American Journal of Plant Sciences
Vol.4 No.3(2013), Article ID:28977,7 pages DOI:10.4236/ajps.2013.43068
Construction and Analysis of SSH-cDNA Library from Leaves of Susceptible Rubber Clone Resistant to Powdery Mildew Induced by BTH*
![]()
Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences/Key Laboratory of Integrated Pest Management of Tropical Crops, Ministry of Agriculture/Key Laboratory of Pests Detection and Control for Tropical Agriculture, Haikou, China.
Email: †fanweed@hotmail.com
Received January 9th, 2013; revised February 19th, 2013; accepted February 26th, 2013
Keywords: Benzothiadiazole; ESTs; Hevea brasiliensis; Induced Resistance; Oidium heveae; Suppression Subtractive Hybridization
ABSTRACT
To understand the mechanism of benzothiadiazole (BTH)-induced susceptible rubber clone resistance to powdery mildew on gene level, a differentially expressed cDNA library was constructed by suppression subtractive hybridization (SSH) with rubber Reyan 7-33-97 clone. The constructed cDNA library was high integrity through detection of the critical processes of SSH, such as efficiency of adaptor connection, subtraction and conversion, as well as the type of recombinant genes. The positive rate was 99% after identification with random 400 white spots. The size of the cDNA clone inserted fragments was various but most in 400 bp - 1000 bp. There were 23 cDNA sequences matching the function of energy and basic metabolism, signal transduction, membrane and transport, secondary metabolism and so on after detection of the 42 positive clone sequences selected randomly from the cDNA library and comparison on nucleic acid sequences in Genbank. 7 ESTs were logged in Genbank and accession numbers were GW873071 and GW874604- GW874610. The results implicated that BTH could effectively induced rubber tree resistance to powdery mildew through increasing expresses of defense-related genes in leaves of rubber tree susceptible clone. It should provide a new approach for rubber disease management.
1. Introduction
Rubber tree (Hevea brasiliensis) is a very important tropical industrial crop. Powdery mildew disease caused by Oidium heveae is one of the most important leaf diseases and impacts severely to the growth and latex production of rubber trees [1]. To control this disease, rubber resistance clone breeding and chemical control were employed generally [2]. Recently, induced rubber resistance to powdery mildew by oligosaccharin [3] and BTH (benzothiadiazole-7-carbothioic acid S-methyl ester or acibenzolar-S-methyl) [4] and to anthracnose (Colletotrichum gloeosporioides) by BTH [5] was studied.
BTH, analogs of salicylic acid, is an excellent chemical inducer that can induce plant resistance to pathogens [6], insects [7], nematodes [8] and parasitic weeds [9]. The mechanism of BTH induced resistance is activation of the plant defense genes and expression resistance-related proteins or enzymes [10]. The defense-related genes of BTH induced diseases resistance to rice, wheat [11], cucumber [12], papaya [13], coffee [14], cocoa [15] and so on were identified and analysed. In rubber tree susceptible clone against the diseases, the peroxidase, phenylalanine aminolyase and β-1,3 glucanase are increasing significantly in leaves after BTH treatment [4,5]. No report has been found so far on BTH induced defense genes in rubber tree susceptible clones against the disease. To understand the mechanism of BTH induced rubber tree resistance to powdery mildew in molecular knowledge, a cDNA library of BTH-induced resistance to the disease of rubber tree was constructed through SSH (suppression subtractive hybridization) and the function of differentially expressed genes induced by BTH was analysed.
2. Materials and Methods
2.1. Plant Growth and Pathogen Collection
Rubber budding seedlings (Reyan 7-33-97 clone, susceptible to powdery mildew [16]) were provided by Rubber Research Institute of CATAS. The seedlings were grown in plastic bags with leaves bronze to light green stage and kept in plant growth chamber with temperature at 25˚C, humidity at 80% and 12 h/12 h at light/dark. Fresh conidia of powdery mildew (Oidium heveae) were collected in suspension from infected rubber tree leaves and adjusted to 5 × 104 spores/ml under a microscope.
2.2. BTH Treatment and the Pathogen Inoculation
The healthy leaves of rubber budding seedlings were sprayed uniformly with BTH (Bion 50%WG, Syngenta) at 250 mg a.i. l−1. The spore suspension was sprayed on the leaves after 5 days of BTH spray. The leaves were collected after 4 days of inoculation. Blank control was the some clone and only inoculated with the pathogen. The leaves were stored at −70˚C till for total RNA extraction.
