American Journal of Plant Sciences
Vol.06 No.12(2015), Article ID:58880,10 pages
10.4236/ajps.2015.612196
Protocol for Artificial MicroRNA Mediated Over-Expression of miR820 in Indica Rice
Neha Sharma, Sandeep Panchal, Neeti Sanan-Mishra*
Plant Molecular Biology Group, International Center for Genetic Engineering and Biotechnology, New Delhi, India
Email: nsharma@icgeb.res.in, panchalsandeep24@gmail.com, *neeti@icgeb.res.in
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 25 June 2015; accepted 16 August 2015; published 19 August 2015
ABSTRACT
In the present study, we illustrate the strategy and protocol required to generate rice transgenics over-expressing the 21-nt form of Osa-miR820. The miR exists in two size variants of 21-nt and 24-nt so the natural precursor cannot be employed for the purpose of miR over-expression as the cellular machinery can process both size variants thereby masking the role of PTGS regulation. Hence, we adopted the artificial miR technology to specifically over-express the 21-nt species in the transgenics. During the course of experiments it was observed that the amiR constructs probably interfered with the regeneration of the transformed callus, necessitating protocol modifications. The results indicate the successful over-expression of the 21-nt miR species. These plants can serve as a useful source for the functional dissection of the role played by the 21-nt Osa-miR820 species. They will also be valuable in highlighting the importance for the existence of a dual mode of miR mediated target regulation.
Keywords:
Osa-miR820, Over-Expression, Agrobacterium, Artificial microRNA (amiR)

1. Introduction
Understanding the genetic framework of plants is an essential prerequisite for developing durable crop cultivars. This is highly recommended to meet the challenges of changing climatic conditions and for achieving the increasing demand for food and other products. Plant transformation is an important tool to understand the fundamental aspects of various genes and their functional roles in plants. This information can then be utilized to generate better crop varieties or can be translated for any other commercial purposes. The Agrobacterium mediated transformation is the method of choice in most cases due to certain inherent advantages such as its high transformation efficiency, low copy T-DNA integration and ability to transfer large DNA segments with minimal transgene rearrangement [1] [2] .
Rice is one of the most important cereal crops, feeding more than half of the world population. Despite being an essential part of the rural economy in Asia, rice is a sensitive crop as its yield is affected by abiotic stresses such as salinity, high temperature, drought, submergence or biotic stresses exerted by its various pathogens and pests. Rice transformation is a challenging task as it is comparatively less amenable to genetic manipulation, especially the indica cultivars [3] . Several methods have been reported for indica rice transformation, but are still unpredictable in terms of transformation efficiency and regenerative capacity of the explants [4] - [7] . This is mainly due to their dependence on the genotype of the plant, age of the callus, type of gene introduced into the genome etc. [3] [8] [9] . The successful regeneration of the callus is also influenced by the modulation of the intrinsic hormonal balance or media components. Moreover, T-DNA integration is affected by inoculation time, age of callus, cell density of Agrobacterium culture and additives such as acetosyringone, cysteine etc. [7] [10] - [13] .
The microRNAs (miRs) represent a class of 21- to 24-nt endogenously transcribed small RNAs, which do not code for proteins. They are transcribed as long primary miR (pri-miR) transcripts by RNA polymerase II [14] . These capped and polyadenylated pri-miRs are then processed by DCL into stem-loop shaped precursor intermediates (pre-miRs) which are in turn processed into the mature miRs. Although the miRs follow the same mode of biogenesis, they differ in the manner in which they function. The canonical 21-nt miRs can regulate gene expression at post-transcriptional level in a sequence-dependent manner [15] [16] . The 22-nt miRs have been implicated in tasiRNA (trans activating small interfering RNA) production from target RNAs [17] while the 24-nt miRs are known to regulate gene expression at the transcriptional levels by controlling DNA methylation [18] . There are several reports on the identification of conserved and novel miRs in rice. The role of these miRs was associated with different aspects of plant development or response to stress [19] - [22] .
One such study described the identification of a rice specific miR, Osa-miR820 from undifferentiated embryogenic callus tissues [23] . It was proposed to be encoded by CACTA transposon family, with its expression being controlled epigenetically at its own locus [24] [25] . Recently, it was shown to be down-regulated under drought [26] , salt [27] and arsenic [28] stress. The pre-miR820 can be processed by either DCL1 or DCL3 to yield 21-nt and 24-nt long mature miRs, respectively [18] . This suggests a dual function of miR820 with the 21-nt species acting in “trans” by directing the AGO1 mediated cleavage of DNA methyltransferase, OsDRM2, whereas the 24-nt species act through AGO4 mediated pathway to establish and/or maintain epigenetic modifications of OsDRM2 as well as its own locus [18] [25] . The presence of two length variants with common targets poses interesting quest regarding the functional significance of this miR in the biology of the rice plant.
In the present study, we illustrate the strategy and protocol required to generate rice transgenics over-express- ing the 21-nt form of Osa-miR820. These plants can serve as a useful source for the functional dissection of the role played by the 21-nt Osa-miR820 species. They will also be valuable in highlighting the importance for the existence of a dual mode of miR mediated target regulation. The natural precursor cannot be employed for the purpose of miR over-expression as it can be processed by the cellular machinery to produce both size variants, thereby masking the role of PTGS (Post Transcriptional Gene Regulation) regulation. Hence, we adopted the artificial miR technology to specifically over-express the 21-nt species in the transgenics. During the course of experiments it was observed that the amiR constructs probably interfered with the regeneration of the transformed callus, necessitating protocol modifications.
