Journal of Biophysical Chemistry
Vol.2 No.4(2011), Article ID:8429,8 pages DOI:10.4236/jbpc.2011.24049
Different behavior of protein B23/nucleophosmin and UBF in HeLa cells during apoptosis
![]()
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry Russian Academy of Sciences, Miklukho-Maklaya, Moscow, Russia. *Corresponding Author: vla.ibch@mail.ru
Received 7 July 2011; revised 11 August 2011; accepted 23 August 2011.
Keywords: TNF-α Induced Apoptosis; Proteolysis of UBF; Monomer-Oligomer State of Nucleophosmin
ABSTRACT
The behavior of UBF (upstream binding factor) and nucleophosmin in HeLa and HeLa-Bcl-2 cells during apoptosis induced by TNF-α, emetine, and their mixture was investigated. A pronounced apoptosis was achieved only in HeLa cells treated with a mixture of the inducers. Immunoblotting analysis of UBF and nucleophosmin in samples containing different portions of cells with apoptotic nuclei was carried out. It showed that UBF was proteolytically cleaved giving a stable 76-kDa fragment. Increasing content of the fragment during apoptosis correlated with the level of cells containing apoptotic nuclei and with a decrease in the content of full-sized UBF. Determination of Nand C-terminal sequences of UBF and 76-kDa fragment allowed us not only to characterize UBF at the protein level, but also to describe the site of the apoptosis-specific proteolysis. Nucleophosmin did not undergo proteolytic cleavage during apoptosis and its content was unchanged even in a sample containing 100% of cells with apoptotic nuclei. However in cells reached terminal stages of apoptosis, the balance between monoand oligomeric forms of nucleophosmin changed due to depletion of monomeric forms and appearance of two additional oligomeric forms with lower molecular weight.
1. INTRODUCTION
Tumor cells are different from normal cells not only by hyperactivity and pleomorphism of the nucleoli, but also by sharply increased contents of some nucleolar proteins, such as argentophil ones, leading to increased resistance of tumor cells to apoptosis [1]. We studied two key argentophil nucleolar proteins—nucleophosmin/B23 and UBF/NOR-90, which are overexpressed in tumor cells and undoubtedly involved in apoptosis [2-5]. Cytotoxicity of many apoptotic agents for tumor cells correlates with their ability to induce translocation of both proteins from the nucleolus into the nucleoplasm or activate proteolysis of UBF and B23. The translocation of nucleophosmin from the nucleolus is the initial stage of nucleolar segregation. In fact, translocation of B23 protein anticipates such events as chromatin condensation and fragmentation of DNA [2].
Overexpression of nucleophosmin leads to a decrease in cell sensitivity to apoptosis through different mechanisms, affecting the activity of key proteins such as factor IRF1, p53 and Arf [6]. The data concerning apoptosis-mediated changes in content of protein B23 is very contradictory. Thus, some authors observed a decrease in protein B23 content associated with either a decrease in the amount of mRNA of nucleophosmin [2] or with its proteolysis [1,7]. And similarly to the case of UBF, this proteolysis may depend not only on caspases, but also on other proteases. In other works [4,5], protein B23 content was shown to be unchanged and only its translocation correlating with the apoptosis degree took place. New literature data have shown that apoptosis-mediated changes in the structure of UBF and B23 trigger the generation of autoimmune antibodies in patients with tumors (e.g. breast cancer, hepatocellular carcinoma) [3]. But what is known about apoptosis-dependent changes in the structure of UBF and nucleophosmin? The majority of available data indicate that in addition to translocation UBF undergoes apoptosis-dependent proteolysis [4,5,8]. Unfortunately in none of these studies the region of UBF proteolysis was characterized. It was shown [3] that cell malignization is accompanied by structural changes in nucleophosmin resulting in appearance of its unique oligomeric forms that are unusually resistant to heating and treatment with SDS, but at present there are virtually no data on the oligomerization state of protein B23 during apoptosis of tumor cells.
The purpose of the present work is not only to evaluate the content of two key nucleolar proteins B23 and UBF, but also at first to analyze the monomer-oligomer state of nucleophosmin and to study structural changes of B23 and UBF on protein level during apoptosis induced by TNF-α in human cervical carcinoma cells (HeLa).
2. MATERIALS AND METHODS
2.1. Materials
We used: 10% fetal calf serum, culture medium DMEM from HyClone (USA); penicillin, streptomycin from Gibco (Great Britain); 5-dimethylamino-1-naphtha-lenesulfonyl chloride (Dns-Cl) from Fluka (Switzerland); emetine, calibrating standard proteins (SDS-6H), NaCl, KCl, Na2HPO4, KH2PO4, EDTA disodium salt, acetonitrile, 3, 3’-diaminobenzidine (DAB), goat antibodies to mouse IgG and IgM conjugated with horseradish peroxidase, sheep antibodies to human IgG conjugated with horseradish peroxidase, 4’6-diamidino-2-phenylindole dihydrochloride (DAPI dihydrochloride), paraformaldehyde (PFA), BSA, PMSF-treated carboxypeptidase A (cpA) from bovine pancreas (with activity of 50 U/mg protein), polyvinylpyrrolidone-40 (PVP-40), monoclonal antibodies to B23 protein (FC82291) from Sigma (USA); reagents for electrophoresis, dry defatted milk, SDS, Tris were from Bio-Rad (USA); immobilon NC and P membranes were from Millipore (USA); Triton X-100, dimethylsulfoxide (DMSO) were from ICN (USA); Mowiol was from Hoechst (USA). Recombinant human TNF-α was kindly presented by the Laboratory of Protein Engineering, Institute of Bioorganic Chemistry, Russian Academy of Sciences; autoimmune serum from a patient with systemic scleroderma was obtained in the Institute of Rheumatology, RAMS. Other reagents were of chemical or special purity qualification (domestic production).
