Advances in Bioscience and Biotechnology
Vol.2 No.3(2011), Article ID:5633,6 pages DOI:10.4236/abb.2011.23025
An effective strategy for cloning the mitochondrial citratecarrier: identification, characterization and tissue distribution in silver eel
![]()
1Department of Biological and Environmental Sciences and Technologies, University of Salento, Lecce, Italy;
2Department of Pharmaco-Biology, University of Calabria, Arcavacata di Rende (CS), Italy.
Email: vdolce@unical.it
Received 9 April 2011; revised 24 May 2011; accepted 27 May 2011.
Keywords: Citrate Carrier; Cloning; Metabolite Transport; Mitochondrial Carrier Family; PCR; Surfactant
ABSTRACT
A general and effective strategy to identify cDNA encoding for mitochondrial silver eel citrate carrier (CIC) has been developed. In particular using primers directed towards the highly conserved signature motif of the ortholog citrate cDNA sequences, the full-length silver eel cDNA from liver was obtained. This is 1193 bp in length with 5’ and 3’ untranslated regions of 90 and 149 bp, respectively. The open reading frame encodes a mature protein of 297 amino acids, preceded by a presequence of 20 amino acids. Additionally the mature CIC overexpressed in Escherichia coli and reconstituted into phospholipids vesicles showed the same substrate specificity of the native protein previously characterized in silver eel. The tissue distribution of silver eel CIC mRNAs was investigated and transcripts were detected at high levels in swim bladder whereas a weaker signal was found in brain, gill, intestine and liver.
1. INTRODUCTION
Mitochondrial carriers are a family of nuclear-coded proteins that are localized in the inner mitochondrial membrane [1-3]. Their function is to provide a link between mitochondria and cytosol by facilitating the flux of a variety of solutes through the mitochondrial membrane. The primary sequence of protein belonging to this family is characterized by the presence of three tandemly repeated domains of about 100 amino acids. Each repeat contains 2 hydrophobic stretches, that span the membrane as alpha-helices, and a characteristic signature sequence motif, P-X-D/E-X-X-R/K-X-R/K, at the end of the odd-numbered transmembrane helices [4,5].
The tricarboxylate (or citrate) carrier (CIC) is a transport protein, which belongs to the mitochondrial carrier family [1,6,7]. CIC is responsible for the exchange of a tricarboxylate for either another tricarboxylate, a dicarboxylate (L-malate) or phosphoenolpyruvate across the mitochondrial inner membrane [7-12]. This carrier protein plays a central role in intermediary metabolism because the citrate efflux from mitochondria provides the carbon source and NADPH + H+ for a number of process such as fatty acid synthesis, sterol biosyntheses, insulin secretion and histone acetylation [13-15]. The tricarboxylate carrier of yellow and silver eels has been purified to homogeneity from liver mitochondria and functionally reconstituted into liposomes [12,16]. The transmembrane organization, the role of cysteine residues and the import pathway of this transport protein have been investigated in silver eel liver mitochondria [17-20].
In this paper we report an effective strategy, used to identify a full-length cDNA sequence of a mitochondrial CIC. The obtained cDNA encodes for a mature and functional protein consisting of 297 amino acids preceded by a presequence of 20 amino acids. Furthermore we provide evidences that the recombinant protein expressed in Escherichia coli shows the same substrate transport specificity of the CIC purified from liver silver eel. We also highlight its different mRNA expression in silver eel tissues.
2. MATERIALS AND METHODS
2.1. Sequence Search and Analysis
The National Center of Biotechnology Information (NCBI) (Washington, D. C.) non-redundant EST databases were screened with Rattus norvegicus, Drosophila melanogaster and Danio rerio citrate sequences (NP_ 059003, NP_569856 and BC056787, respectively) using TBLASTN program. The Caenorhabditis elegans genome databases at the Sanger Center (Xinton, United Kingdom) and at the NCBI were screened with the sequence of the silver eel (AJ744769) using BLASTP [21]. Amino acid sequences were aligned with ClustalW (version 1.7).
