Beverly, MA) were added to the culture and incubated for 45 min at 37˚C without shaking and 45 min at 37˚C with shaking. The culture was centrifuged, and the cells were resuspended in 2XTY medium containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. The culture was incubated overnight at 30˚C with shaking. The next day, phage particles were precipitated using polyethylene glycol-6000 (1 M PEG 6000 and 2.5 M NaCl) and then centrifuged for 40 min at 25000 × g. The phage particle pellet was resuspended in 1 ml of phosphatebuffered saline (PBS; 3.2 mM Na2HPO4 × 2H2O, 1.4 mM KH2PO4, 2.7 mM KCl and 137 mM NaCl) and then filter sterilized using a 0.45-μm Millex-HV membrane (Millipore, Billerica, MA).
2.4. ELISA for HBsAg-Displaying Phage Particles
Two different ELISA methods have been used to detect the HBsAg displayed on phage particles. Each assay was conducted in triplicate.
2.4.1. First ELISA Method
Phage particles that did (recombinant phage) or did not (wild-type phage) display HBsAg were used to coat each ELISA plate well at 1010 phage particles per well overnight. The next day, the wells were washed with TPBS (PBS with 0.1% Tween-20) and then blocked with TPBS containing 1% BSA. Polyclonal rabbit anti-HBsAg antibody (1/200) was used as a primary antibody, and goat anti-rabbit alkaline phosphatase (Thermo Scientific, Rockford, Illinois, USA) conjugate (1/1000) was used as a secondary antibody. A para-nitrophenylphosphate substrate solution was used as a chromogen. The absorbance was measured at 405 nm using an ELISA reader (BioRad).
2.4.2. Second ELISA Method
ELISA plate wells were coated overnight with rabbit anti-HBsAg polyclonal antibody (1/200). The next day, the wells were washed with TPBS (PBS with 0.1% Tween-20) and then blocked with TPBS containing 1% BSA. Phage particles (1010) displaying HBsAg or wildtype phage were then added to each well. MouseantiM13 horseradish peroxidase (GE Healthcare, Freiburg, Germany) conjugate (1/1000) was used as a secondary antibody. The absorbance was measured at 410 nm using an ELISA reader (Bio-Rad) after the addition of a 2,2’- azino-bis (3-ethylbenzothiazoline-6-sulphonic acid (ABTS) substrate solution.
2.5. Mouse Immunization Assay
Immunization assays were conducted with 8-week-old male immunocompetant BALB/cJ mice. Six different mice groups containing 3 mice each were subjected to different injections without any adjuvant. The first 3 groups received 300 μl of solutions with different concentrations of phage displaying HBsAg (1010, 1011 and 1012 phage particles diluted in PBS). The fourth and fifth groups received 1011 or 1012 phage particles without HBsAg in 300 μl of PBS. The mice in group 6 received 300 μl of PBS as a control. All immunizations were performed in triplicate at 3-week intervals. Intradermal injections were conducted using 50 μl under each front leg and 100 μl under each back leg pit. Blood samples were collected from the tail of each mouse before each injection; the last bleed was performed 4 d after the last injection. Blood samples were centrifuged, and the supernatants were stored for ELISA assays.
2.6. ELISA of Mice Blood after Immunization
ELISA plate wells were coated overnight with 200 ng of HBsAg (Ad/Ay, 100 ng each) (Fitzgerald Industries International, North Acton, MA, USA). The next day, the wells were washed with TPBS (PBS with 0.1% Tween- 20) and then blocked with TPBS containing 1% BSA. The mice sera (1/50) were then incubated for 1 h at room temperature. The wells were washed with TPBS, and goat anti-mouse HRP conjugate (Thermo Scientific, Rockford, Illinois, USA) was added (1/1000) and incubated for 1 h. After the addition of ABTS substrate solution, the absorbance was measured at 410 nm with an ELISA reader (Bio-Rad).
2.7. Statistical Analysis
The IBM SPSS Statistics (SPSS), version 17 for Windows was used for statistical analysis. The differences in BALB/cJ mice humoral immune response between groups were analyzed using mixed model ANOVA with Tukey post hoc test. The significant difference between groups and within groups at different injection periods were analyzed using univariate General Linear Model with Tukey post hoc test. p values of <0.05 were considered statistically significant.