2.2. Cell Cultures
Human cell cultures HeLa and HeLa-Bcl-2 obtained in the Institute of Poliomyelitis and Viral Encephalitis, RAMS, by transfection of HeLa cells with the pLPS-bcl-2 vector were used. The cells were grown in culture flasks or on cover slips on DMEM medium supplemented with 10% fetal calf serum, penicillin (10 µg/ml), and streptomycin (10 µg/ml), at 37˚C in the presence of 5% CO2. In the case of HeLa-Bcl-2 cells, the medium also contained puromycin (1 µg/ml).
2.3. Apoptosis
Experiments on apoptosis were performed on the day after replanting, in the exponential growth phase of the cells. The cells were incubated in the presence of the protein synthesis inhibitor emetine (1 µg/ml), or TNF-α (10 ng/ml), or their mixture at 37˚C for 2 h and 6 h. The control cells were incubated under the same conditions, but without emetine and TNF-α. The extent of apoptosis was quantified by counting apoptotic cells in three randomly chosen microscopic fields with a 40X objective before and after exposure to the corresponding agents. The cells starting apoptosis were identified visually with a light microscope and with a fluorescent microscope after staining of cells with DAPI (by the presence of characteristic morphological changes, such as vesicular structures on the membranes—blebbing, nuclear fragmentation, and appearance of apoptotic bodies). Results of determination of the percent of apoptotic cells are presented as the mean ± standard deviation, calculated for ~ 500 cells in each of three independent experiments. Treated and untreated cell monolayers were washed from the medium in phosphate-buffered saline (PBS) (pH 7.2), containing 2.7 mM KCl, 140 mM NaCl, 8.1 mM Na2HPO4, and 1.5 mM KH2PO4. The cells were harvested from the culture flask surface with Versene solution (0.2% EDTA in PBS) for 5 min at 37˚C and precipitated by centrifugation for 10 min at 1672 rpm (500 g) at 4˚C using a 5804R centrifuge from Eppendorf (Germany). Cells exposed to TNF-α/emetine mixture for 6 h were analyzed as two fractions. The floating cell fraction was collected together with the culture medium. The culture flask was washed with PBS, and the wash-outs were combined with the floating cell fraction and then centrifuged for 20 min at 1672 rpm (500 g) at 4˚C. The cells adhered to the flask surface were washed with PBS, harvested with Versene solution, and centrifuged under the same conditions. Pellets of these two fractions were washed twice with PBS and centrifuged under the same conditions. The pelleted cells were resuspended in PBS, protein concentration was determined by the Lowry method [9] and than electrophoretic and immunochemical analysis were performed.
2.4. Staining of Cells with DAPI
Cells grown on cover slips were washed with PBS and fixed with 3% paraformaldehyde solution in PBS for 20 min at room temperature. The fixed cells were washed three times for 5 min with PBS, stained with DAPI in PBS (0.1 µg/ml), and mounted into Mowiol. A part of the floating cells (after washing with PBS) was precipitated by centrifugation for 5 min at 2048 rpm (750 g) at 4˚C using the 5804R centrifuge. The cells remaining on the flask surface were washed in PBS, removed with Versene solution, and centrifuged under the same conditions. The resulting cell pellets were supplemented each with 5 ml PBS containing 3% PFA, carefully resuspended by shaking, and maintained for 20 min at room temperature. The cells were collected by centrifugation for 5 min at 2048 rpm (750 g) at 4˚C, and then the pellets were washed three times for 5 min in PBS and placed onto cover slips. The cell-carrying slips were covered with PBS containing 0.1% gelatin and dried overnight at room temperature. Afterwards, the cells were stained with DAPI in PBS (0.1 µg/ml) and mounted into Mowiol. All preparations were examined with an Axiovert 200 microscope (Carl Zeiss, Germany). The images were recorded with a 12- bit monochrome CCD CoolSnapcf camera (Roper Scientific, USA) and processed using the Adobe Photoshop version 7.0 program.