2.2. Silver Eel CIC cDNA
Total silver eel RNA from liver was extracted with triazol kit (Invitrogen), following the supplier’s protocol. 2.5 µg of total RNA were reverse-transcribed with the Gene Amp RNA PCR Core kit (PerkinElmer Life Sciences) using oligo (dT)16 as primers (final volume, 40 µL). A reverse-transcribed RNA was tailed with a run of A residues at its 5’ end as described previously [22]. A partial cDNA sequence was obtained by PCR reactions using primers (Table 1) designed on the conserved signature sequence motif of Rattus norvegicus (NP_ 059003) and Danio rerio (BC056787). The products, with the expected molecular weight (about 300 base pairs), were cloned in a Topo TA vector (Invitrogen) and sequenced. The obtained partial fragments, of the putative silver eel CIC, were extended in both 5’ and 3’ directions. The PCR reactions were performed as previously described [23].
2.3. Bacteria Overexpression of the Silver Eel CIC
The mature coding region of the silver eel CIC (Accession number AJ744769) was cloned into the pMW7 vector for expression in E. coli. The mature coding sequence of CIC was amplified from eel liver cDNA by PCR using primers corresponding to the nt 151 - 168 and nt 1016 - 1041 of the eel cDNA (Accession number AJ744769) with additional NdeI and HindIII sites. The overproduction of the CIC as inclusion bodies in the cytosol of E. coli was accomplished as described previously [24]. Control cultures carrying empty vector were processed in parallel. Inclusion bodies were isolated, and CIC was purified as described previously [25].
2.4. Reconstitution into Liposomes and Transport Assays
Recombinant protein in sarkosyl was reconstituted into liposomes in the presence of substrates [26]. The external substrate was removed from proteoliposomes by Sephadex G-75 columns pre-equilibrated with 50 mM NaCl and 10 mM PIPES at pH 7.0 [26]. The transport was started by addition of labelled citrate at the indicated concentrations. The carrier-mediated transport was terminated by addition of 30 mM pyridoxal 5’-phosphate and 10 mM bathophenanthroline. In control samples the inhibitors were added at time 0 according to the inhibitor stop method [26]. The assay temperature was 25˚C. All transport measurements were carried out at the same internal and external pH values (PIPES 10mM, pH 7.0). Finally, the external substrate was removed, and the radioactivity in the liposomes was measured [26].
2.5. Expression Analysis by RT-PCR
Total RNA from various tissues of silver eel was extracted and reverse transcribed as described in paragraph 2.2. A 470-bp fragment of the CIC cDNA was amplified by 30 cycles of PCR using oligonucleotides 5’-GAGAG GGCCAACCCGCCA-3’ and 5’-CTCACTCCGTGGA AGAAGCCCC-3’ as forward and reverse primers, respectively. As a control, a 309-bp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) fragment was amplified in the same reaction tube using forward and reverse primers based on the NCBI silver eel EST AAO6 5598.