3. RESULTS
3.1. Construction of HBsAg-Displaying Phages
The HBsAg gene provided by GATA [18] was amplified by polymerase chain reaction (PCR), and the product was run on a 1.2% agarose gel. A single ~700 bp band was observed as expected (data not shown). The PCR product was then cloned into the pCANTAB5E phagemid vector. The recombinant phagemid vector was then transferred into TG1 E. coli cells. Twenty randomly selected transformed cells were used for colony PCR. Three colonies identified as positive PCR product clones were selected and prepared for dye-terminated DNA sequence analysis. The resulting sequences were translated using the San Diego Supercomputer Center (SDSC) Biology Workbench Internet website. The protein sequences resulting from translation were compared with HBsAg and pIII protein sequences by CLUSTALW multiple sequence alignment. Sequencing showed that HBsAg was properly cloned into the phagemid vector and fused to pIII without any frame shift.
3.2. Phage ELISA
Phages displaying HBsAg were collected by phage rescue using an M13 helper phage. The titer of the phage particles purified by PEG precipitations was estimated. A phage concentration of approximately 4 × 1012 phage particles/ml was obtained after each phage rescue step. Two different ELISA methods were used to verify the HBsAg display on the phage particles (Figure 2). In the first method, ELISA plate wells were coated either with phage particles displaying HBsAg or wild-type phages. Then, a rabbit anti-HBsAg polyclonal antibody was used as a primary antibody, and goat anti-rabbit AP conjugate was used as the secondary antibody. The ELISA results had a 9-fold higher binding signal for HBsAg-displaying phages relative to the wild-type phages (Figure 3). In the second method, polyclonal rabbit anti-HBsAg antibody was used to coat the wells. In this assay, either phage particles displaying HBsAg or wild-type phages were used as the primary antibody, and a mouse anti-M13/ HRP conjugate was used as the secondary antibody. The binding signal for HBsAg-displaying phages was 7.7 times higher than the binding signal for the wild-type phages (Figure 3).
3.3. ELISA of Mice Sera
Immunizations of 8-week-old male BALB/cJ mice were performed by intradermal injections of the phage nanoparticles at different concentrations (1010, 1011 or 1012 phage particles). The immunizations used either HBsAgdisplaying recombinant phage nanoparticles, wild-type phage nanoparticles or PBS.
Three injections were performed at three-week intervals, and mouse blood was collected prior to each injecttion. For determining the immune response developed against HBsAg an ELISA test against anti-HBsAg anti-
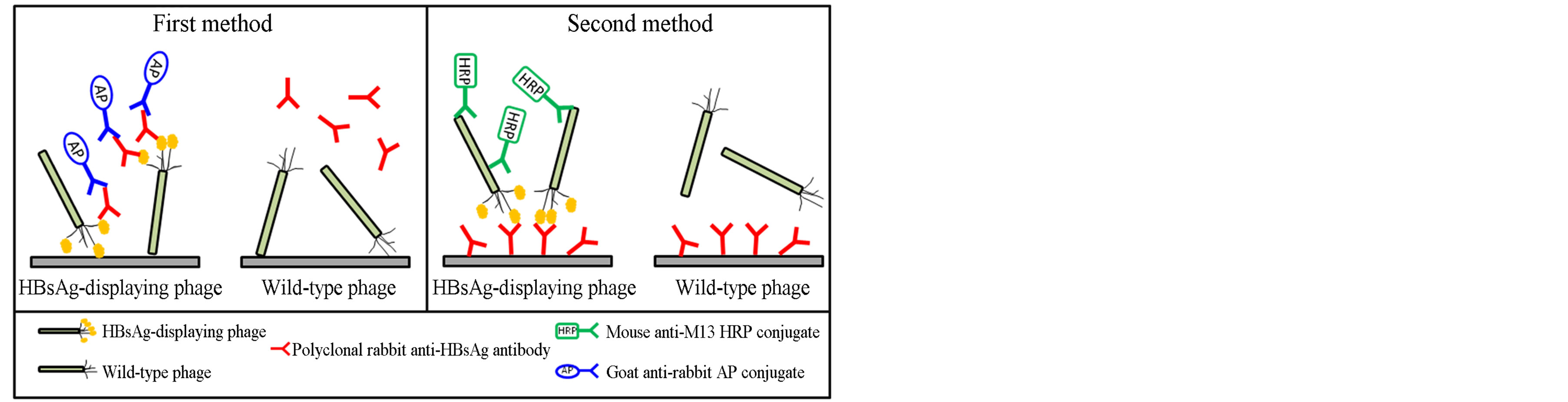
Figure 2. Schematic representation of phage ELISA methods for the detection of HBsAg displayed on M13 phage surfaces. First method: Phage particles with or without (wild-type phage) HBsAg display were used to coat ELISA plate wells. Polyclonal rabbit anti-HBsAg antibody (1/200) was used as a primary antibody, and goat anti-rabbit alkaline phosphatase conjugate was used as a secondary antibody. The absorbance was measured at 405 nm. Second method: ELISA plate wells were coated with rabbit anti-HBsAg polyclonal antibody. Phage particles displaying HBsAg or wild-type phage were then added to each well. Mouse anti-M13 horseradish peroxidase conjugate was used as a secondary antibody. The absorbance was measured at 410 nm.