2. Methods
2.1. Plant Materials
Seeds of Pusa Basmati 1 (PB1) rice were used in this study. Mature dehusked seeds were surface sterilized by treating with 70% ethanol for 1 min followed by 10% sodium hypochlorite with a drop of Tween-20 for 30 minutes with constant slow agitation. The seeds are thoroughly washed repeatedly with sterile distilled water and then blot dry for subsequent use. For agroinfiltration assay, Nicotiana tabacum L. cv. Xanthi leaves from a young plant were selected.
2.2. Plasmid Construct
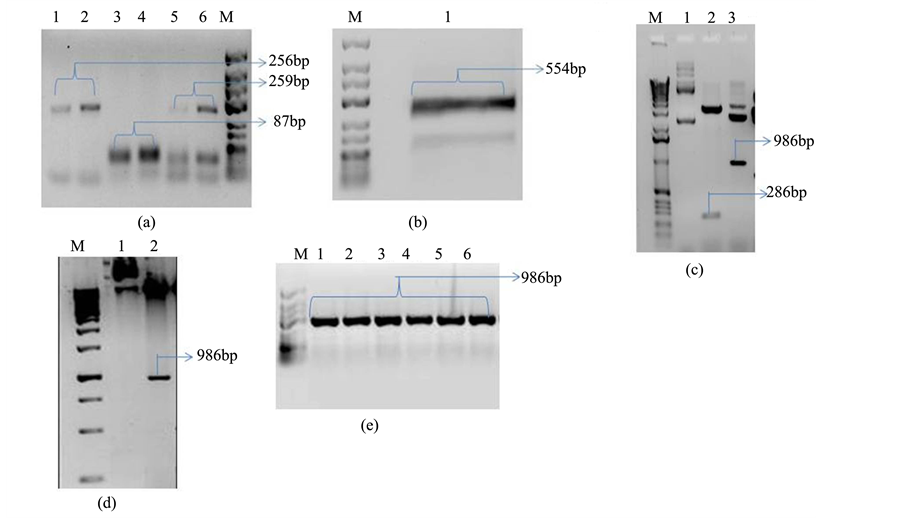
The conserved stem-loop backbone of Osa-miR528 in the amiR vector, pNW55, was used to generate the artificial pre-miR820. The primers (Table 1) specific to mature sequence of Osa-miR820 were designed using WMD3 (Web MicroRNA Designer 3) [29] and were used to replace the miR528 with our sequence of interest. The different combinations of primers were used to generate three sets of PCR fragments, of sizes 256 bp, 87 bp and 259 bp, respectively. These individual fragments were gel purified and fused via overlapping PCR using G3468 and G3469 universal primers to obtain an amplification product of 554 bp. These were trimmed to 286 bp using specific restriction sites of EcoRI and BamHI, as a part of forced cloning strategy (Figure 1).
Figure 1. Generation and cloning of amiR820 construct. (a) Amplification products of three individual PCRs required for construction of the amiR820; (b) Fusion PCR product of the three fragments to produce the 554 bp long artificial pre-miR; (c) Restriction digestion to confirm the pRT101-amiR820 plasmid, lane 1: uncut plasmid, lane 2: pre-miR insert released by EcoRI/BamHI digestion, lane 3: cassette specific insert released by HindIII digestion; (d) Restriction digestion of pCAMBIA1300-amiR820 plasmid, lane 1: uncut plasmid, lane 2: cassette specific insert released by HindIII digestion; (e) Colony PCR to confirm Agrobacterium transformation. Lanes 1-5: individual positive colonies. M: marker. Blue arrow indicates the size of 286 bp and red arrows indicate the size of 986 bp.
Cloning of amiR820 into Agrobacterium tumefaciens binary vector, pCAMBIA1300 was accomplished in three steps. In the first step the amplified PCR product of 286 bp was cloned into plant expression vector pRT101, using EcoRI and BamHI sites for directional cloning, under CaMV-35S promoter. The positive colonies were subsequently confirmed by double digestion with EcoRI/BamHI. The whole cassette was excised out from pRT101-amiR820 by restriction digestion with HindIII and transformed into pCAMBIA1300 binary vector using the same restriction enzyme. The recombinant pCAMBIA1300-amiR820 was confirmed by PCR and restriction digestion. The plasmid was transformed into Agrobacterium tumefaciens strain EHA105 and LBA4404 and positive colonies were screened and selected for rice transformation.
2.3. Agroinfiltration
Agrobacterium-strain containing the binary plasmid construct, pCAMBIA1300-amiR820, was checked for its functional performance in Nicotiana tabacum L. cv. Xanthi leaves. The agroinfiltration was achieved through pressure infiltration, as described previously [30] . Briefly, Agrobacterium culture was grown overnight in LB medium with the appropriate antibiotics and 20 µM acetosyringone. The cells were pelleted with a brief centri-
Table 1. List of primers used in the study.
fugation and re-suspended in MMA medium (MS salts, 10 mM MES, pH 5.6, 200 µM acetosyringone). The cells were incubated for at least 1 h at 28˚C and subsequently, diluted in MES buffer to get a OD600 of about 0.3 - 0.4. The homogenous culture mixture was infiltrated in the young leaves with the help of needleless syringe by generating a vacuum with the help of finger on the dorsal side of the leaf and mouth of the syringe on the ventral side. After 3, 5, 7 and 10 days post infiltration (dpi) the infiltrated zone was analyzed for miR820 expression.