2.5. SDS-PAGE and Electroblotting
Samples were supplemented with PBS and glycerol to adjust the protein concentration to 1 mg/ml and glycerol to 30%. The lysing solution which contained SDS, 2- mercaptoethanol and EDTA, was added to samples to the concentration of 5%, 2.5%, and 1 mM, respectively. The mixture was usually heated at 100˚C for 1 min or at 100˚C for 10 min (to provide for degradation of oligomeric form of B23 protein). Equal quantities of protein samples (~30 µg) were separated by SDS-PAGE at 12˚C, transferred onto Immobilon NC membranes for immunostaining or Immobilon P membranes for sequencing and hydrolysis of C-terminal amino acid residues with carboxypeptidase A. The electrotransfer was performed under conditions described in [10]: 0.025 M sodium bicarbonate buffer (pH 9.0) containing 20% CH3OH and 0.1% SDS. The electrotransfer was done at 12˚C and constant current of ~400 mA for 4 h. For more efficient transfer of proteins with different molecular weights (as in case of simultaneous UBF and B23 analysis), a threestage differentiated electrotransfer was used. For this, after immunoblotting for 3 h, the gel region containing proteins with molecular weights <60 kDa and the corresponding region of the membrane were cut off, and the upper part of the gel carrying high-molecular-weight proteins was electrotransferred additionally for 1 h. Then the gel region containing proteins with molecular weights <120 kDa and the corresponding region of the membrane were cut off, and the upper part of the gel with proteins of higher molecular weights was subjected to additional electrotransfer for 10 - 12 h (depending on the percentage and on the thickness of the gel). The completeness of electrotransfer was monitored by staining the gels (after the blotting) with 0.1% Coomassie G-250 and marker proteins on the membranes with 0.1% Amido Black 10B solution in 45% methanol. Two-stage electrotransfer (for analysis of monomeroligomer state of B23) was also used in the present work: the proteins with molecular weights <60 kDa (the lower region of the gel) were electrotransferred for 3 h, and the proteins with the higher molecular weights (the upper region of the gel) were transferred for 12 h additionally. Immunostaining was performed with monoclonal antibodies specific to protein B23 or with autoimmune antibodies specific to UBF from a patient with systemic scleroderma as described in [11]. The appropriate conjugated with horseradish peroxidase secondary antibodies (goat antibodies to mouse IgG + IgM for B23 protein or sheep antibodies to human IgG for UBF) were used.
2.6. Analysis of N-Terminal Amino Acid Sequence
Immobilon P membranes containing proteins were used for sequencing. Prior to sequencing of full-size UBF (100 kDa), the formyl protection was removed from immobilized on Immobilon P membrane protein as described in [12]. Automated Edman degradation was performed with Procise 491 Protein Sequencing System gas phase sequencer (Applied Biosystems, USA). Phenylhydantoin derivatives of amino acids were identified with 785 A PTH-analyzer (Applied Biosystems).
2.7. Two Steps Protein Hydrolysis by Carboxypeptidase A and Analysis of C-Terminal Sequences
Proteins (UBF and its 76 kDa-fragment) were hydrolyzed by carboxypeptidase A directly on the Immobilon P membrane. To prevent nonspecific adsorption of carboxypeptidase, the membrane was treated with PVP-40 as described in [11]. Hydrolysis was performed in two stages. In the first stage, the membrane was supplemented with 30 µl of 0.2 M N-ethylmorpholine acetate buffer (pH 8.3) containing 1 µg cpA. The mixture was incubated for 120 min at 37˚C. An aliquot of the supernatant (~1/5 V) was removed for analysis. In the second stage, the buffer was acidified with CH3COOH to pH 6.0, then 1 µg cpA was added, and hydrolysis was continued at 37˚C for 120 min. After termination of the reaction, the supernatant was collected, and the membrane was washed successively by 20 μl portions of buffer (pH 6.0), Milli-Q water, and methanol. All wash-outs were combined with the supernatant and evaporated. The detached amino acids were analyzed as dansyl derivatives by twodimensional chromatography on silica gel plates as described in [10].
3. RESULTS AND DISCUSSION
3.1. Effect of Apoptosis Inducers TNF-α and Emetine on HeLa ana HeLa-Bcl-2 Cells
TNF-α is well-studied and efficient physiological inducer of apoptosis in animal cells [13]. Cytotoxic effects of TNF-α are balanced by defense mechanisms in some cell lines, including the HeLa tumor cells used in this work. Because anti-apoptotic defense mechanisms depend on protein synthesis, TNF-α induced apoptosis can occur only in the presence of the protein synthesis inhibitors [13]. As an inhibitor we have chosen emetine, which blocks translation and translocation [14]. Besides HeLa cells we also used HeLa-Bcl-2 cells transfected with a construct containing cDNA of the anti-apoptotic protein Bcl-2.
Incubation of HeLa cells with TNF-α or emetine separately did not induce apoptosis and the number of nuclei with morphological signs of apoptosis was low, comparable with untreated control cells (~2% - 3%). The number of such nuclei increased to 25% on exposure for 2 h to mixture of TNF-α and emetine (Figure 1); these nuclei had irregular shape and distinct heterochromatin fragmentation. Nuclear fragmentation is the main sign of the cell starting apoptosis [15]. According to our findings and the published data [16], similar schemes of apoptosis induction by the 6 h exposure of HeLa cells to the TNF-α and emetine mixture resulted in appearance of a large number of cells with pronounced signs of apoptosis. We found that many of those cells treated for 6 h with the TNF-α and emetine mixture lost their adhesiveness and detached (floated) into the culture medium. Therefore, we analyzed two fractions of the cells: the detached (floating) and adhered to the culture flask surface. After the exposure to apoptosis inducers for 6 h, 45% of the adhered cells and 100% of the floating cells had fragmented nuclei (Figure 1). Similarly to results of work [16] in our experiments Bcl-2 had a protective effect in all apoptosis induction experiments, and the number of HeLa-Bcl-2 cells with apoptotic nuclei was comparable to that in the control, even after 6 h exposure to the mixture of TNF-α and emetine (Figures 1(a)-(b)).