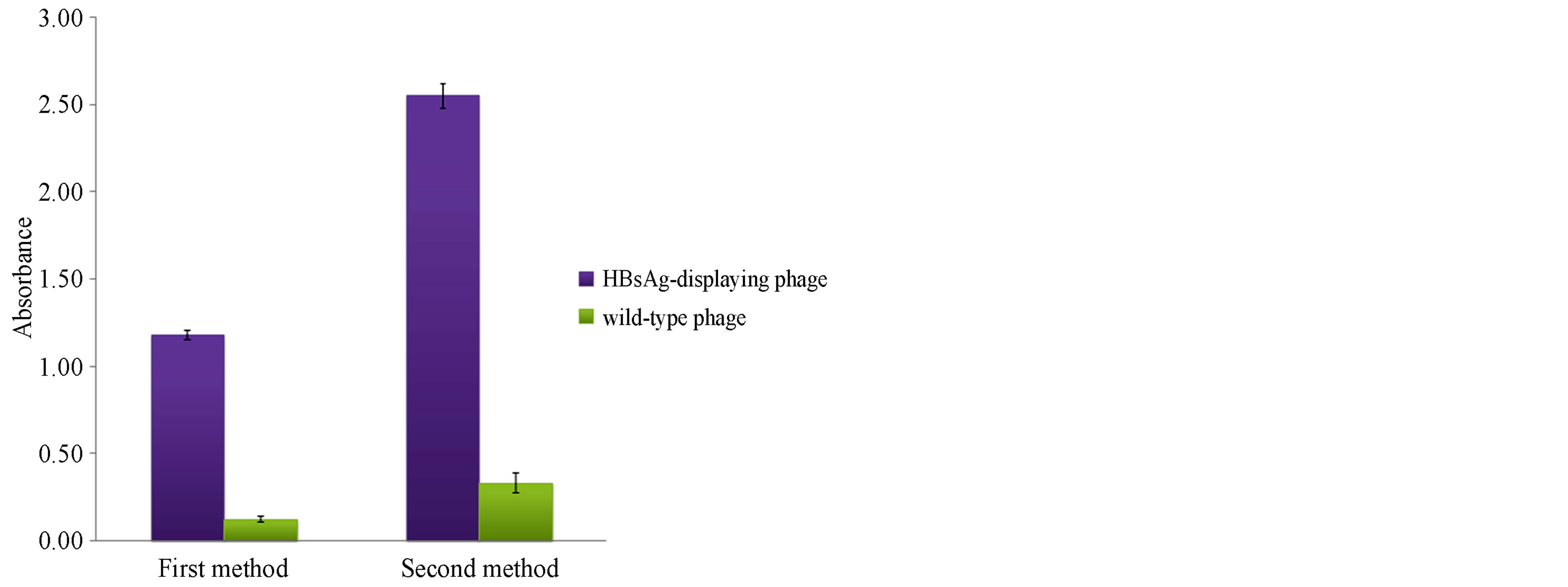
Figure 3. The binding profile of HBsAg displayed on phages with anti-HBsAg polyclonal antibodies. Wavelengths of 405 nm and 410 nm were used to measure the optical absorbance for the first and second ELISA methods, respectively.