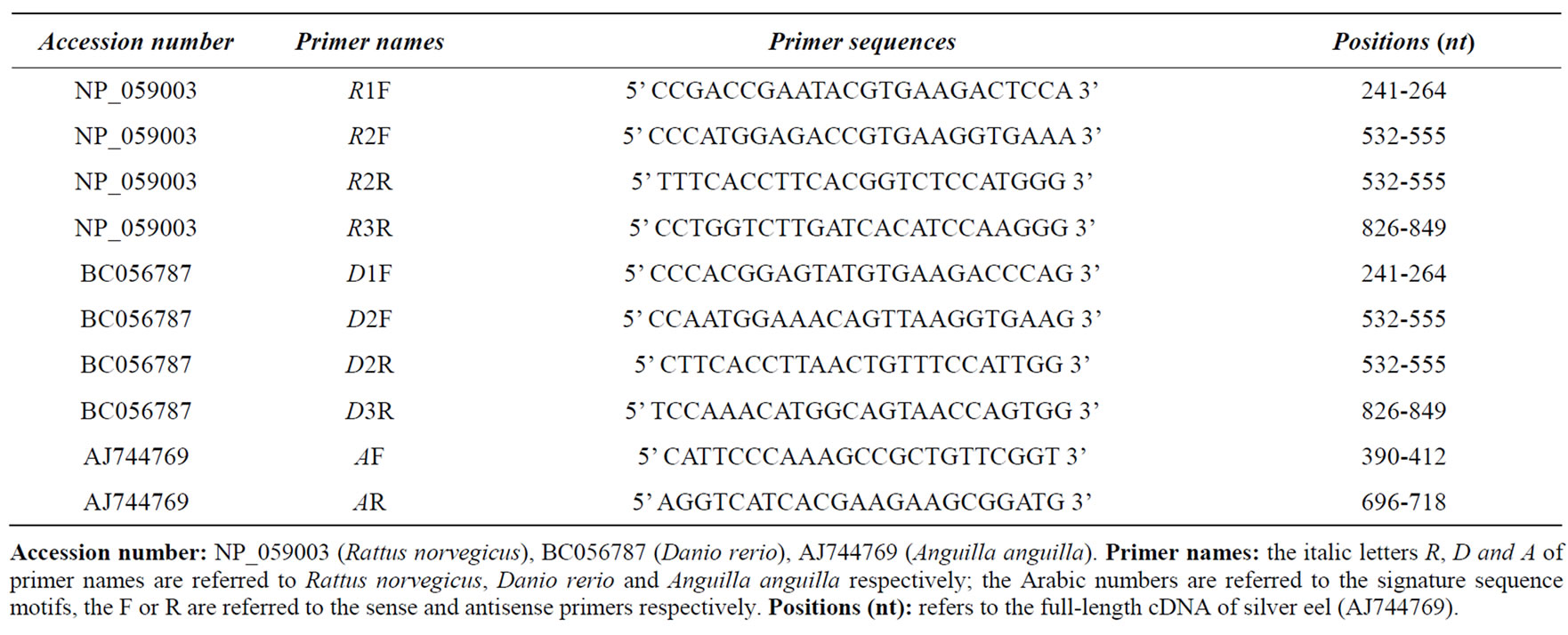
Table 1. Oligonucleotide Primers used in PCR reactions for amplifying the cDNA of CIC silver eel.
3. RESULTS AND DISCUSSION
3.1. Cloning of the Silver Eel Liver Mitochondrial Citrate Carrier
A systematic screening of databases available on-line, with the nucleotide and protein sequences of known citrate carriers from various organisms such as yeast [6], rat [27], ox [28] and fruit fly [21] did not retrieve any result for silver eel ortholog. Therefore in order to identify the CIC of silver eel, PCR amplifications were performed using primers based on the citrate nucleotide sequences of Rattus norvegicus and/or Danio rerio; in particular the primers were chosen in the nucleotide sequences coding for the three tandemly repeated signature sequence motif (P-X-D/E-X-X-R/K-X-R/K) of mitochondrial carrier family [4,5]. Several different reaction PCRs were then carried out on silver eel liver first strand cDNA, using all possible combinations of primer pairs of Rattus norvegicus and Danio rerio (Table 1). Among these, the only two reactions which succeeded were the PCR conducted with the rat forward primer R1F and the Danio rerio reverse primer D2R (see Table 1 and Figure 1) and the PCR performed with the rat forward primer R2F and the rat reverse primer R3R (see Table 1 and Figure 1). These partial sequences were extended at 5’ and 3’ end respectively, using a primer pairs oligodT/AR and AF/oligo-dT (see Table 1 and Figure 1) designed according to the cDNA sequence of silver eel CIC we obtained before.
The full-length silver eel cDNA sequence is 1193 bp in length with 5’ and 3’ untraslated regions of 90 and 149 bp, respectively. The methionine initiation codon (nucleotides 91 - 93) can be assigned based on the following considerations: 1) this is the only methionine in the region immediately preceding the amino-terminal sequence that was deduced when samples of the native purified silver eel liver protein were subjected to the Edman degradation [19]; 2) this is preceded by an in-phase stop codon (nucleotides 70 - 72); 3) there are three in-frame stop codons between this initiation codon and the next in-frame upstream ATG codon. Although a stop codon is clearly evident, no polyadenylation signal was found in the 3’-untranslated region, our inability to extend the sequence further forwards 3’ might be due to the presence of an A-rich region that hybridised with the oligo-dT reverse primer used in 3’ PCR extension.