2.3. Total RNA Extraction and mRNA Purification
The BTH treatment and the pathogen inoculation were as the tester and the only pathogen inoculation was as the driver. The total RNAs of tester and driver leaves were extracted with the modified Bugos extract [100 mmol·l−1 Tris, 200 mmol·l−1 Nacl, 15 mmol·l−1 EDTA, 0.5% (W/V) SDS]. The total RNA was dissolved in DEPC-H2O after treatment with DNase I (TaKaRa, Dalian, China) for 30 min at 37˚C. The extracted RNA was kept in cryopreservation for integrity examination with 1.2% agarose gel electrophoresis and amount and purity detection with UV at A260/280. mRNAs of tester and driver from the RNAs were purified with OligotexTM-dT30
2.4. Differentially Expressed cDNA Library Construction, Adaptor Ligation Detection and Transformation
The differentially expressed cDNA library of BTH induced resistance to the pathogen was constructed with PCR-SelectTM cDNA Subtraction kit (TaKaRa, Dalian, China). The adaptor ligation efficiency before and after SSH was detected by primer of Actin gene [17] (a housekeeping gene of rubber, designed as ACTIN-F: 5’- CAGTGGTCGTACAACTGGTAT-3’ and ACTIN-R: 5’- ATCCTCCAATCCAGACACTGT-3’, synthesized by SBS Genetech, Beijing, China). Adaptor ligation 1 or 2 was used as a template for the tester cDNA. PCR amplification was conducted respectively with the primer Actin 3’ as one side and Actin 5’ or adaptor ligation 1 as another side. The purpose cDNA fragments 3 μl, pMD18-T Vector (TaKaRa, Dalian, China) 1 μl, Ligation Solution I 5 μl and ddH2O 1 μl were mixed in microcentrifuge tube and slightly centrifuged, then transformed into E. coli DH5α. The droplets on the tube wall were put down on the bottom of test tube and overnight at 16˚C. 50 μl of competence was poured into 1.5 ml centrifuge tube with 10 μl ligation product. The tube was placed on ice bath for 20 min, shocked at 42˚C for 90 sec and immediately placed on the ice bath for 2 min again. The tube was shook at 37˚C for 45 min after addition of 700 μl LB medium.
2.5. cDNA Library Reorganization Rate Detection
The conversion products were coated on LB ampicillin plates with X-gal and IPTG and cultured at 37˚C under dark conditions for 12 - 16 h. The positive clones were identified after coloration at 4˚C when the colony size was suitable. Recombination rate of cDNA library was calculated.
2.6. Fragment Length Recognization and Recombinant Sequence
A 25 μl reaction system [10 × Buffer (Mg2+) 2.5 μl, dNTP (2.5 M) 2 μl, rTaq (5 U/μl) (Sangon Biotech, Shanghai, China) 0.25 μl, primer 1 (20 pm) 1.5 μl, primer 2 (20 pm) 1.5 μl, template 0.5 μl, sterile water 16.75 μl] was prepared in 0.2ml PCR tube and immediately accessed to the following cycle after centrifugation: 4.5 min at 94˚C, 35 sec at 94˚C, 30 sec at 66˚C for 30 cycles; then 1.5 min at 72˚C and 5 min at 72˚C. The reactant of 5 μl was loaded on 1.2% agarose gel electrophoresis. The inserted fragments of positive clones identified by PCR were sequenced.
3. Results
3.1. The Total RNA Extraction
2 bright bands of total RNAs were appeared after agarose gel electrophoresis, corresponding to 28S and 18S rRNA with a ratio of intensity at 2:1 (Figure 1). The UV absorbance of the RNAs was at 1.89 - 2.03 (common value at 1.9 - 2.1). This indicated that the extracted total RNAs were high quality, and corresponding to build library.
3.2. The Adaptor Ligation Efficiency
The connection efficiency of the adaptors 1 and 2 was more than 25% (Figure 2). The efficiency met the requirement.
3.3. cDNA Library Subtractive Efficiency
There was appearance of lighter bands after the 18 cycles on the samples before subtraction and after the 28 cycles on the samples after subtraction (Figure 3). The more than 10 cycles meant that the most of expressed constitutive genes were effectively removed. The subtractive library was high quality.