bodies in mice sera was conducted. The injections gave no signal for mice immunized with 1010 and 1011 phage particles (data not shown). However, a three-fold increase in absorbance was observed in the sera of mice immunized with 1012 recombinant phage nanoparticles displaying HBsAg. A mixed model ANOVA with Tukey post hoc test was conducted to assess the immune response difference between different treatment groups (phage particles displaying HBsAg, phage particles without HBsAg and PBS) across different injection periods (Before injection, first injection, second injection and third injection). There was a statistically significant interaction between the treatment groups and the injections on the immune response, F(6,18) = 80.208, p < 0.0005, partial eta squared = 0.964. Then univariate General Linear Model with Tukey post hoc analysis were conducted to determine the differences of immune responses between groups at each level of injection periods. There immune response was statistically lower in the PBS group (M = 0.39, SE = 0.017, p < 0.0005) and the phage particles without HBsAg group (M = 0.38, SE = 0.017, p < 0.0005) compared to the phage particles dispaying HBsAg group (Figure 4).
4. DISCUSSION
The manufacture of proteins is the critical component in diverse fields such as agriculture, biotechnology and health care. Many different processes and methods have been developed to produce and purify proteins of economic importance, although these protocols can be difficult, time consuming and expensive [20]. Natural proteins can be purified from their host cells, but the resulting pure protein yields can be very low. Therefore, recombinant protein technology has revolutionized the target protein expression protocols.
HBsAg is a key antigen used either for HBV infection diagnosis or HBV vaccines. HBsAg has been produced in many different mammalian, microbial and plant cell systems [21-23]. Although large amounts of HBsAg are produced by these systems, the procedures for purifying HBsAg remain costly and time consuming.
Phage display technology is a powerful approach for the expression of proteins, peptides, enzymes and antibodies on phage surfaces [9,24,25]. However, phage display allows for the expression of the target protein only as a fusion with the cell surface or coat proteins. Even if proteins expressed on the phage surface are not “pure”, they are easy to produce and purify using a simple PEG precipitation step followed by centrifugation. These properties make them suitable for immunization purposes because phages are antigenic and immunogenic and are capable of inducing antigen-specific antibodies by recruiting helper T cells without the requirement of an adjuvant [26,27]. As a result, phages have been used to elicit immune responses in animal models [28,29] with the intent of using phage nanoparticles in vaccine development. Several studies on HIV-1 [30], hepatitis C virus (HCV) [31], Alzheimer’s disease [32] and cancer [17] have demonstrated the potential use of phage-displayed antigens in the development of vaccines in mice and rabbits. The first phage display-based vaccine system for large animals was recently developed against Taenia solium pig cysticercosis [33].