Investigation of protein states in apoptotic cells is of special interest. Apoptosis-induced changes in the protein structure are usually studied in the total population of cells exposed to apoptotic agents, although such population includes both apoptotic and non-apoptotic cells. We know only two works, where pure populations of apoptotic cells were analyzed [7,17]. The apoptotic fractions
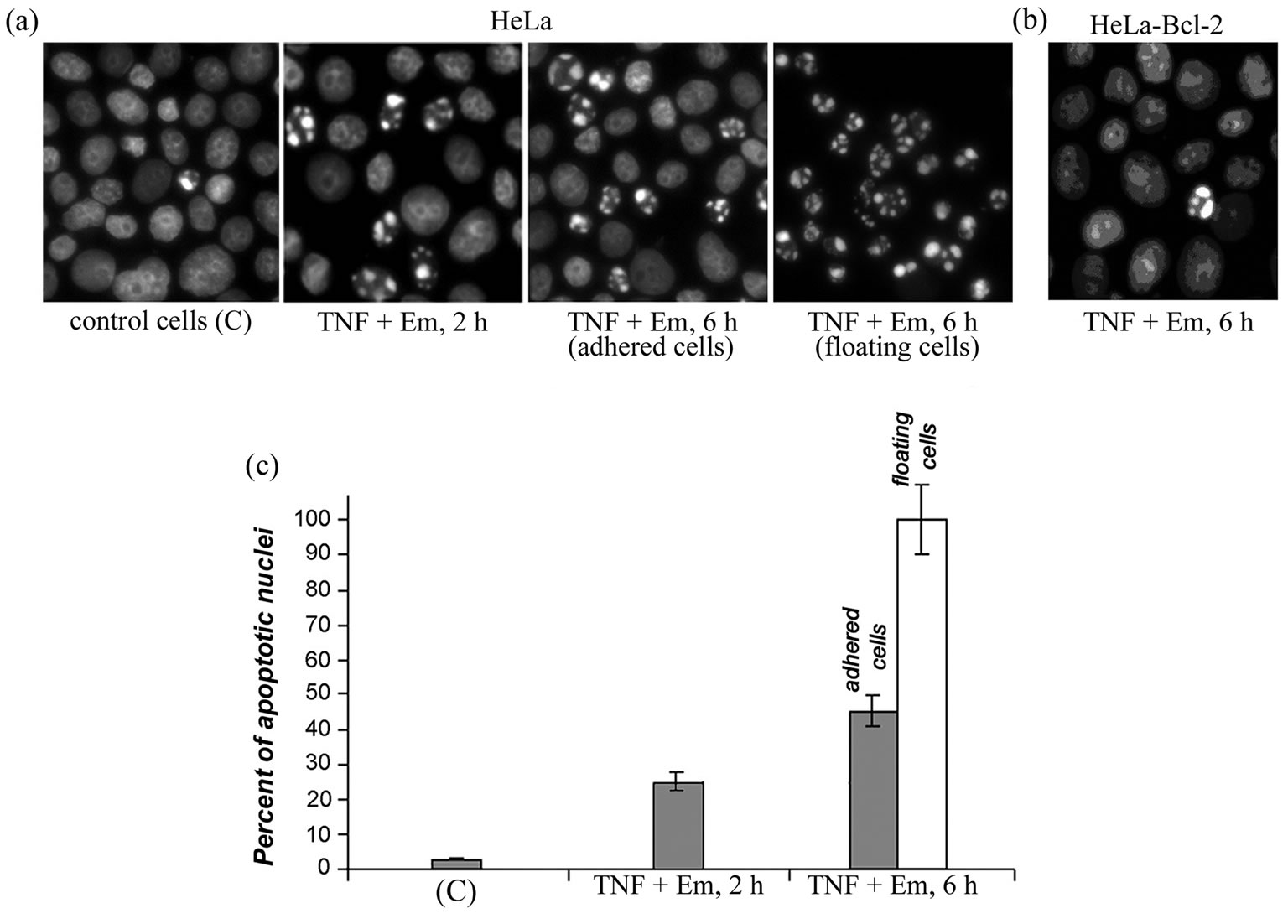
Figure 1. Effect of mixture apoptosis inducers TNF-α and emetine (TNF + Em) on HeLa cells (a), (c) and HeLa-Bcl-2 cells (b). Staining with DAPI (a), (b). Percent of apoptotic nuclei in treated HeLa cells (c). Two types of the cells were analyzed: those adhered to the surface of the culture flask (dark rectangles) and those floating (light rectangles). Untreated control cells (C).
were analyzed electrophoretically (Figure 2(a)). The protein composition of the apoptotic and control cells occurred to be strikingly alike, changes being record only in the content of some proteins as described also in work [7]. So, in the electrophoregram of the floating cell fraction the band corresponding to 66-kDa proteins was markedly strengthened (Figure 2(a), lane 4). We identified in this band BSA (according MALDI-MS analysis of its tryptic peptides). This fraction of apoptotic cells was capable of increased uptake and/or sorption of BSA from the culture medium.
It is very important to asses the state of the whole complex of nucleolar proteins during apoptotic death of cells. We decided to study apoptosis-induced changes in the structure of two key nucleolar proteins—nucleophosmin and UBF, undoubtedly involved in apoptosis but having different intranucleolar location, dynamics of translocation from nucleolus into nucleoplasm during apoptosis and functions [4,5].