2.4. RNA Isolation and Stem-Loop Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from infiltrated leaves using the modified Guanidium isothiocyanate extraction method [20] . The expression levels of mature miR820 were analysed using stem-loop end point PCR as described earlier [27] . Presence of miR820 was confirmed by sequencing, proving the functionality of this construct in vivo system. The 18S rRNA gene was used as a constitutive internal standard for equal loading. The amplification products were analyzed on 3% agarose gel.
2.5. DNA Extraction and Genomic DNA PCR
Genomic DNA was extracted from young leaves of putative transgenic and wild type plants using the CTAB based extraction method [31] . Briefly, 1 g of plant material was crushed in liquid nitrogen and homogenized using CTAB extraction buffer. The homogenized sample was kept at 65˚C for 30 minutes. Thereafter, the mixture was centrifuged at 10,000 rpm for 5 min and the supernatant was collected in a fresh tube. Equal volume of chloroform: isoamylalcohol (24:1) was added and the mixture was again centrifuged as mentioned above. Genomic DNA was precipitated by adding 2/3rd volume of isopropanol. The pellet was obtained after an incubation of 30 mins and washed twice with chilled 70% ethanol. The final pellet was air dried at room temperature and dissolved in TE buffer.100 ng of genomic DNA was used for PCR analysis in a 20 μl reaction mixture containing 2.0 μl 10X buffer, 0.3 μl of 10 mM dNTPs, 0.4 μl each of 10 mM primers (Table 1), 1U Taq polymerase, with a PCR program of 94˚C for 5 min, followed by 35 cycles of 94˚C for 30 s, 55˚C for 30 s, and 72˚C for 30 s, with a final extension of 72˚C for 5 min. The amplification product (286 bp) was checked on 2 % agarose gel and photographed.
2.6. Southern Blot
20 µg of genomic DNA from PCR-positive rice lines was digested with KpnI, run on 0.8% agarose gel and blotted on HybondN+ membranes (Amersham Pharmacia). Non-radioactive DIG-labelled 445 bp of hygromycin gene was used as a probe and the experiment was performed as per manufacturer’s guide (Roche, Life Sciences, USA). Hybridization was performed at 62˚C for 12 hours and after washing it was imaged on an X-ray film using standard procedures.
2.7. Callus Induction
Surface-sterilized seeds of rice cultivar Pusa Basmati 1 (PB1) were dried on autoclaved Whatman paper and incubated on callus induction media (CIM) in dark at 25˚C - 28˚C. The CIM favoured the development of the scutellar region into a highly regenerative calli within a period of 3 weeks. The calli were excised and sub-cul- tured onto fresh CIM in dark for 7 days.
For co-infection, the sub-cultured calli were immersed in the Agrobacterium suspension for 35 minutes with continuous slow shaking. After infection, calli were blot dried on sterile filter papers and then were incubated on filter paper moistened with co-cultivation medium (CCM) at 26˚C - 28˚C for two days, in dark. After co-culti- vation, the calli were washed with autoclaved distilled water twice for 30 min each containing both carbenicillin and cefotaxime (250 mg/ml each). In order to dilute the toxic effect of antibiotics, final wash was given by N6 liquid media (without any sugar) for 5 - 7 minutes and then it was kept on resting media (RM) for 7-10 days so that it regains the proliferating capacity.
Thereafter, the calli were washed and dried on sterile filter papers and cultured on callus selection medium (CSM) in dark for selecting transgenic calli. After first round of selection for 20 days, brownish or black colored calli were discarded and white calli were transferred to fresh CSM medium for second selection cycle for 15 days. This step allowed the proliferation of micro calli and when micro calli started growing on the mother calli, each micro callus was gently separated from the mother calli and transferred to fresh CSM medium for the third selection for 15 days. Healthy calli were selected for regeneration.
After the third selection, healthy calli were transferred to the specific regeneration medium (RM1-4) and incubated in dark in culture room for 7 days. After which they were transferred to fresh regeneration medium and incubated at 26˚C - 28˚C under light. After 1 - 2 weeks, green buds were seen arising from the calli. The green buds developed into shoots and were transferred to rooting medium (RoM) in presence of hygromycin (50 mg/l) under light for 20 days. The whole plants were transferred to vermiculite pots before being transferred to the soil pots and grown in the green house.
The CIM, CCM, CSM and RoM media compositions used are detailed below. These were adapted from Sahoo, et al. 2011 [11] with some modification.
CIM: MS salts with Vitamin B5 + 30 g/l sucrose + 0.3 g casein hydrolysate + 2.0 mg/l 2,4-D + 0.5 g proline + 0.3% phytagel, pH 5.8;
CCM: Liquid N6 media + 3 mg/ml 2,4-D + 100 µM acetosyringone, pH 5.2;
RM: CIM with no antibiotic selection;
CSM: CIM + 250 cefotaxime + 50 mg/l hygromycin;
RM1: MS salts with Vitamin B5 + 500 mg proline + 0.3 g casein hydrolysate + 30 g/l sorbitol + 1.0 mg/l BAP + 2.0 mg/l Kinetin + 0.5 mg/l NAA, pH 5.8;
RoM: MS salts with Vitamin B5 + 30 g sucrose + 3g/l phytagel, pH 5.8.