In most phage display immunization studies, antigenic peptides were displayed on the PVIII major coat proteins [34,35]. This method is used mainly because of the high copy number of PVIII coat proteins (~2500 copies) compared to the PIII minor coat protein number, which varies
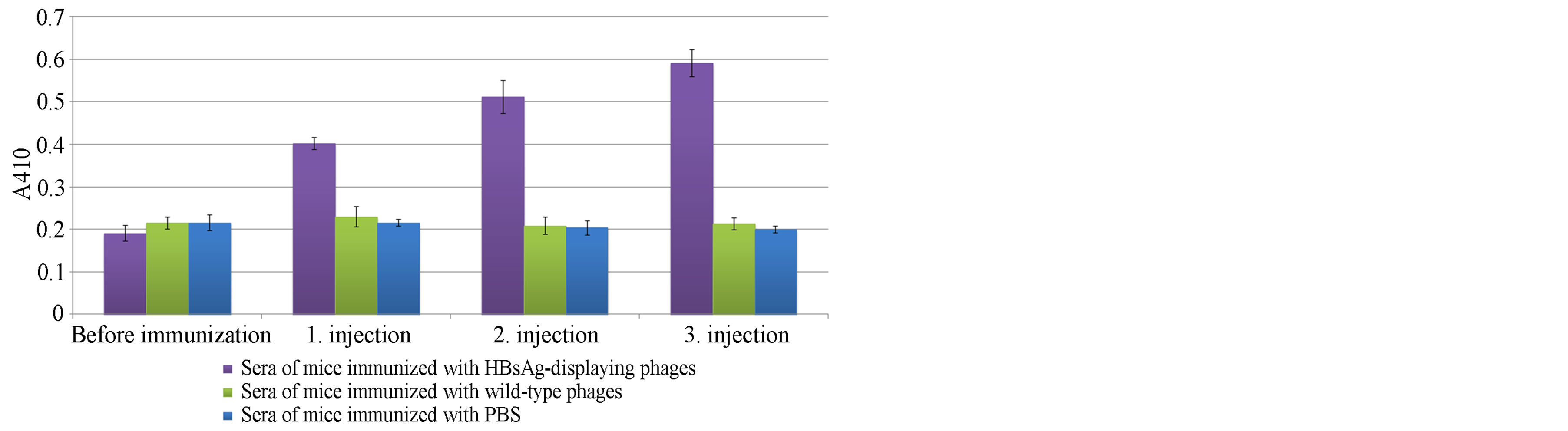
Figure 4. ELISA results for mice immunized with 1012 phage nanoparticles before the first injection and after each injections. ELISA plate wells were coated with 200 ng of HBsAg, and the wells were then treated with 1/50 diluted mouse serum. Antimouse HRP conjugate was used as the detection antibody. The absorbance was measured at 410 nm.
from three to five copies [36]. Prior to this present study, the HBsAg immunogenic epitope (HBsAg111-156) or the hepatitis virus B pre-S1 region (47 and 49 amino acids regions of PRES1) has been displayed on T7 bacteriophage nanoparticles [6,15] and used for immunization and vaccination purposes. In these studies, T7 bacteriophage nanoparticles were used because the displayed peptide size is too large to use on the PVIII coat proteins of filamentous phages. Another reason is the high copy number of 10B capsid proteins (415 copies). The expression of a high peptide antigen copy number is an important criterion for vaccine application because a substantial immune response can be obtained, although this response is generated against a single epitope [37]. Certain epitopes are known to be common to different antigens, and thus, a single epitope vaccination can cause unexpected immune responses against antigens with the same epitope. Therefore, immunizations containing the entire antigen generate a more specific response against the target [38,39]. Recently, a full-length HBV core antigen (HBcAg) was displayed on the phage surface as a pIII fusion [40]. In addition, Ozdemir-Bahadir and colleagues successfully obtained anti-HBcAg monoclonal antibodies from BALB/c mice immunized with HBcAg-displaying phages.
The goal of this present study was to express the full-length small HBsAg on the surface of an M13 filamentous phage as a PIII fusion protein. The gene coding the full-length HBsAg was cloned into a pCANTAB5E phagemid vector for this purpose. After the gene insertion was confirmed by DNA sequence analysis, infective phages displaying HBsAg were obtained. Small HBsAg expression was confirmed by two different ELISA methods, as illustrated in Figure 2. In both methods, a binding signal was expected if HBsAg was displayed on phage surfaces, and if no HBsAg was displayed (wildtype phages), then no signal was expected. The ELISA results from both methods clearly showed a specific binding between phages displaying HBsAg and polyclonal anti-HBsAg antibodies. In contrast, no binding for the wild-type phages to the polyclonal anti-HBsAg antibodies was detected, as expected. Therefore, the expression of HBsAg on the phage surfaces was confirmed.
The HBsAg-displaying phage nanoparticles were then used for immunization assays. The results showed that injections of 1010 and 1011 phage nanoparticles were not sufficient to generate an immune response against HBsAg but were sufficient for generating an immune response against an M13 phage (data not shown). A significant (p < 0.05) anti-HBsAg immune response was observed in mice immunized with 1012 recombinant phage nanoparticles after the third immunization. Therefore, our results demonstrated that at least 1012 recombinant phage nanoparticles are necessary to generate an immune response against HBsAg.