3.2. Changes in UBF during Apoptosis
In some works [4,5,8] UBF was shown to be an early target of the apoptosis-induced proteolysis, which depends on both cell types and apoptosis inducers. We decided to assess the behavior of UBF during apoptosis of HeLa cells induced by TNF-α and emetine mixture. By Western-blot analysis a partial proteolysis of UBF resulting in a 76-kDa fragment already after 2-h treatment with the TNF-α and emetine mixture was revealed, when the number of cells with apoptotic nuclei was ~25% (Figure 2(b), lane 2). Prolongation of the treatment time to 6 h increased the number of apoptotic cells and enhanced the proteolysis of UBF associated with production of this fragment (Figure 2(b), lanes 3 and 4). The cleavage was practically complete in the floating cells in the terminal stages of apoptosis (lane 4). Thus, the TNF- α and emetine mixture-induced apoptosis of HeLa cells in these conditions was associated with a characteristic proteolytic fragmentation of UBF, and the content of this fragment increased in correlation with a decrease in the content of full-size UBF.
The amino acid sequence of UBF has been deduced from the analysis of the cDNA structure [18], but there is no data concerning Nand C-terminal protein processing. To characterize the structure of UBF and its 76-kDa fragment, we cut off two protein strips from the immobilon P membrane (similar to shown on Figure 2(b)). One of these strips corresponded to the full-size protein UBF and the other to its 76-kDa fragment. The amino acid sequence of the full-size UBF was determined by automated degradation according to Edman method after the removal of formyl group from the protein. The sequence of its twelve N-terminal amino acid residues coincided with the sequence derived from the cDNA sequence of protein beginning from Met1 residue. For determination of C-terminal sequences of UBF and its 76-kDa fragment we used hydrolysis by carboxypeptidase A, which was performed in two stages. In the first stage (at pH 8.3) asparagine and serine (N > S) were detected in the supernatant fraction of both samples. In the second stage (at pH 6.0) apart from asparagine and serine, asparaginic acid was detected, thereby S > N > D. Analysis of the C-terminal sequences of UBF and its 76-kDa fragment allowed us not only to determine the C-terminal sequence of UBF, but also to establish that the 76-kDa fragment was the C-terminal fragment of the protein. Sequencing of the N-terminal amino acid sequence of the 76-kDa fragment of UBF allowed us to reveal the point of the apoptosis-specific proteolysis. The amino acid sequence of its twelve N-terminal amino acid residues coincided with the sequence of UBF: (186) LIQNAKKSDIPE(197). So the region of UBF proteolysis was for the first time characterized on the protein level. Analysis of the specific apoptosis-induced proteolytic fragmentation of the UBF polypeptide chain resulting in the stable 76-kDa fragment may be very informative for the subsequent identification of the protease responsible for hydrolysis of nucleolar proteins during apoptosis. Although the proteolysis site (182)DHPD↓(186)L corresponds, for instance, to the specificity of caspases 2 and 3 (DXXD↓), we could not now specify exactly the type of caspase responsible for the proteolysis of UBF, because of contradictions or absence in literature of the data on the substrate specificity of some caspases. It is important that apoptosis-induced proteolysis of UBF in this region of the polypeptide chain resulted in detachment of N-terminal fragment, which contained a domain extremely important for both UBF dimerization and functioning as a transcription factor and one of six HMG-boxes. The apoptosis-induced proteolysis of UBF was also observed in HL-60 cells treated with camptothecin and in Jurkat cells treated with CD-95 (Fas/APO-1).
3.3. Changes in B23 during Apoptosis
We assessed the monomer-oligomer state of protein B23 during apoptosis, because only its monomeric forms had been characterized in the majority of published works. However, analysis of only monomeric forms is insufficient, first, because both nucleophosmin isoforms (B23.1 and B23.2) can in vitro and in vivo produce homoand heterooligomers (~210 - 230 kDa). Second, tumor cells have been shown to contain unique oligomers with an unusually high resistance to SDS treatment and heating [3,12,19,20]. The absence of data about protein B23 monomer-oligomer state is due, in particular, to the absence of methods for concurrent detection and
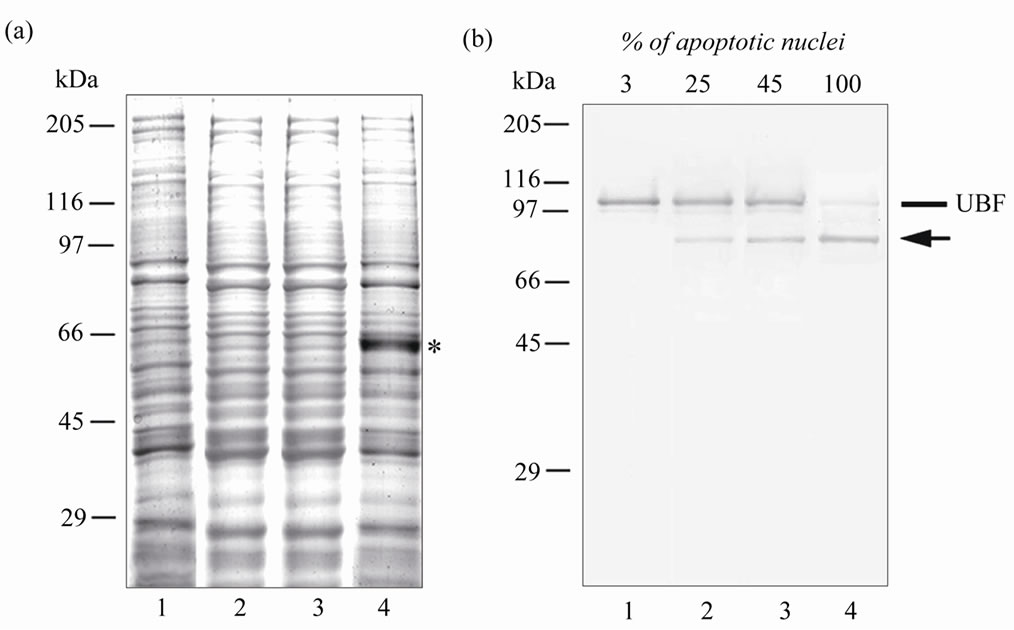
Figure 2. Electrophoregram (a) and immunoblot (b) of control HeLa cells (lane 1) and HeLa cells treated with TNF-α and emetine mixture for 2 h (lane 2), or 6 h (adhered cells (lane 3), floating cells (lane 4)). The electrophoresis was performed in 12.5% SDS-PAG. For Western blotting analysis of UBF autoimmune antibodies were used. The samples for electrophoresis were heated for 1 min at 100˚C. The proteins were electrotransferred onto Immobilon membrane NC (or P) in one stage (4 h). The intact UBF is shown by line, the 76-kDa fragment – by arrow. Positions of the marker proteins are shown on the left. At the top of (b) the percentage of cells with apoptotic nuclei is indicated.