3.4. Differentially Expressed cDNA Examination
A total of 18,350 clones were collected in cDNA library,
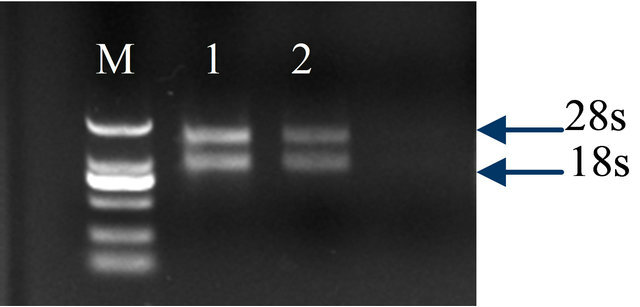
Figure 1. Total RNAs integrity from rubber tree leaves; Lane 1: tester, Lane 2: driver, Lane M: Marker DGL2000 (TakaRa).
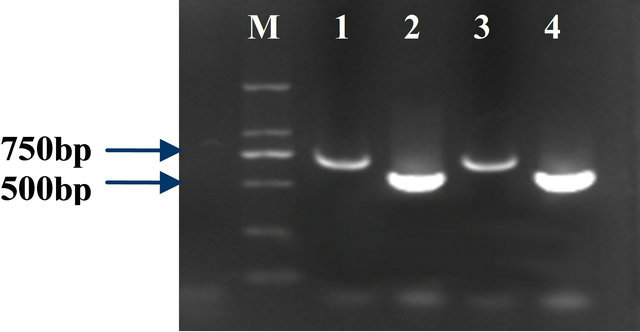
Figure 2. Efficiency of the adaptor ligation with Actin; Lane 1: tester1-1 as the template and the actin3’ primer, PCR primer1; Lane 2: tester1-1 (adaptor1-ligated-cDNA fragment) as the template and the actin3’, 5’ primer; Lane 3: tester1-2 as the template and the actin 3’ primer, PCR primer1; Lane 4: tester1-2(adaptor 2R ligated-cDNA fragments) as the template and the actin 3’, 5’ primer; Lane M: Marker DGL2000.
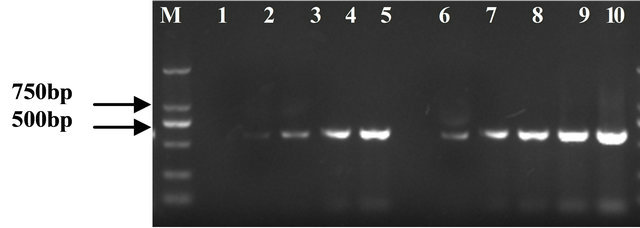
Figure 3. Evaluation of subtraction efficiency; Lanes 1-5: Subtracted samples after 18, 23, 28, 33 and 38 cycles of amplification; Lanes 6-10: Unsubtracted samples after 18, 23, 28, 33 and 38 cycles of amplification; Lane M: Marker DL2000.
thereinto, 15,180 clones was white spots. The positive rate was 99% after PCR identification with random 400 white spots. The recombination rate was 82.7%. The cDNA library met the general requirement.
The size of cDNA inserted fragment was different and most in 400 to 1000 bp after PCR amplification with the random differentially expressed cDNA clones. The constructed cDNA library insert size was consistent after electrophoresis (Figure 4).
3.5. The Sequence Analysis and Gene Functional Annotation
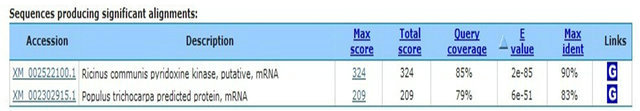
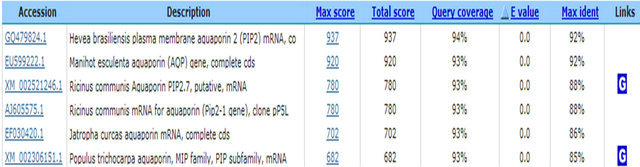
There were 23 cDNA sequences and 2 repeats known as function as energy and basic metabolism, signal transduction, membrane and transport, secondary metabolism and so on, 13 sequences unknown of function, 2 sequences no significant match and 2 clones no needed sequences after detection of the sequences of 42 positive clones randomly selected from the cDNA library and comparison on nucleic acid sequences in Genbank (Table 1). 7 ESTs were logged in Genbank and accession numbers were GW873071 and GW874604-GW874610. Parts of the sequences and their functions were attached in Appendix, including disease resistance and defense response gene—zinc finger protein, signal transduction protein— kinase and ion channels—membrane water channel protein.