The present results support the concept that M13 phages are good antigen carriers built upon the inexpensive and easy phage display method to express HBsAg for immunization. Furtherour results demonstrate that fulllength HBV small surface antigen displayed on M13 phage minor pIII coat proteins can activate an anti-HBsAg immune response in BALB/cJ mice, even at a very low pIII copy number.
5. CONCLUSION
To date, phage nanoparticles have been used as potential vaccine delivery vehicles, and several immunization studies against HBsAg have been conducted using phage display. However, these studies used short regions of antigens displayed on M13 PVIII major coat proteins. This present study describes the first successful expression of the small HBsAg on an M13 filamentous phage surface as a pIII fusion protein and the use of these recombinant phage nanoparticles as an immunization agent without the need of any additional adjuvant.
ACKNOWLEDGEMENTS
This work was partly supported by a grant from TUBITAK (TUBITAK KAMAG 1007 – Project no. 105G056). We are grateful to Aydin Bahar and Sakir Sekmen for their excellent technical assistance.
REFERENCES
- Francois, G., Hallauer, J. and Van Damme, P. (2002) Hepatitis B vaccination: How to reach risk groups. Vaccine, 21, 1-4.
- Rantala, M. and van de Laar, M.J. (2008) Surveillance and epidemiology of hepatitis B and C in Europe—A review. Euro Surveill, 13.
- Szmuness, W., Stevens, C.E., Harley, E.J., Zang, E.A., Oleszko, W.R., William, D.C., Sadovsky, R., Morrison, J.M. and Kellner, A. (1980) Hepatitis B vaccine: Demonstration of efficacy in a controlled clinical trial in. The New England Journal of Medicine, 303, 833-841. http://dx.doi.org/10.1056/NEJM198010093031501
- Valenzuela, P., Medina, A. and Rutter, W. (1982) Synthesis and assembly of Hepatitis-B virus surface-antigen particles in yeast. Nature, 298, 347-350. http://dx.doi.org/10.1038/298347a0
- Qin, S., Tang, H., Zhao, L., He, F., Lin, Y., Liu, L. and He, X. (2003) Cloning of HBsAg-encoded genes in different vectors and their expression in eukaryotic cells. World Journal of Gastroenterology, 9, 1111-1113.
- Tan, G., Yusoff, K., Seow, H. and Tan, W. (2005) Antigenicity and immunogenicity of the immunodominant region of hepatitis B surface antigen displayed on bacteriophage T7. Journal of Medical Virology, 77, 475-480. http://dx.doi.org/10.1002/jmv.20479
- Smith, G. (1985) Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science, 228, 1315-1317. http://dx.doi.org/10.1126/science.4001944
- Smith, G. and Petrenko, V. (1997) Phage display. Chemical Reviews, 97, 391-410. http://dx.doi.org/10.1021/cr960065d
- Vidova, B., Godany, A. and Sturdik, E. (2008) Phage display—A tool for detection and Prevention against pathogens. Nova Biotechnologica, 8, 23-33.
- Erdag, B., Balcioglu, K., Kumbasar, A., Celikbicak, O., Zeder-Lutz, G., Altschuh, D., Salih, B. and Baysal, K. (2007) Novel short peptides isolated from phage display library inhibit vascular endothelial growth factor activity. Molecular Biotechnology, 35, 51-63. http://dx.doi.org/10.1385/MB:35:1:51
- Prachasuphap, A., University, K., Kittigul, C., University, K., Sunthoranandh, P., University, K., Dhepakson, P., Center, M.B., Buddhirakkul, N., Center, M.B., Balachandra, K. and Medical Biotechnology Center, M.o.P.H. (2006) Construction of recombinant monoclonal antibody against hepatitis b surface antigen by phage display. Animal Cell Technology: Basic & Applied Aspects, 14, 227- 232. http://dx.doi.org/10.1007/1-4020-4457-7_31
- Erdag, B., Balcioglu, B., Bahadir, A., Serhatli, M., Kacar, O., Bahar, A., Seker, U., Akgun, E., Ozkan, A., Kilic, T., Tamerler, C. and Baysal, K. (2011) Identification of novel neutralizing single-chain antibodies against vascular endothelial growth factor receptor 2. Biotechnology and Applied Biochemistry, 58, 412-422. http://dx.doi.org/10.1002/bab.61
- Tamerler, C., Khatayevich, D., Gungormus, M., Kacar, T., Oren, E.E., Hnilova, M. and Sarikaya, M. (2010) Molecular biomimetics: GEPI-based biological routes to technology. Biopolymers, 94, 78-94. http://dx.doi.org/10.1002/bip.21368
- Bayrovic, K., Erdag, B., Atalay, O.E. and Cirakoglu, B. (2001) Full resistance to tobacco mosaic virus infection conferred by the transgenic expression of a recombinant antibody in tobacco. Biotechnology & Biotechnology Equipment, 15, 21-27.