3. Results and Discussion
3.1. Construction and Validation of Artificial Osa-miR820
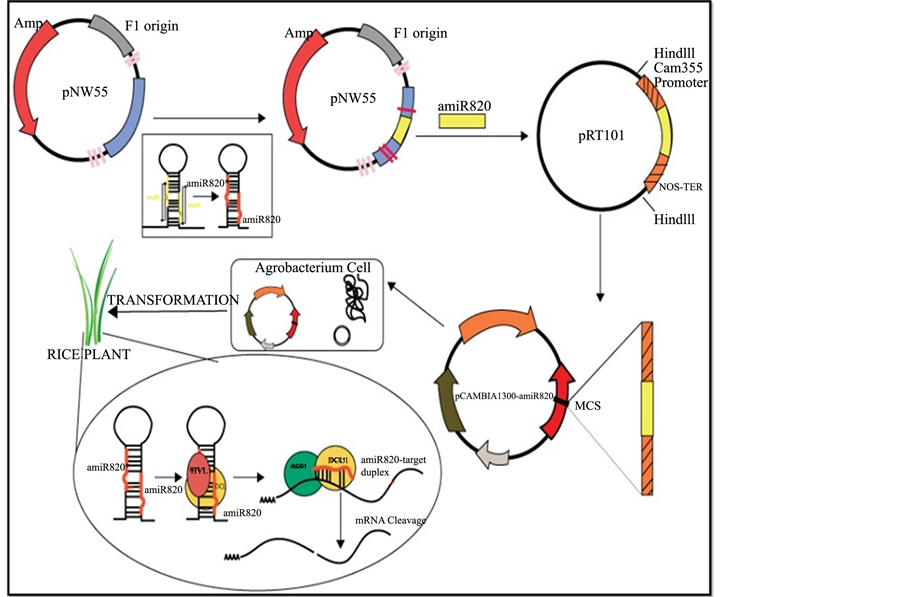
Artificial miRs (amiRs) are exclusively engineered pre-miR molecules which can produce the desired mature miR sequences when expressed in vivo. The amiRs have been widely used in directed gene silencing technology to specifically suppress the expression of target mRNAs in wide variety of plants [29] [32] [33] . In this study we have applied the amiR technology as a novel alternative tool to over-express the 21-nt form of the Osa-miR820. This miR is encoded as two length variants of 21-nt and 24-nt from five different loci [25] . While there is absolute similarity between the mature Osa-mR820 sequences, their precursor sequences share more than 90% similarity. As reported earlier the digital expression status of Osa-miR820 family, in the leaf and panicle tissues of PB1, revealed that the 24-nt species accumulated to a higher level as compared to the 21-nt species [27] . Therefore, to decipher the functional role of the 21-nt Osa-miR820 species we adopted the amiR approach, to allow its over-expression in indica rice over and above the endogenous levels. The schematic representation of the amiR820 over expression in rice is shown in Figure 2.
To confirm whether the amiR820 construct was processing the correct 21-nt mature miR an in planta assay was performed by agroinfiltrating the pCAMBIA1300-amiR820 construct in tobacco leaves (Figure 3(a)). The infiltrated zones were analyzed for Osa-miR820 expression at 3, 5, 7 and 10 dpi, while using the uninfiltrated leaf as control. Stem-loop RT-PCR revealed that there was gradual increase in the levels of Osa-miR820 from 3 dpi to 7dpi with maximum expression observed at 7 dpi (Figure 3(b)). Beyond this, the levels started to decrease, as expected, because the transient expression allowed the miR biogenesis machinery to work for a specified time in planta in the infiltrated zones. The amplified bands were sequenced to confirm the presence of the 21-nt Osa-miR820 sequence.
3.2. Production of Osa-miR820 Over-Expressing Rice
Agrobacterium mediated transformation of callus is the preferred method for stable transformation of genes in rice [34] . However, the efficiency of callus induction and proliferation is largely genotype based and is also influenced by other factors such as hormonal and amino acids supplements [35] . In the present study, mature embryo was used as the choice of explant to develop callus, as it is reported to be most responsive. The highly regenerative callus obtained was used for infection with Agrobacterium containing pCAMBIA1300-amiR820 constructs.
Both EHA105 and LBA4404 Agrobacterium-strains were used for evaluating the transformation efficiency, but a greater number of positive transformants were obtained with LBA4404 as compared to EHA105. It was ob-
Figure 2. The schematic representation of strategy employed for amiR820 generation and subsequent rice transformation.
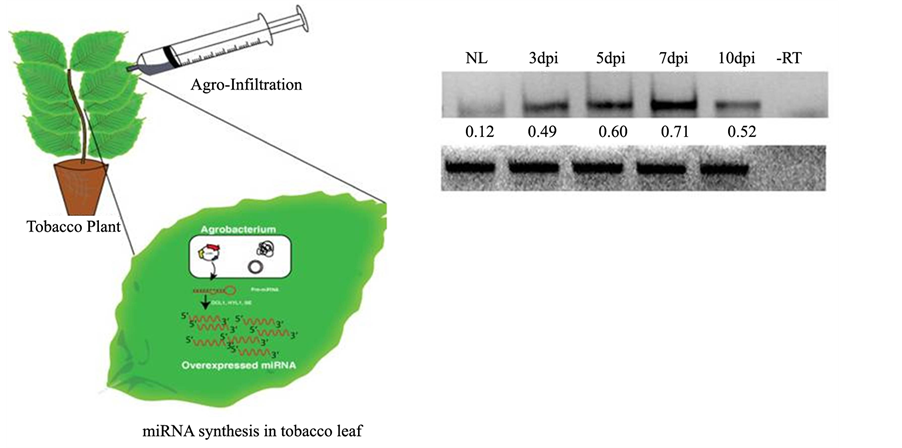
Figure 3. Agro-infiltration assay to check the functionality of amiR820. (a) Overview of the in planta agro- infiltration assay; (b) Stem-loop RT PCR for amplifying the mature sequence of amiR820 from un-infiltrated (NL: normal leaf) and infiltrated regions of tobacco. Leaves at 3, 5, 7 and 10 days post inoculation (dpi). 18S rRNA gene is used as an internal loading control. ?RT: negative control. Normalized intensity values are given below each lane.
served that EHA105 being a super virulent strain [36] leads to overgrowth of Agrobacterium in intial stages of selection which made subsequent handling tedious while, LBA4404, worked in a balanced manner providing effective infection with minimum amount of Agrobacterium contamination during the selection process. This decrease in the Agrobacterium growth reduced the use of excess antibiotics in the selection media and the frequency of sub-culturing to fresh selection media and may indirectly contribute to an enhanced number of regenerative calli.