The open reading frame encodes a mature protein of 297 amino acids, preceded by a presequence of 20 amino acids, and the amino terminus of the mature CIC was assigned on the basis of the direct protein sequencing

Figure 1. Nucleotide and deduced amino acid sequence of a full-length cDNA encoding the precursor of the silver eel liver mitochondrial CIC. The amino acids are numbered from –20 to 297. The vertical arrow above the protein sequence indicates the first amino acid of the mature CIC. An asterisk denotes the stop codon. The boxed sequences indicate the primers employed for the PCR. Horizontal arrows pointing to the left or right indicate that the primers were synthesized as either the sequence shown or its complementary, respectively.
data [19]. The molecular weight of the mature protein calculated from the deduced sequence is 32.158 kDa, a value that is in agreement with the previously estimated molecular weight of 30.400 kDa, obtained by the analysis of the purified protein on SDS-PAGE [29]. A search in databases revealed that the CIC (AJ744769) silver eel is 87, 79, 66, 65 and 36% identical to Rattus norvegicus (NP_059003), Danio rerio (BC056787), Drosophila melanogaster (NP_569856), Caenorhabditis elegans (NP_4 99187) and Saccharomyces cerevisiae (NP_009850) mature proteins, respectively (Figure 2).
As previously found for other mitochondrial transport protein the silver eel CIC shows a tripartite structure made of related sequences of about 100 amino acids in length, each repeat contains 2 hydrophobic stretches, that span the membrane as alpha-helices, and a characteristic signature sequence motif, P-X-D/E-X-X-R/K-XR/K, at the end of the odd-numbered transmembrane helices [4,5]. Furthermore, the hydropathy analysis (data not shown) of the silver eel CIC suggests the presence of two membrane-spanning segments for each repeat, as shown for the ADP/ATP carrier [30].