- Tang, K., Yusoff, K. and Tan, W. (2009) Display of Hepatitis B Virus PreS1 peptide on bacteriophage T7 and its potential in gene delivery into HepG2 cells. Journal of Virological Methods, 159, 194-199. http://dx.doi.org/10.1016/j.jviromet.2009.03.015
- Solomon, B. (2005) Generation of anti-beta-amyloid antibodies via phage display technology towards Alzheimer’s disease vaccination. Vaccine, 23, 2327-2330. http://dx.doi.org/10.1016/j.vaccine.2005.01.034
- Fang, J., Wang, G., Yang, Q., Song, J., Wang, Y. and Wang, L. (2005) The potential of phage display virions expressing malignant tumor specific antigen MAGE-A1 epitope in murine model. Vaccine, 23, 4860-4866. http://dx.doi.org/10.1016/j.vaccine.2005.05.024
- Kubar, A., Yapar, M., Ozyurt, M., Haznedaroglu, T. and Gun, H. (1988) Cloning of hepatitis B virus surface gene region to Escherichia coli. Flora, 6, 108-113.
- Sambrook, J., Fritsch, E.E. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Lab. Press, New York.
- Sorensen, H. and Mortensen, K. (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microbial Cell Factories, 4. http://dx.doi.org/10.1016/j.vaccine.2005.05.024
- Lunin, V.G., Gol’dberg, E.Z., Grigor’ev, V.G., Mikhailov, M.M., Khudiakov, I.E., Skripkin, E.A., Smirnov, V.D., Naroditsky, B.S., Ketiladze, E.S. and Tikhonenko, T.I. (1983) Cloning and ex-pression of gene coding the hepatitis B virus surface antigen (HBsag) in Escherichia coli. Doklady Akademii Nauk Sssr, 268, 496-498.
- Guan, Z., Guo, B., Huo, Y., Guan, Z. and Wei, Y. (2010) Overview of expression of hepatitis B surface antigen in transgenic plants. Vaccine, 28, 7351-7362. http://dx.doi.org/10.1016/j.vaccine.2010.08.100
- Vianna, C.O., da Silva e Mouta Junior, S., de Oliveira da Silva, G., da Silva Freire, M. and de Moraes, M.T. (2003) Screening of CHO cell clones expressing histidine-tagged major S hepatitis B surface protein using a semi-quantitative PCR protocol. Journal of Virological Methods, 114, 171-174. http://dx.doi.org/10.1016/j.jviromet.2003.08.007
- Barbas, C.F.I., Burton, D.R., Scott, J.K. and Silverman, G.J. (2001) Phage display: A laboratory manual. Cold Spring Harbor Lab. Press, New York.
- Rakonjac, J., Bennett, N., Spagnuolo, J., Gagic, D. and Russel, M. (2011) Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Current Issues in Molecular Biology, 13, 51-75.
- Irving, M.B., Pan, O. and Scott, J.K. (2001) Randompeptide libraries and antigen fragment libraries for epitope mapping and the development of vaccines and diagnostics. Current Opinion in Chemical Biology, 5, 314- 324. http://dx.doi.org/10.1016/S1367-5931(00)00208-8
- Willis, A., Perham, R. and Wraith, D. (1993) Immunological properties of foreign peptides in multiple display on a filamentous bacteriophage. Gene, 128, 79-83. http://dx.doi.org/10.1016/0378-1119(93)90156-W
- Molenaar, T., Michon, I., de Haas, S., van Berkel, T., Kuiper, J. and Biessen, E. (2002) Uptake and processing of modified bacteriophage M13 in mice: Implications for phage display. Virology, 293, 182-191. http://dx.doi.org/10.1006/viro.2001.1254
- Wang, L. and Yu, M. (2004) Epitope identification and discovery using phage display libraries: Applications in vaccine development and diagnostics. Current Drug Targets, 5, 1-15. http://dx.doi.org/10.2174/1389450043490668
- Scala, G., Chen, X., Liu, W., Telles, J.N., Cohen, O.J., Vaccarezza, M., Igarashi, T. and Fauci, A.S. (1999) Selection of HIV-specific immunogenic epitopes by screening random peptide. Journal of Immunology, 162, 6155- 6161.