Co-cultivation of Agrobacteria with the callus helps for better penetration of the adhered bacterium inside the totipotent cells. Phenolic compounds such as acetosyringone are known to induce greater Agrobacterium infection into the callus [13] . In this protocol, a standard final concentration of 100 µM acetosyringone was added just prior to inoculation. After infection, callus were blot dry and kept on filter paper wicks rather than solid media, based on earlier reports that filter paper wicks increase Agrobacterium infection efficiency [37] [38] . After co-cultivation, callus is kept on resting media to avoid over-growth for 7 - 10 days in the callus induction media without hygromycin selection so that callus regains their proliferating capacity. Such a scheme was earlier proposed in rice and ryegrass, but unlike in our case, they used resting media along with the selection antibiotic [10] . The proliferating callus is then put on selection medium for 3 weeks with regular changing of media as and when required.
Regeneration of callus into “green mass” which has the capacity to develop into shootlets and plantlets is another criticial step in rice plant transformation. It is significant to point here that though several excellent protocols are available for regeneration from rice callus, but the frequency of regenerants varies drastically. This indicates that regeneration frequency is dependent on multiple factors such as genotype, type of explants, concentration of growth regulators and presence of osmolytes. The nature of construct that is used for transformation also influences regeneration potential of the callus [7] . This may be attributed to the fact that some genes may have an additional advantage for the survival of the callus over and above its inherited function. It was observed that using the available protocols we were unable to get regenerants for pCAMBIA1300-amiR820 constructs though wild type and pCAMBIA1300 vector backbones supported regeneration. This indicated that the nature of the construct might be responsible for such recalcitrance.
A careful analysis of the targets of Osa-miR820 using psRNA target finder identified a putative NAC domain protein 77 as a potential target [27] . This indicated a disturbance in the auxin pathway. Hence the composition of the RM1 was suitably modified to induce green calli (Table 2). Regeneration was observed with a combination of high cytokinins supplemented with 30 g/l sorbitol concentration in a 1% agarose based media [9] . Sorbitol was used as the main carbon source during regeneration for pCAMBIA1300-amiR820 calli as compared to maltose that was being previously reported as the preferred sugar for indica rice transformation [5] [39] . Interestingly, 30 g/l of sorbitiol was found to inhibit the regeneration in two recalcitrant upland rice cultivars, Kusan and Siam. Like other sugars, it acts as an osmotic regulator during callus tissue culture and is known to initiate desiccation in callus to induce a higher proliferating rate. It has now been proved by several studies that sorbitol has a greater role to play in rice regeneration as a carbon source or an osmotic regulator as it is showed that sorbitol regulates the expression of several genes, acting as a chemical signalling molecule to divert important cellular processes and signal transduction machinery to favour regeneration of callus by inducing organogeneisis [40] - [42] .
The modified protocol supported the development of green callus in 7 days for the pCAMBIA1300-amiR820 constructs as compared to 4 - 5 days for the wild type and pCAMBIA1300 vector constructs. The subsequent shoot formation was delayed by around 3 days for the pCAMBIA1300-amiR820 constructs. This method supported a 27% transformation efficiency of amiR820 which was less than in case of control (>50%), but it is far better than that reported for other constructs in various rice cultivars ranging from 9% to 12% [43] - [45] . Regenerated shoots were then put on RoM and acclimatized to greenhouse conditions by transferring them to vermiculite pots.
3.3. Molecular Analysis of Osa-miR820 Over-Expressing Rice
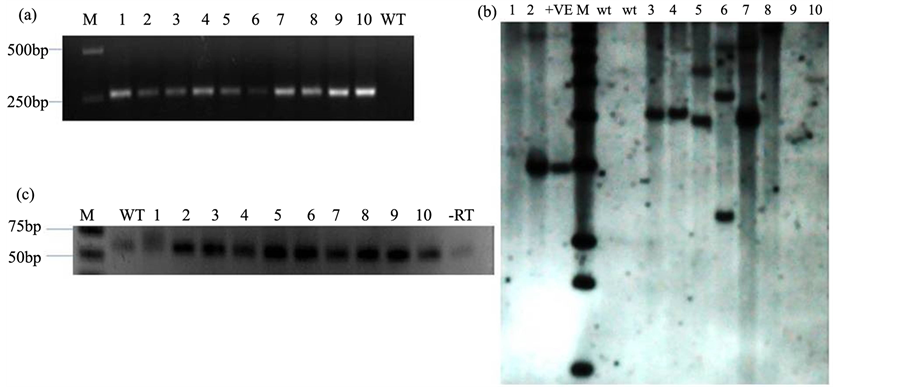
The putative transgenic lines were screened for transgene integration by genomic PCR using specific primers. A fragment of 286 bp was amplified in all transgenic events except for in wild type. pCAMBIA1300-amiR820 plasmid was used as a positive control (Figure 4(a)). The copy number of transgenic rice lines was also confirmed by Southern blot analysis using probe specific for the hygromycin gene (Figure 4(b)). It was observed that both single copy and multiple copy integrations were present in the different lines.