3.2. Transport Substrate Specificity
The substrate specificity of recombinant silver eel CIC expressed in E. coli and investigated by measuring the uptake of [14C]citrate into reconstituted liposomes preloaded with various substrates. As shown in Table 2, [14C] citrate was efficiently taken up by proteoliposomes containing citrate, cis-aconitate, threo-isocitrate, phosphenolpyruvate and L-malate. To a much lesser extent some succinate was exchanged for external [14C] citrate, whereas a very low exchange was measured for transaconitate, 1, 2, 3 benzenetricarboxylate, malonate and maleate. Virtually no exchange was observed with internal 1, 2, 5 pentanetricarboxylate or substrates of other mitochondrial carriers like fumarate, pyruvate, phosphate, 2-oxoglutarate, glutamate and ADP. All these data clearly demonstrates that the protein identified with this strategy corresponds to the mitochondrial citrate carrier previously characterized from liver silver eel mitochondria [12,16].
3.3. Expression of CIC in Various Tissues
The tissue distribution of mRNAs for the silver eel CIC was studied by RT-PCR performed on total RNA, using primers based on the full-length cDNA previously obtained. A PCR product of the predicted size has been detected at high levels in swim bladder, a weaker but significant signal was found in brain, gill, intestine and liver (Figure 3), whereas no band was visible in heart and skeletal muscle. A control RT-PCR has been carried out using specific primers for GAPDH (Figure 3). The expression pattern of the CIC is substantially in agree-
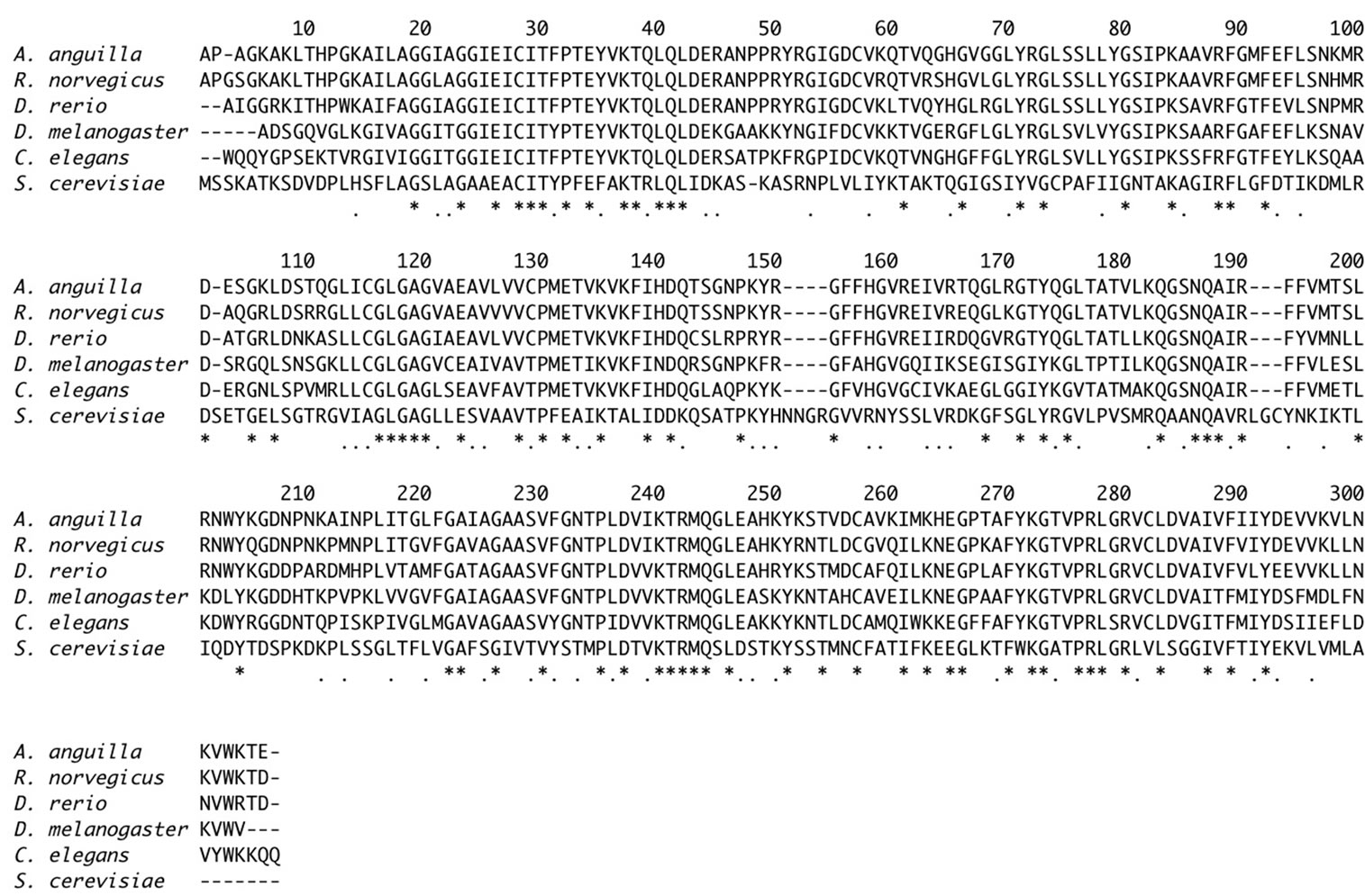
Figure 2. Alignment of A. anguilla, R. norvegicus, D. rerio, D. melanogaster, C. elegans and S. cerevisiae CICs. Asterisks and dots indicate identity and conservation, respectively, of residues in all six sequences.