4. Discussion
During the plant growth process, it would be attacked by many pathogen microorganisms, appearing as disease susceptible or resistant. The process of disease resistant of plant is an outcome by a series of signal recognition, signal transduction, defense reaction activation and the coordinate action of defense gene expression. SSH technology has obvious advantage in gene expression of concentration difference. Therefore, it has been used broadly for understanding the gene differences of plant
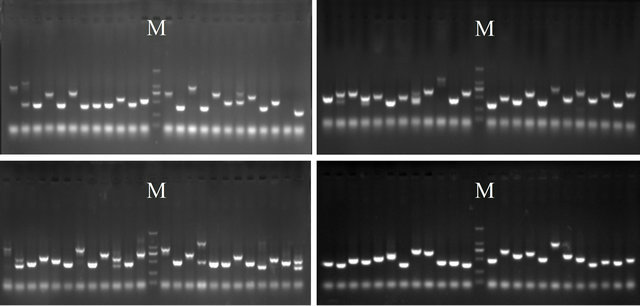
Figure 4. Identification of inserted fragments in plasmids of subtractive cDNA library by PCR; lane M: Marker DGL- 2000, Others: inserted fragments in subtractive cDNA library.
Table 1. Sequence alignment and annotation of gene function.
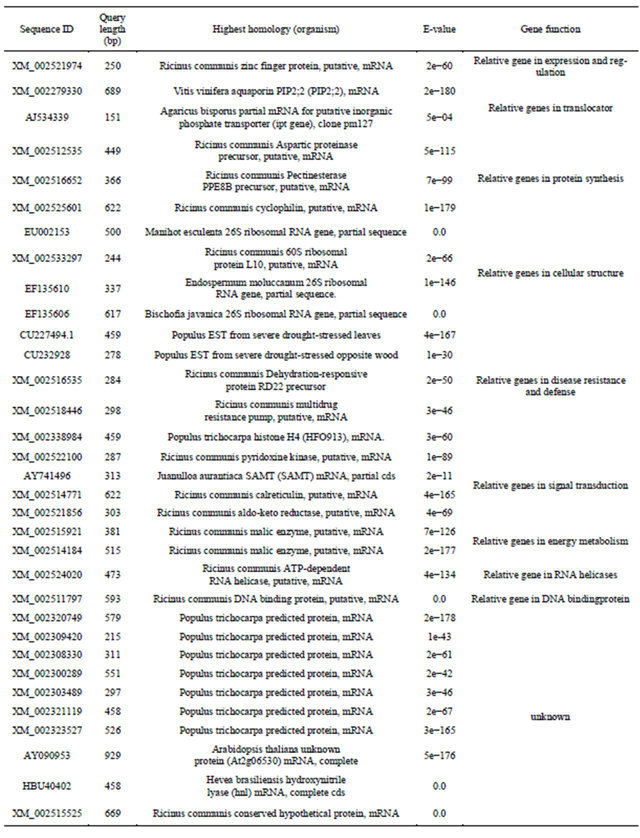
Continued
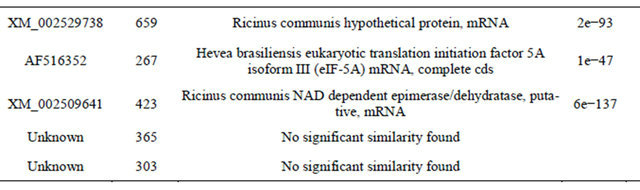
disease resistance [10] and environment adversity stress [18]. The present study we constricted BTH inducing positive and negative SSH-cDNA library of the rubber blade. There were 13 sequences that their function was unknown. It would possibly imply some important genes related to the disease resistant. There were 2 sequences that had no obvious match. It might be represented novel unknown genes or high variable cDNA noncoding region sequences. All these are valuable to be further study.