quantitative evaluation of its oligomeric and monomeric forms.
We have analyzed the structural state of nucleophosmin during apoptosis using immunoblotting scheme developed by us earlier [12]. This scheme includes some modifications at the stages of preparing samples for electrophoresis (as compared to the Laemmli method) and in the mode of electroblotting (described in “Materials and Methods”). Differential electrotransfer of proteins in two or three stages onto immobilon membranes allowed us to quantitatively transfer proteins of a wide range of molecular weights including oligomeric (180 - 230 kDa) and monomeric (38 - 40 kDa) B23 forms, and to compare the states of proteins B23 and UBF during apoptosis (in the same lane of the gel). Immunoblots of the proteins from HeLa cells treated only with emetine or TNF-α (not shown) were identical to the immunoblots of the proteins from the control cells (Figure 3(a), lane 1): bands could be detected which corresponded to the full-size UBF, and monomeric and three oligomeric forms of B23 protein. Analysis of the immunoblots allowed us to show for the first time that in the fractions with a high percentage of apoptotic nuclei including the cells in its terminal stages (with full proteolysis of UBF) SDS-resistant oligomeric forms of nucleophosmin were retained (Figure 3(a)). Moreover, we detected some specific features of nucleophosmin in apoptotic cells. Thus, in lysates from the cells in the terminal stages of apoptosis (Figure 3(a), lane 4) the ratio of monomeric and oligomeric forms of B23 protein was changed: the amount of monomers was decreased and two additional (as compared to the lysates from control cells) bands appeared in the zone of oligomers with a lower (~180 - 200 kDa) molecular weight, and their intensity was maximal in floating cells.
In some works, apoptosis-dependent decrease in nucleophosmin level was shown to be associated with caspase-mediated [1] or caspase-independent proteolysis [3, 7] resulting in formation of a ~21 - 22 kDa proteolysisresistant fragment. By immunochemical analysis of proteins of the floating apoptotic cells (after their separation in 15% PAG) we demonstrated the absence of protein B23 cleavage products of >6 kDa (Figure 3(b)). We suggested that the decrease in the content of B23 monomers during apoptosis could be caused not only by proteolysis but also by shift in the monomer-oligomer state towards oligomers. For estimation of the total protein B23 content we used thermal treatment conditions developed by us earlier [11] (10 min, 100˚C) leading to destruction of B23 oligomers. The proteins were separated by electrophoresis in 10% SDS-PAG (the optimum conditions for separation of nucleophosmin monomers) (Figure 3(c)). Under these conditions, oligomeric forms of nucleophosmin were virtually completely destroyed in both the control and treated cells. However, even in cells in the terminal stages of apoptosis the intensity of the nucleophosmin band in the zone of monomers was comparable to its intensity of band in the control cells (lanes 1 and 4). These data combined with our earlier results
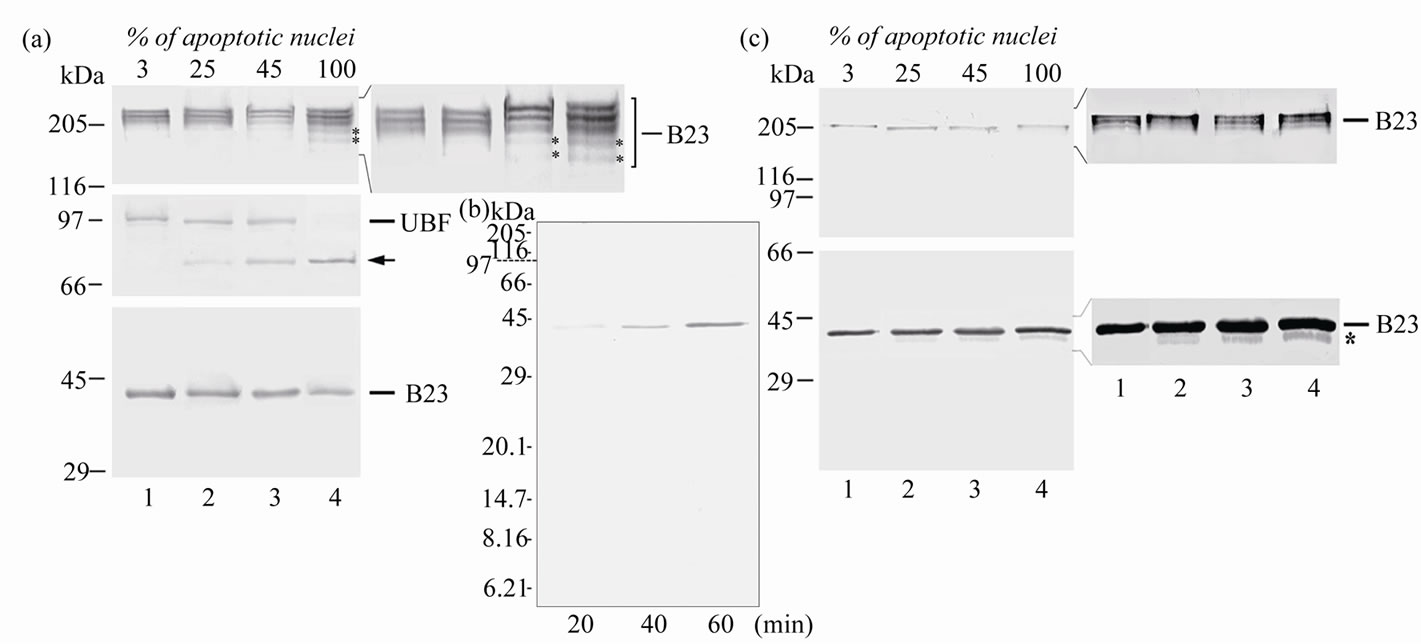