To analyse the extent of over-expression of mature miR820 in putative transgenic lines, endpoint stem-loop
Table 2. Effect of different media on the regeneration of calli transformed with pCAMBIA1300-amiR820 constructs.
Figure 4. Molecular analysis of putative transgenic rice plants. (a) Genomic PCR using specific primers, lane 1 - 10: transgenic lines, WT-untransformed wild type PB1; (b) Southern blot for putative rice transgenic plants. Lanes 1 - 10: putative transgenic lines, +ve: pCAMBIA1300-amiR820 plasmid, wt: untransformed wild type plant; (c) Stem-loop RT-PCR to identify over-expressing lines. Lanes 1 - 10: putative transgenic lines, wt: wild type untransformed plant, −RT: negative control, M: marker.
RT PCR was done to capture expression profiles of miR820 in these lines. As shown in Figure 4(c), most of the putative transgenic events were over-expressing miR820 as compared to the wild type (untransformed) PB1 plants. Hence, it was confirmed that amiR820 was successfully transformed in PB1 rice plants with an over-ex- pression of the 21-nt Osa-miR820 in vivo.
3.4. Conclusion
In the present work, we report a successful transformation of rice with artificial miR construct to over-express the 21-nt species of Osa-miR820. It was observed that the construct interfered with the regeneration potential of the callus warranting hormonal modifications in the protocol. These transgenics will be subjected to further analysis to unravel the functional role and requirement of the 21-nt Osa-miR820 species in regulating the plant biology.
Acknowledgements
The authors would like to thank Prof. Detlef Weigel for providing the pNW55 construct. NS acknowledges the fellowship received from CSIR, India. The research was supported by financial grants received from the Department of Biotechnology, Government of India.
Cite this paper
NehaSharma,SandeepPanchal,NeetiSanan-Mishra, (2015) Protocol for Artificial MicroRNA Mediated Over-Expression of miR820 in Indica Rice. American Journal of Plant Sciences,06,1951-1961. doi: 10.4236/ajps.2015.612196
References
- 1. Birch, R.G. (1997) Plant Transformation: Problems and Strategies for Practical Application. Annual Review of Plant Biology, 48, 297-326.
http://dx.doi.org/10.1146/annurev.arplant.48.1.297 - 2. Hansen, G. and Wright, M.S. (1999) Recent Advances in the Transformation of Plants. Trends in Plant Science, 4, 226-231.
http://dx.doi.org/10.1016/S1360-1385(99)01412-0 - 3. Zaidi, M., Narayanan, M., Sardana, R., Taga, I., Postel, S., Johns, R., McNulty, M., Mottiar, Y., Mao, J. and Loit, E. (2006) Optimizing Tissue Culture Media for Efficient Transformation of Different Indica Rice Genotypes. Agronomy Research, 4, 563-575.
- 4. Hiei, Y. and Komari, T. (2006) Improved Protocols for Transformation of Indica Rice Mediated by Agrobacterium tumefaciens. Plant Cell, Tissue and Organ Culture, 85, 271-283.
http://dx.doi.org/10.1007/s11240-005-9069-8 - 5. Lin, Y.J. and Zhang, Q. (2005) Optimising the Tissue Culture Conditions for High Efficiency Transformation of Indica Rice. Plant Cell Reports, 23, 540-547.
http://dx.doi.org/10.1007/s00299-004-0843-6 - 6. Mohanty, A., Kathuria, H., Ferjani, A., Sakamoto, A., Mohanty, P., Murata, N. and Tyagi, A. (2002) Transgenics of an Elite Indica Rice Variety Pusa Basmati 1 Harbouring the codA Gene Are Highly Tolerant to Salt Stress. Theoretical and Applied Genetics, 106, 51-57.
- 7. Sah, S., Kaur, A., Kaur, G. and Cheema, G. (2014) Genetic Transformation of Rice: Problems, Progress and Prospects. Journal of Rice Research, 3, 2.
- 8. Ge, X., Chu, Z., Lin, Y. and Wang, S. (2006) A Tissue Culture System for Different Germplasms of Indica Rice. Plant Cell Reports, 25, 392-402.
http://dx.doi.org/10.1007/s00299-005-0100-7 - 9. Kant, P., Kant, S., Jain, R. and Chaudhury, V. (2007) Agrobacterium-Mediated High Frequency Transformation in Dwarf Recalcitrant Rice Cultivars. Biologia Plantarum, 51, 61-68.
http://dx.doi.org/10.1007/s10535-007-0012-1 - 10. Patel, M., Dewey, R. and Qu, R. (2013) Enhancing Agrobacterium tumefaciens-Mediated Transformation Efficiency of Perennial Ryegrass and Rice Using Heat and High Maltose Treatments during Bacterial Infection. Plant Cell, Tissue and Organ Culture, 114, 19-29.
http://dx.doi.org/10.1007/s11240-013-0301-7 - 11. Sahoo, K.K., Tripathi, A.K., Pareek, A., Sopory, S.K. and Singla-Pareek, S.L. (2011) An Improved Protocol for Efficient Transformation and Regeneration of Diverse Indica Rice Cultivars. Plant Methods, 7, 49.
http://dx.doi.org/10.1186/1746-4811-7-49 - 12. Shri, M., Rai, A., Verma, P., Misra, P., Dubey, S., Kumar, S., Verma, S., Gautam, N., Tripathi, R., Trivedi, P. and Chakrabarty, D. (2013) An Improved Agrobacterium-Mediated Transformation of Recalcitrant Indica Rice (Oryza sativa L.) Cultivars. Protoplasma, 250, 631-636.
http://dx.doi.org/10.1007/s00709-012-0439-x - 13. Tripathi, R., Bisht, H. and Singh, R. (2010) Effect of Acetosyringone and Callus Age on Transformation for Scutellum-Derived Callus of Rice. International Journal of Pharma and Bio Sciences, 4, 163-170.