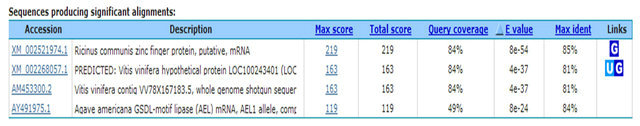
In the present study, constructed SSH-cDNA library has coded zinc finger protein gene occurring (ESTs accession numbers: GW874607). Zinc finger protein recognizes transcription factor structure of special alkaline residue sequence. The function of lant C2H2 zinc finger protein is possible related to plant bio-organism or biological stress. The C3H group of RAR1 and HvRar1 zinc finger protein genes from Arabidopsis thaliana has been proved to be located in the relative tight squeezed location of disease resistance [19-21]. In constructed SSHcDNA library, it also has water channel protein gene (ESTs accession numbers: GW874608) appearing. Water channel protein belongs to membrane intrinsic protein. Its expression is influenced by many hormones (ABA, GA) and environment factors (blue light, water stress and pathological infection). Yamada et al. (1995) [22] found that under high salt stress, level of mRNA expressed water channel protein in ice plant declines rapidly. The mRNA level would gradually recover to its original level or much higher level as along with accumulation of osmosis active substances (such as sucrose and polyamine) in the cell. So, it can enhance resistance after expression of inducing water channel protein [23]. In addition, we fund that inorganic phosphate transferring and light harvesting proteins were excited. It is sure after BTH induction, the transferring activity of the host is more active, the function of substances transferring is enhanced, even discover of the presence of inorganic phosphate transfering protein and light harvesting proteins are unable make sure their genes relating to disease resistant directly. In this study, the sequences encoding aspartic protease precursor, calreticulin and cyclophilin were appertaining. These proteins all have disease resistant role in animals and plant, as well as in human [24,25]. We presume these protein genes likely participate in the reaction of rubber resistant to powdery mildew. We also fund some genes (The factors causing stress include cold, salt, drought, and pathogenic bacteria) relating to biological and nonbiological stress. For example, the genes of coded stress response protein are considered to play a role in coordination of resistant outside stress in plant.
In conclusion, BTH induces resistant to rubber powdery mildew disease relating to many ways including recognizing process of pathogen and host, translocating process of signal substances, starting various disease resistance pathways in host and final producing large amount of resistant-related substances against powdery mildew invasion and hypha extension. Our results demonstrated BTH could effectively induce resistant to rubber powdery mildew disease, it provides a new way for controlling such disease, and also provides an useful information for further study on the expression and signal transduction of rubber defense genes.
5. Acknowledgements
We thank Mr. Cheng Bai (EPPI, CATAS) for the correction.
REFERENCES
- G. C. Mondal and K. Jacob, “Effect of Powdery Mildew Disease on Yield of Rubber in Northern Part of West Bengal,” Proceedings of Placrosym, 2002, pp. 531-534.
- J. Liu, “Recent Advances in Rubber Powdery Mildew Research,” Tropical Agricultural Science & Technology, Vol. 33, No. 3, 2010, pp. 1-5.
- J. L. Shan, Q. C. Xiao, Z. T. Yu, et al., “A Preliminary Study on Mechanism of Oligosaccharin Inducing Rubber Tree Resistance to Powdery Mildew,” Subtropical Plant Science, Vol. 34, No. 1, 2005, pp. 31-32. doi:10.1086/425207
- C. J. Luo, Z. W. Fan, Y. D. Shen, H. T. Cheng and L. Z. Liu, “Effects of Rubber Tree Resistance Induced by BTH to Oidium heveae and Assay of Resistance-Related Enzymes,” Chinese Journal of Tropical Crop, Vol. 32, No. 3, 2011, pp. 475-479.
- Z. Sun and F. C. Zheng, “Effect of BTH on Resistance Induction in Rubber against Colletotrichum gloeosporioides Disease,” Guangdong Agricultural Science, No. 7, 2008, pp. 76-77.
- J. Goerlach, S. Volrath, G. Knauf-Beiter, et al., “Benzothiadiazole, a Nevel Class of Inducers of Systemic Acquired Resistance, Activates Genes Expression and Disease Resistance in Wheat,” The Plant Cell, Vol. 8, 1996, pp. 629-643.