Figure 3. Immunoblots (a)-(c). Immunochemical analysis of B23 (a)-(c) and UBF (a) in HeLa cells in apoptosis. The electrophoresis was performed in 7.5% (a), 15% (b) and 10% (c) SDS-PAG. Samples used: lysates of control cells (lane 1), cells treated with mixture of TNF-α and emetine for 2 h (lane 2), or 6 h (adhered cells (lane 3), floating cells (lane 4)). The samples for electrophoresis were heated for 1 min at 100˚C. (a), (b) or for 10 min at 100˚C (c). The membranes were immunostained with monoclonal antibodies specific to B23 (a-c), or with autoimmune antibodies to UBF (a). The proteins were electrotransferred onto Immobilon NC membrane in one (b), two (c) or three (a) stages. Intact proteins are marked by lines, the proteolytic fragment of UBF—by arrow, apoptosis-specific oligomeric and monomeric forms of B23—by asterisks. Positions of the marker proteins are shown on the left.
make unlikely the considerable proteolysis of nucleophosmin (in contrast to UBF) during the apoptosis of HeLa cells induced by the TNF-α and emetine mixture. However, in apoptotic cells with pronounced proteolysis of UBF, destruction (in our experiment) of nucleophosmin oligomers was accompanied by appearance on immunoblots of an additiona band with higher electrophoretic mobility in the zone of monomers (Figure 3(c)). The intensity of this band was low but increase proportionally to the fraction of the cells with apoptotic nuclei, and this seemed to indicate the appearance of an apoptosis-specific form of B23 protein. Possibly, just this new form of B23 protein is responsible for changes in the ratio of monomeric and oligomeric forms and appearance of additional oligomers of B23 protein in the fraction of apoptotic cells, especially in the fraction of fully apoptotic ones where its content is the highest. This new apoptosis-specific form was not characterized in the present work (because of its low content), but it is clearly of interest. The appearance of the apoptosis-specific form of nucleophosmin could be associated with chemical modifications of the polypeptide chain (acetylation, phosphorylation etc.) or N/C-terminal processing or proteolysis.
4. CONCLUSIONS
The findings of the present work are consistent with the data on camptothecin-induced of HeLa cells [5] and CD-95-induced apoptosis of Jurkat cells [4,8]. In these works the cleavage of UBF and the absence of B23 proteolysis were recorded, and total protein B23 content was unchanged even upon complete proteolysis of UBF [8]. In this work we are the first to show that the cascade of apoptotic reactions induced by TNF-α in HeLa cells is accompanied not only by proteolysis of proteins (as in the case of UBF) but also by fine structural changes resulting in new, apoptosis-specific forms of protein, changes in B23 oligomerization state, without significant changes in the total content of the protein. We suggested that in some proteins present as several isoforms and prone to formation of homoand heterooligomers with different location and properties (as in the case of nucleophosmin) structural changes may concern not the total protein fraction but its distinct structural forms. It can be proposed also that different structural forms of such a complex proteins as nucleophosmin may be involved in different apoptotic cascades.
5. ACKNOWLEDGEMENTS
This work was supported by the gtants of the Russian Foundation for Basic Research (project No. 09-04-00713-a) and of the Program “Fundamental Sciences for Medicine-2011”.
REFERENCES
- Horky, M., Wurzer, G., Kotala, V., Anton, M., Vojtesek, B., Vacha, J. and Wesierska-Gadek, J. (2001) Segregation of nucleolar components coincides with caspase-3 activation in cisplatin-treated HeLa cells. Journal of Cell Science, 114, 663-670.