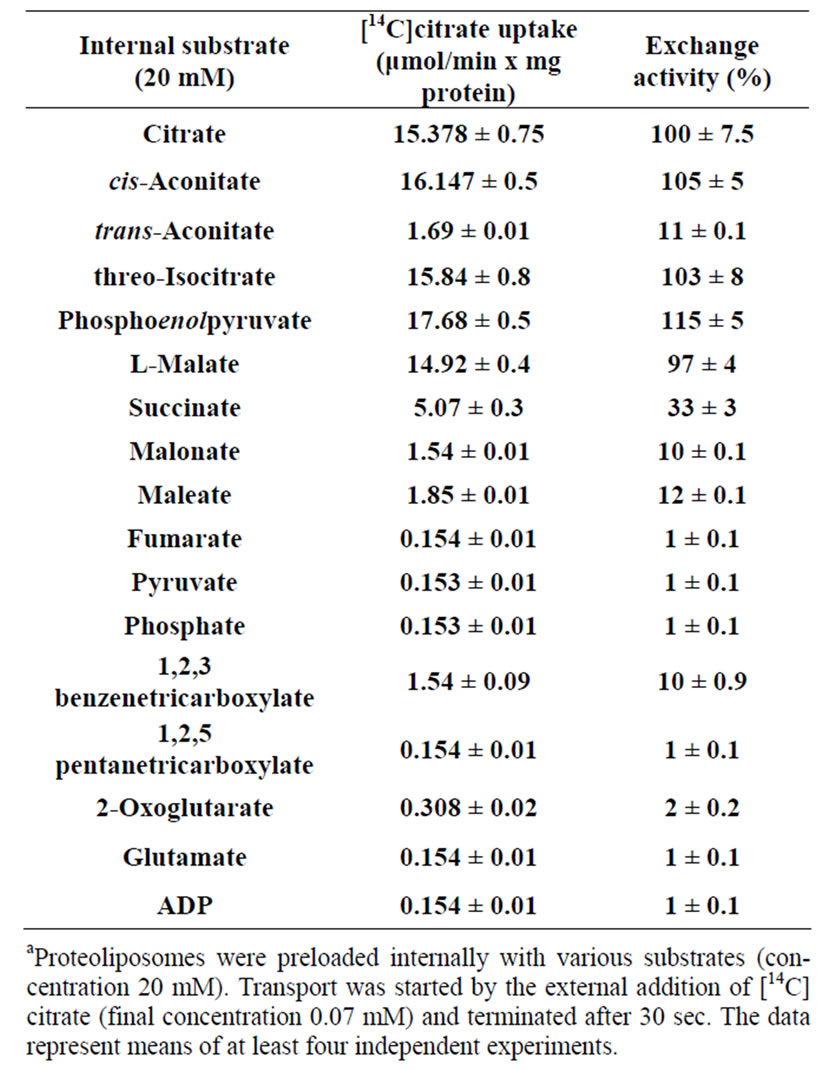
Table 2. Transport substrate specificitya.
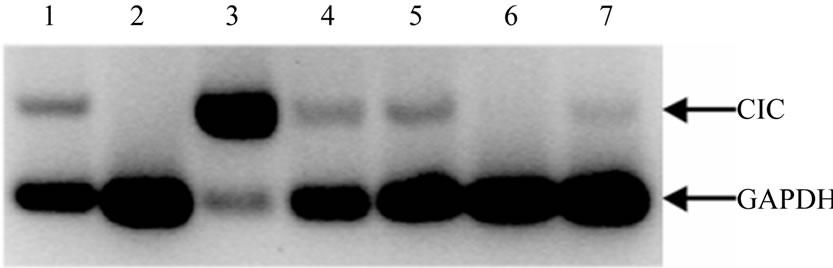
Figure 3. Expression of the CIC in various silver eel tissues. Ethidium bromide staining of the RT-PCR conducted on first strand cDNA from silver eel (lanes 1-7) brain, heart, swim bladder, gill, intestine, skeletal muscle and liver. The relative quantification of silver eel mitochondrial citrate carrier was performed by amplifying CIC and GADPH (internal control) in the same reaction vial.
ment with previously published data [31] and with the metabolic and environmental necessity of silver eel. The huge level of expression observed in swim bladder suggests that this carrier is also necessary to reduce the surface tension and/or to fill this organ [32]. On the other hand the undetectable mRNA in heart and in skeletal muscle may be explained by the very low activity of fatty acid synthesis in these tissues.
REFERENCES
- Palmieri, F. (2004) The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflügers Archiv European Journal of Physiology, 447, 689-709. doi:10.1007/s00424-003-1099-7
- Palmieri, F. (2008) Diseases caused by defects of mitochondrial carriers: A review. Biochimica et Biophysica Acta, 1777, 564-578. doi:10.1016/j.bbabio.2008.03.008
- Palmieri, F. and Pierri, C.L. (2010) Mitochondrial metabolite transport. Essays in Biochemistry, 47, 37-52. doi:10.1042/bse0470037
- Palmieri, F. and Pierri, C.L. (2010) Structure and function of mitochondrial carriers—Role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Letters, 584, 1931-1939. doi:10.1016/j.febslet.2009.10.063
- Palmieri, F., Pierri, C.L., De Grassi, A., Nunes-Nesi, A. and Fernie, A.R. (2010) Evolution, structure and function of mitochondrial carriers: A review with new insights. The Plant Journal, 66, 161-181. doi:10.1111/j.1365-313X.2011.04516.x
- Kaplan, R.S., Mayor, J.A. and Wood, D.O. (1993) The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. The Journal of Biological Chemistry, 268, 13682-13690.