- C. Nombela, S. Pascual, M. Aviles, et al., “Benzothiadiazole Induces Local Resistance to Bemisia tabaci (Hemiptera: Aleyrodidae) in Tomato Plants,” Journal of Economic Entomology, Vol. 98, No. 6, 2005, pp. 2266-2271. doi:10.1603/0022-0493-98.6.2266
- B. Chinnasri, B. S. Sipes and D. P. Schmitt, “Effects of Acibenzolar-S-Methyl Application to Rotylenchulus reniformis and Meloidogyne javanica,” Journal of Nematology, Vol. 35, No. 1, 2003, pp. 110-114.
- Z. W. Fan, H. Buschmann and J. Sauerborn, “Main Effects and Interactions among Acibenzolar-S-Methyl, a Biocontrol Fungus and Sunflower Cultivar on Control of Orobanche cumana Wallr.,” Journal of Plant Diseases and Protection, Vol. 114, No. 2, 2007, pp. 76-81.
- L. Z. Xiong, M. W. Lee, M. Qi and Y. N. Yang, “Identification of Defense-Related Rice Genes by Suppression Subtractive Hybridization and Differential Screening,” Molecular Plant-Microbe Interaction, Vol. 14, No. 5, 2001, pp. 685-692. doi:10.1094/MPMI.2001.14.5.685
- K.-H. Kogel and G. Langen, “Induced Disease Resistance and Gene Expression in Cereals,” Cellular Microbiology, Vol. 7, No. 11, 2005, pp. 1555-1564. doi:10.1111/j.1462-5822.2005.00592.x
- C. Bovie, M. Ongena, P. Thonart, et al., “Cloning and Expression Analysis of cDNAs Corresponding to Genes Activated in Cucumber Showing Systemic Acquired Resistance after BTH Treatment,” BMC Plant Biology, Vol. 4, 2004, pp. 15-26. doi:10.1186/1471-2229-4-15
- X. H. Qiu, P. Z. Guan, M.-L. Wang, et al., “Identification and Expression Analysis of BTH Induced Genes in Papaya,” Physiological and Molecular Plant Pathology, Vol. 65, 2004, pp. 21-30. doi:10.1016/j.pmpp.2004.11.004
- B. De Nardi, R. Dreos, L. Del Terra, et al., “Differential Responses of Coffea arabica L. Leaves and Roots to Chemically Induced Systemic Acquired Resistance,” Genome, Vol. 49, 2006, pp. 1594-1605. doi:10.1139/g06-125
- J. A. Verica, S. N. Maximova, M. D. Strem, et al., “Isolation of ESTs from Cacao (Theobroma cacao L.) Leaves Treated with Inducers of the Defense Response,” Plant Cell Reports, Vol. 23, No. 6, 2004, pp. 404-413. doi:10.1007/s00299-004-0852-5
- S. C. Wang, “Preliminary Evaluation of Resistance of Novel Rubber Clones to Powdery Mildew,” Chinese Journal of Tropical Agriculture, Vol. 23, No. 5, 2003, pp. 1-4.
- Y. Yang, Z. L. Zhang, K. C. Liu, et al., “Clone and Characteristics of a Novel Gene HbUEP from Latex in Hevea brasiliensis,” Journal of Agricultural Biotechnology, Vol. 16, No. 2, 2008, pp. 305-308.
- B. Ouyang, T. Yang, H. X. Li, L. Zhang, Y. Y. Zhang, J. H. Zhang, Z. J. Fei and Z. B. Ye, “Identification of Early Salt Stress Response Genes in Tomato Root by Suppression Subtractive Hybridization and Microarray Analysis,” Journal of Experimental Botany, Vol. 58, No. 3, 2007, pp. 507-520. doi:10.1093/jxb/erl258
- K. Shirasu, T. Lahaye and M. W. Tan, “A Novel Class of Eukaryotic Zinc-Binding Protein Is Required for Disease Resistance Signaling in Barley and Development in C. elegans,” Cell, Vol. 99, No. 4, 1999, pp. 355-366. doi:10.1016/S0092-8674(00)81522-6
- P. R. Muskett, K. Kahn, M. J. Austin, et al., “Arabidopsis RARl Exerts Rate-Limiting Control of R Gene-Mediated Defenses against Multiple Pathogens,” The Plant Cell, Vol. 14, No. 5, 2002, pp. 979-992. doi:10.1105/tpc.001040
- C. Azevedo, A. Sadanandom, K. Kitagawa, et al., “The RARl Interactor SGT1: An Essential Component of R Gene Triggered Disease Resistance,” Science, Vol. 295, No. 5562, 2002, pp. 2073-2076. doi:10.1126/science.1067554
- S. Yamada, M. Katsuhara, W. B. Kelly, C. B. Michalowski and H. J. Bohnert, “A Family of Transcripts Encoding Water Channel Proteins: Tissue-Specific Expression in the Common Ice Plant,” The Plant Cell, Vol. 7, No. 8, 1995, pp. 1129-1142.