- Puntoriero, G., Meola, A., Lahm, A., Zucchelli, S., Ercole, B.B., Tafi, R., Pezzanera, M., Mondelli, M.U., Cortese, R., Tramontano, A., Galfre, G. and Nicosia, A. (1998) Towards a solution for hepatitis C virus hypervariability: mimotopes of the. The EMBO Journal, 17, 3521-3533. http://dx.doi.org/10.1093/emboj/17.13.3521
- Frenkel, D., Katz, O. and Solomon, B. (2000) Immunization against Alzheimer’s beta-amyloid plaques via EFRH phage. Proceedings of the National Academy of Sciences of the United Stated of America, 97, 11455-11459. http://dx.doi.org/10.1073/pnas.97.21.11455
- Manoutcharian, K., Diaz-Orea, A., Gevorkian, G., Fragoso, G., Acero, G., Gonzalez, E., de Aluja, A., Villalobos, N., Gomez-Conde, E. and Sciutto, E. (2004) Recombinant bacteriophage-based multiepitope vaccine against Taenia solium pig cysticercosis. Veterinary Immunology and Immunopathology, 99, 11-24. http://dx.doi.org/10.1016/j.vetimm.2003.12.009
- Solomon, B. (2007) Active immunization against Alzheimer’s beta-amyloid peptide using phage display technology. Vaccine, 25, 3053-3056. http://dx.doi.org/10.1016/j.vaccine.2007.01.069
- Morales, J., Martinez, J., Manoutcharian, K., Hernandez, M., Fleury, A., Gevorkian, G., Acero, G., Blancas, A., Toledo, A., Cervantes, J., Maza, V., Quet, F., Bonnabau, H., de Aluja, A., Fragoso, G., Larralde, C. and Sciutto, E. (2008) Inexpensive anti-cysticercosis vaccine: S3Pvac expressed in heat inactivated M13 filamentous phage proves effective against naturally acquired Taenia solium porcine cysticercosis. Vaccine, 26, 2899-2905. http://dx.doi.org/10.1016/j.vaccine.2008.03.042
- Greenwood, J., Willis, A.E. and Perham, R.N. (1991) Multiple display of foreign peptides on a filamentous bacteriophage. Peptides from Plasmodium falciparum circumsporozoite protein as antigens. Journal of Molecular Biology, 220, 821-827. http://dx.doi.org/10.1016/0022-2836(91)90354-9
- Mirano-Bascos, D., Tary-Lehmann, M. and Landry, S. (2008) Antigen structure influences helper T-cell epitope dominance in the human immune response to HIV envelope glycoprotein gp120. European Journal of Immunology, 38, 1231-1237. http://dx.doi.org/10.1002/eji.200738011
- Chiang, C., Benencia, F. and Coukos, G. (2010) Whole tumor antigen vaccines. Seminars in Immunology, 22, 132-143. http://dx.doi.org/10.1016/j.smim.2010.02.004
- Kolodziej, P. and Young, R. (1991) Epitope tagging and protein surveillance. Methods in Enzymology, 194, 508- 519. http://dx.doi.org/10.1016/0076-6879(91)94038-E
- Ozdemir-Bahadir, A., Balcioglu, B., Uzyol, K., Hatipoglu, I., Sogut, I., Basalp, A. and Erdag, B. (2011) Phage Displayed HBV Core Antigen with Immunogenic Activity. Applied Biochemistry and Biotechnology, 165, 1437-1447. http://dx.doi.org/10.1007/s12010-011-9365-1