- 14. Lee, Y., Kim, M., Han, J., Yeom, K.-H., Lee, S., Baek, S.H. and Kim, V.N. (2004) MicroRNA Genes Are Transcribed by RNA Polymerase II. The EMBO Journal, 23, 4051-4060.
http://dx.doi.org/10.1038/sj.emboj.7600385 - 15. Rogers, K. and Chen, X. (2013) Biogenesis, Turnover, and Mode of Action of Plant microRNAs. The Plant Cell, 25, 2383-2399.
http://dx.doi.org/10.1105/tpc.113.113159 - 16. Voinnet, O. (2009) Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell, 136, 669-687.
http://dx.doi.org/10.1016/j.cell.2009.01.046 - 17. Cuperus, J.T., Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Burke, R.T., Takeda, A., Sullivan, C.M., Gilbert, S.D., Montgomery, T.A. and Carrington, J.C. (2010) Unique Functionality of 22-nt miRNAs in Triggering RDR6-Dependent siRNA Biogenesis from Target Transcripts in Arabidopsis. Nature Structural & Molecular Biology, 17, 997-1003.
http://dx.doi.org/10.1038/nsmb.1866 - 18. Wu, L., Zhou, H., Zhang, Q., Zhang, J., Ni, F., Liu, C. and Qi, Y. (2010) DNA Methylation Mediated by a microRNA Pathway. Molecular Cell, 38, 465-475.
http://dx.doi.org/10.1016/j.molcel.2010.03.008 - 19. Jones-Rhoades, M.W. and Bartel, D.P. (2004) Computational Identification of Plant microRNAs and Their Targets, including a Stress-Induced miRNA. Molecular Cell, 14, 787-799.
http://dx.doi.org/10.1016/j.molcel.2004.05.027 - 20. Mittal, D., Mukherjee, S.K., Vasudevan, M. and Mishra, N.S. (2013) Identification of Tissue-Preferential Expression Patterns of Rice miRNAs. Journal of Cellular Biochemistry, 114, 2071-2081.
http://dx.doi.org/10.1002/jcb.24552 - 21. Sanan-Mishra, N., Kumar, V., Sopory, S.K. and Mukherjee, S.K. (2009) Cloning and Validation of Novel miRNA from Basmati Rice Indicates Cross Talk between Abiotic and Biotic Stresses. Molecular Genetics and Genomics, 282, 463-474.
http://dx.doi.org/10.1007/s00438-009-0478-y - 22. Sunkar, R., Chinnusamy, V., Zhu, J. and Zhu, J.K. (2007) Small RNAs as Big Players in Plant Abiotic Stress Responses and Nutrient Deprivation. Trends in Plant Sciences, 12, 301-309.
http://dx.doi.org/10.1016/j.tplants.2007.05.001 - 23. Luo, Y.-C., Zhou, H., Li, Y., Chen, J.-Y., Yang, J.-H., Chen, Y.-Q. and Qu, L.-H. (2006) Rice Embryogenic Calli Express a Unique Set of microRNAs, Suggesting Regulatory Roles of microRNAs in Plant Post-Embryogenic Development. FEBS Letters, 580, 5111-5116.
http://dx.doi.org/10.1016/j.febslet.2006.08.046 - 24. Nosaka, M., Ishiwata, A., Shimizu-Sato, S., Ono, A., Ishimoto, K., Noda, Y. and Sato, Y. (2013) The Copy Number of Rice CACTA DNA Transposons Carrying MIR820 Does Not Correlate with MIR820 Expression. Plant Signaling & Behavior, 8, Article ID: e25169.
http://dx.doi.org/10.4161/psb.25169 - 25. Nosaka, M., Ono, A., Ishiwata, A., Shimizu-Sato, S., Ishimoto, K., Noda, Y. and Sato, Y. (2013) Expression of the Rice microRNA miR820 Is Associated with Epigenetic Modifications at Its Own Locus. Genes & Genetic Systems, 88, 105-112.
http://dx.doi.org/10.1266/ggs.88.105 - 26. Jeong, D.-H., Park, S., Zhai, J., Gurazada, S.G.R., De Paoli, E., Meyers, B.C. and Green, P.J. (2011) Massive Analysis of Rice Small RNAs: Mechanistic Implications of Regulated microRNAs and Variants for Differential Target RNA Cleavage. The Plant Cell, 23, 4185-4207.
http://dx.doi.org/10.1105/tpc.111.089045 - 27. Sharma, N., Tripathi, A. and Sanan-Mishra, N. (2015) Profiling the Expression Domains of a Rice-Specific microRNA under Stress. Frontiers in Plant Science, 6, 333.
http://dx.doi.org/10.3389/fpls.2015.00333 - 28. Sharma, D., Tiwari, M., Lakhwani, D., Tripathi, R.D. and Trivedi, P.K. (2015) Differential Expression of microRNAs by Arsenate and Arsenite Stress in Natural Accessions of Rice. Metallomics, 7, 174-187.
http://dx.doi.org/10.1039/C4MT00264D - 29. Warthmann, N., Chen, H., Ossowski, S., Weigel, D. and Hervé, P. (2008) Highly Specific Gene Silencing by Artificial miRNAs in Rice. PLoS ONE, 3, e1829.