- Liu, W.H. and Yung, B.Y. (1998) Mortalization of human promyelocytic leukemia HL-60 cells to be more susceptible to sodium butyrate-induced apoptosis and inhibition of telomerase activity by down-regulation of nucleophosmin/B23. Oncogene, 17, 3055-3064. doi:10.1038/sj.onc.1202234
- Ulanet, D.B., Torbenson, M., Dang, C.V., Casciola-Rosen, L. and Rosen, A. (2003) Unique conformation of cancer autoantigen B23 in hepatoma: A mechanism for specificity in the autoimmune response. Proceedings of the National Academy of Sciences of the United States of America, 100, 12361-12366. doi:10.1073/pnas.2035245100
- Casiano, C.A. and Tan, E.M. (1996) Antinuclear autoantibodies: Probes for defining proteolytic events associated with apoptosis. Molecular Biology Reports, 23, 211-216.
- Martelli, A.M., Robuffo, I., Bortul, R., Ochs, R.L., Luchetti, F., Cocco, L., Zweyer, M., Bareggi, R. and Falcieri, E. (2000) Behavior of nucleolar proteins during the course of apoptosis in camptothecin-treated HL60 cells. Journal of Cellular Biochemistry, 78, 264-277. doi:10.1002/(SICI)1097-4644(20000801)78:2<264::AID-JCB9>3.3.CO;2-V
- Grisendi, S., Mecucci, C., Falini, B. and Pandolfi, P.P. (2006) Nucleophosmin and cancer. Nature Reviews. Cancer, 6, 493-505. doi:10.1038/nrc1885
- Patterson, S.D., Grossman, J.S., D’Andrea, P. and Latter, G.I. (1995) Reduced numatrin/B23/nucleophosmin labeling in apoptotic Jurkat T-lymphoblasts. The Journal of Biological Chemistry, 270, 9429-9436.
- Torres-Montaner, A., Bolivar, J., Astola, A., GimenezMas, J.A., Brieva, J.A. and Valdivia, M.M. (2000) Immunohistochemical detection of ribosomal transcription factor UBF and AgNOR staining identify apoptotic events in neoplastic cells of Hodgkin’s disease and in other lymphoid cells. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society, 48, 1521-1530. doi:10.1177/002215540004801109
- Lowry, O.H., Rosebrough, N.J., Farr, A.L. and Randall, R.J. (1951) Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry, 193, 265- 275.
- Vladimirova, N.M., Potapenko, N.A., Sachs, G. and Modyanov, N.N. (1995) Determination of the sidedness of the carboxy-terminus of the Na+/K(+)ATPase alphasubunit using lactoperoxidase iodination. Biochimica et Biophysica Acta, 1233, 175-184.
- Sautkina, E.N., Potapenko, N.A. and Vladimirova, N.M. (2006) State of nucleolar proteins B23/nucleophosmin and UBF in HeLa cells during apoptosis induced by tumor necrosis factor. Biochemistry (Moscow), 71, 634- 643. doi:10.1134/S0006297906060071
- Vladimirova, N.M., Sautkina, E.N., Tabdanov, E.D., et al. (2004) Identification and characterization of monomeric and oligomeric isoforms of a major nucleolar protein B23/ nucleophosmin in HeLa cells. Biological Membrany (Moscow), 21, 94-101.
- Rath, P.C. and Aggarwal, B.B. (1999) TNF-induced signaling in apoptosis. Journal of Clinical Immunology, 19, 350-364.
- Schweighoffer, T., Schweighoffer, E., Sviresheva, M., Apati, A., Balogh, A., Csuka, I., Luat, N.N. and Banfalvi, G. (1993) Oncogene expression in thymocytes after emetine treatment of mice. Cellular and Molecular Biology Research, 39, 647-656.
- Lazebnik, Y.A., Cole, S., Cooke, C.A., Nelson, W.G. and Earnshaw, W.C. (1993) Nuclear events of apoptosis in vitro in cell-free mitotic extracts: A model system for analysis of the active phase of apoptosis. The Journal of Cell Biology, 123, 7-22. doi:10.1083/jcb.123.1.7
- Domnina, L.V., Ivanova, O.Y., Cherniak, B.V., Skulachev, V. P. and Vasiliev, J.M. (2002) Effects of the inhibitors of dynamics of cytoskeletal structures on the development of apoptosis induced by the tumor necrosis factor. Biochemistry (Moscow), 67, 737-746.
- Robaye, B., Doskeland, A.P., Suarez-Huerta, N., Doskeland, S.O. and Dumont, J.E. (1994) Apoptotic cell death analyzed at the molecular level by two-dimensional gel electrophoresis. Electrophoresis, 15, 503-510. doi:10.1002/elps.1150150168
- Jantzen, H.M., Admon, A., Bell, S.P. and Tjian, R. (1990) Nucleolar transcription factor hUBF contains a DNAbinding motif with homology to HMG proteins. Nature, 344, 830-836. doi:10.1038/344830a0
- Chan, P.K. and Chan, F.Y. (1995) Nucleophosmin/B23 (NPM) oligomer is a major and stable entity in HeLa cells. Biochimica et Biophysica acta, 1262, 37-42. doi:10.1016/0167-4781(95)00044-H
- Vladimirova, N.M., Lobanova, N.V. and Potapenko, N.A. (2010) State of oncomarker protein B23/nucleophosmin in HeLa cells. Biochemistry (Moscow), 75, 851-860. doi:10.1134/S0006297910070060