- Bisaccia, F., De Palma, A., Prezioso, G. and Palmieri, F. (1990) Kinetic characterization of the reconstituted tricarboxylate carrier from rat liver mitochondria. Biochimica et Biophysica Acta, 1019, 250-256. doi:10.1016/0005-2728(90)90201-E
- Palmieri, F., Stipani, I., Quagliariello, E. and Klingenberg, M. (1972) Kinetic study of the tricarboxylate carrier in rat liver mitochondria. European Journal of Biochemistry, 26, 587-594. doi:10.1111/j.1432-1033.1972.tb01801.x
- Meijer, A.J. and Van Dam, K. (1974) The metabolic significance of anion transport in mitochondria. Biochimica et Biophysica Acta, 346, 213-244.
- Bisaccia, F., De Palma, A. and Palmieri, F. (1989) Identification and purification of the tricarboxylate carrier from rat liver mitochondria. Biochimica et Biophysica Acta, 977, 171-176. doi:10.1016/S0005-2728(89)80068-4
- Bisaccia, F., De Palma, A., Dierks, T., Kramer, R. and Palmieri, F. (1993) Reaction mechanism of the reconstituted tricarboxylate carrier from rat liver mitochondria. Biochimica et Biophysica Acta, 1142, 139-145. doi:10.1016/0005-2728(93)90095-W
- Zara, V., Iacobazzi, V., Siculella, L., Gnoni, G.V. and Palmieri, F. (1996) Purification and characterization of the tricarboxylate carrier from eel liver mitochondria. Biochemical and Biophysical Research Communications, 223, 508-513. doi:10.1006/bbrc.1996.0925
- Morciano, P., Carrisi, C., Capobianco, L., Mannini, L., Burgio, G., Cestra, G., De Benedetto, G.E., Corona, D.F., Musio, A. and Cenci, G. (2009) A conserved role for the mitochondrial citrate transporter Sea/SLC25A1 in the maintenance of chromosome integrity. Human Molecular Genetics, 18, 4180-4188. doi:10.1093/hmg/ddp370
- Conover, T.E. (1987) Does citrate transport supply both acetyl groups and NADPH for cytoplasmic fatty acid synthesis? Trends in Biochemical Sciences, 12, 88-89. doi:10.1016/0968-0004(87)90042-9
- Joseph, J.W., Jensen, M.V., Ilkayeva, O., Palmieri, F., Alarcon, C., Rhodes, C.J. and Newgard, C.B. (2006) The mitochondrial citrate/isocitrate carrier plays a regulatory role in glucose-stimulated insulin secretion. The Journal of Biological Chemistry, 281, 35624-35632. doi:10.1074/jbc.M602606200
- Zara, V., Palmieri, L., Giudetti, A., Ferramosca, A., Capobianco, L. and Gnoni, G.V. (2000) The mitochondrial tricarboxylate carrier: unexpected increased activity in starved silver eels. Biochemical and Biophysical Research Communications, 276, 893-898. doi:10.1006/bbrc.2000.3579
- Capobianco, L., Ferramosca, A. and Zara, V. (2002) The mitochondrial tricarboxylate carrier of silver eel: Dimeric structure and cytosolic exposure of both Nand C-termini. Journal of Protein Chemistry, 21, 515-521. doi:10.1023/A:1022473504904
- Capobianco, L., Impagnatiello, T., Ferramosca, A. and Zara, V. (2004) The mitochondrial tricarboxylate carrier of silver eel: Chemical modification by sulfhydryl reagents. Journal of Biochemistry and Molecular Biology, 37, 515-521. doi:10.5483/BMBRep.2004.37.5.515
- Zara, V., Dolce, V., Capobianco, L., Ferramosca, A., Papatheodorou, P., Rassow, J. and Palmieri, F. (2007) Biogenesis of eel liver citrate carrier (CIC): Negative charges can substitute for positive charges in the presequence. Journal of Molecular Biology, 365, 958-967. doi:10.1016/j.jmb.2006.10.077
- Capobianco, L., Bisaccia, F., Michel, A., Sluse, F.E. and Palmieri, F. (1995) The Nand C-termini of the tricarboxylate carrier are exposed to the cytoplasmic side of the inner mitochondrial membrane. FEBS Letters, 357, 297-300. doi:10.1016/0014-5793(94)01379-F