- 30. Karjee, S., Islam, M. and Mukherjee, S. (2008) Screening and Identification of Virus-Encoded RNA Silencing Suppressors. In: Barik, S., Ed., RNAi, Humana Press, New York, 187-203.
- 31. Murray, M.G. and Thompson, W.F. (1980) Rapid Isolation of High Molecular Weight Plant DNA. Nucleic Acids Research, 8, 4321-4325.
http://dx.doi.org/10.1093/nar/8.19.4321 - 32. Khraiwesh, B., Ossowski, S., Weigel, D., Reski, R. and Frank, W. (2008) Specific Gene Silencing by Artificial microRNAs in Physcomitrella patens: An Alternative to Targeted Gene Knockouts. Plant Physiology, 148, 684-693.
http://dx.doi.org/10.1104/pp.108.128025 - 33. Schwab, R., Ossowski, S., Riester, M., Warthmann, N. and Weigel, D. (2006) Highly Specific Gene Silencing by Artificial microRNAs in Arabidopsis. The Plant Cell, 18, 1121-1133.
http://dx.doi.org/10.1105/tpc.105.039834 - 34. Hiei, Y., Ohta, S., Komari, T. and Kumashiro, T. (1994) Efficient Transformation of Rice (Oryza sativa L.) Mediated by Agrobacterium and Sequence Analysis of the Boundaries of the T-DNA. The Plant Journal, 6, 271-282.
http://dx.doi.org/10.1046/j.1365-313X.1994.6020271.x - 35. Niroula, R., Sah, B., Bimb, H. and Nayak, S. (2007) Effect of Genotype and Culture Media on Callus Induction and Plant Regeneration from Matured Rice Grain Culture. Journal of the Institute of Agriculture and Animal Science, 26, 21-26.
- 36. Hood, E., Gelvin, S., Melchers, L. and Hoekema, A. (1993) New Agrobacterium Helper Plasmids for Gene Transfer to Plants. Transgenic Research, 2, 208-218.
http://dx.doi.org/10.1007/BF01977351 - 37. Nanasato, Y., Konagaya, K.-I., Okuzaki, A., Tsuda, M. and Tabei, Y. (2011) Agrobacterium-Mediated Transformation of Kabocha Squash (Cucurbita moschata Duch) Induced by Wounding with Aluminum Borate Whiskers. Plant Cell Reports, 30, 1455-1464.
http://dx.doi.org/10.1007/s00299-011-1054-6 - 38. Nanasato, Y., Konagaya, K.-I., Okuzaki, A., Tsuda, M. and Tabei, Y. (2013) Improvement of Agrobacterium-Mediated Transformation of Cucumber (Cucumis sativus L.) by Combination of Vacuum Infiltration and Co-Cultivation on Filter Paper Wicks. Plant Biotechnology Reports, 7, 267-276.
http://dx.doi.org/10.1007/s11816-012-0260-1 - 39. Jain, R.K., Davey, M.R., Cocking, E.C. and Wu, R. (1997) Carbohydrate and Osmotic Requirements for High-Frequency Plant Regeneration from Protoplast-Derived Colonies of Indica and Japonica Rice Varieties. Journal of Experimental Botany, 48, 751-758.
http://dx.doi.org/10.1093/jxb/48.3.751 - 40. Feng, X., Zhao, P., Hao, J., Hu, J., Kang, D. and Wang, H. (2011) Effects of Sorbitol on Expression of Genes Involved in Regeneration of Upland Rice (Oryza sativa L.). Plant Cell, Tissue and Organ Culture, 106, 455-463.
http://dx.doi.org/10.1007/s11240-011-9943-5 - 41. Geng, P., La, H., Wang, H. and Stevens, E.J. (2008) Effect of Sorbitol Concentration on Regeneration of Embryogenic Calli in Upland Rice Varieties (Oryza sativa L.). Plant Cell, Tissue and Organ Culture, 92, 303-313.
http://dx.doi.org/10.1007/s11240-007-9321-5 - 42. Steinitz, B. (1999) Sugar Alcohols Display Nonosmotic Roles in Regulating Morphogenesis and Metabolism in Plants That Do Not Produce Polyols as Primary Photosynthetic Products. Journal of Plant Physiology, 155, 1-8.
http://dx.doi.org/10.1016/S0176-1617(99)80133-3 - 43. Azhakanandam, K., McCabe, M.S., Power, J.B., Lowe, K.C., Cocking, E.C. and Davey, M.R. (2000) T-DNA Transfer, Integration, Expression and Inheritance in Rice: Effects of Plant Genotype and Agrobacterium Super-Virulence. Journal of Plant Physiology, 157, 429-439.
http://dx.doi.org/10.1016/S0176-1617(00)80028-0 - 44. Khanna, H.K. and Raina, S.K. (1999) Agrobacterium-Mediated Transformation of Indica Rice Cultivars Using Binary and Superbinary Vectors. Functional Plant Biology, 26, 311-324.
http://dx.doi.org/10.1071/pp98160 - 45. Zhang, J., Xu, R.J., Elliott, M.C. and Chen, D.F. (1997) Agrobacterium-Mediated Transformation of Elite Indica and Japonica Rice Cultivars. Molecular Biotechnology, 8, 223-231.
http://dx.doi.org/10.1007/BF02760776
NOTES
*Corresponding author.