- Carrisi, C., Madeo, M., Morciano, P., Dolce, V., Cenci, G., Cappello, A.R., Mazzeo, G., Iacopetta, D. and Capobianco, L. (2008) Identification of the Drosophila melanogaster mitochondrial citrate carrier: bacterial expression, reconstitution, functional characterization and developmental distribution. The Journal of Biochemistry, 144, 389-392. doi:10.1093/jb/mvn076
- Dolce, V., Scarcia, P., Iacopetta, D. and Palmieri, F. (2005) A fourth ADP/ATP carrier isoform in man: Identification, bacterial expression, functional characterization and tissue distribution. FEBS Letters, 579, 633-637. doi:10.1016/j.febslet.2004.12.034
- Dolce, V., Iacobazzi, V., Palmieri, F. and Walker, J.E. (1994) The sequences of human and bovine genes of the phosphate carrier from mitochondria contain evidence of alternatively spliced forms. The Journal of Biological Chemistry, 269, 10451-10460.
- Dolce, V., Fiermonte, G., Runswick, M.J., Palmieri, F. and Walker, J.E. (2001) The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. The Proceedings of the National Academy of Sciences Online (U.S.), 98, 2284-2288. doi:10.1073/pnas.031430998
- Fiermonte, G., Walker, J.E. and Palmieri, F. (1993) Abundant bacterial expression and reconstitution of an intrinsic membrane-transport protein from bovine mitochondria. Biochemical Journal, 294, 293-299.
- Palmieri, F., Indiveri, C., Bisaccia, F. and Iacobazzi, V. (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods in Enzymology, 260, 349-369. doi:10.1016/0076-6879(95)60150-3
- Xu, Y., Mayor, J.A., Gremse, D., Wood, D.O. and Kaplan, R.S. (1995) High-yield bacterial expression, purification, and functional reconstitution of the tricarboxylate transport protein from rat liver mitochondria. Biochemical and Biophysical Research Communications, 207, 783- 789. doi:10.1006/bbrc.1995.1255
- Iacobazzi, V., De Palma, A. and Palmieri, F. (1996) Cloning and sequencing of the bovine cDNA encoding the mitochondrial tricarboxylate carrier protein. Biochimica et Biophysica Acta, 1284, 9-12. doi:10.1016/0005-2736(96)00115-0
- Zara, V., Palmieri, L., Franco, M.R., Perrone, M., Gnoni, G.V. and Palmieri, F. (1998) Kinetics of the reconstituted tricarboxylate carrier from eel liver mitochondria. Journal of Bioenergetics and Biomembranes, 30, 555-563. doi:10.1023/A:1020532500749
- Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn, R., Trezeguet, V., Lauquin, G.J. and Brandolin, G. (2003) Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature, 426, 39-44. doi:10.1038/nature02056
- Huizing, M., Ruitenbeek, W., van den Heuvel, L.P., Dolce, V., Iacobazzi, V., Smeitink, J.A., Palmieri, F. and Trijbels, J.M. (1998) Human mitochondrial transmembrane metabolite carriers: Tissue distribution and its implication for mitochondrial disorders. Journal of Bioenergetics and Biomembranes, 30, 277-284. doi:10.1023/A:1020501021222
- Zwerger, P., Nimeth, K., Wurtz, J., Salvenmoser, W. and Pelster, B. (2002) Development of the swimbladder in the European eel (Anguilla anguilla). Cell and Tissue Research, 307, 155-164. doi:10.1007/s00441-001-0488-5
ABBREVIATIONS
CIC, citrate carrier; EST, expressed sequence tag; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PCR, polymerase chain reaction; PIPES, 1,4–piperazinediethanesulfonic acid; SDS-PAGE; polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate; RT-PCR, reverse transcription polymerase chain reaction.