- C. Maurel, “Plant Aquaporins: Novel Functions and Regulation Properties,” FEBS Letters, Vol. 581, No. 12, 2007, pp. 2227-2236. doi:10.1016/j.febslet.2007.03.021
- H. W. Chuang, T. F. Hsieh, M. Duval and T. L. Thomas, “Genomic Analysis of Arabidopsis Gene Expression in Response to a Systemic Fungicide,” In: H. J. Bohnert and R. A. Prade, Eds., Genomics of Plants and Fungi, CRC Press, Boca Raton, 2003, pp. 237-253. doi:10.1201/9780203912249.ch7
- Y. J. Xia, H. Suzuki, J. Borevitz, J. Blount, Z. J. Guo, K. Patel, et al., “An Extracellular Aspartic Protease Functions in Arabidopsis Disease Resistance Signaling,” The EMBO Journal, Vol. 23, 2004, pp. 980-988. doi:10.1038/sj.emboj.7600086
Appendix
Parts of the Sequences and Their Functions
1. Disease Resistance and Defense Response Gene— Zinc Finger Protein TCGAGCGGCCGCCCGGGCAGGTACAAGCAATTGGAATACTTCCAACAATACCAGCAAAGGGTTACTGGGCTTATTGGAGCTGAGCAAACTCAGAGATTAGTTAATGAAGCACTTGTCCTTATGACCGTGGGAGGCAATGACTTTGTTAACAACTACTATTTGGTCCCCTTCTCTGCTAGATCTCGCCAATTCTCCCTCCCAGACTATGTAGTCTACGTCATCTCCGAGTACCTCGGCCGCGACCACGCTA
2. Signal Transduction Protein—Kinase TAGCGTGGTCGCGGCCGAGGTACATAAAGCTTCCCTTCATCACCCATTACTGGATCACAAACATATGTAAGTTTGGGATTTATGAAGCGAAGCTTGTTGACAACTTCCAATACAGTGTCCAAAAATGAAACTGAACCAATATAACCTGTTAACAAATGAGTATAATACAGCAAGTCATTTGCTTCAAGGCCTTCTATTAAATCCCATAGTTGCTGTCCATTCAAAACTTGGCCTTTAAAGGAAGGATATCCTGTGTGATTTGAGACCTGCCCGGGCGGCCGCTCGAA
3. Ion Channels—Membrane Water Channel Protein TAGCGTGGTCGCGGCCGAGGTAAGGAGGTGAGTGAAGAAACGCAGCCTACCCATGGGAAGGACTATGTTGATCCACCACCAGCTCCTCTCATTGACGTGGCTGAGCTCAAGCTCTGGTCTTTCTACCGTGCTCTTATAGCTGAGTTCATAGCCACTCTTCTTTTCCTCTACATCACTGTAGCTACTGTAATTGGCTACAAGAAACAAGCTGACCCTTGTGGCGGAGTTGGGCTTCTGGGTATTGCATGGGCCTTTGGTGGCATGATTTTTATCCTTGTTTACTGCACTGCTGGTATCTCTGGTGGTCATATTAACCCAGCGGTCACTTTTGGACTTTTCTTGGCGAGGAAGGTGTCACTGATTAGGGCAGTGGCTTACATGGTGGCTCAGTGCTTGGGTGCAATCTGTGGTGTTGGGTTGGTGAAGGCATTTATGAAGCATCCATATAATGCTCTTGGAGGCGGTGCTAACTCCGTGGCTCATGGTTACAACAAAGGCACCGCTTTGGGTGCTGAGATCATAGGCACTTTTGTGCTTGTCTACACTGTTTTCTCTGCAACTGACCCTAAGAGGAGTGCACGTGACTCTCACGTCCCTGTGTTGGCTCCTCTTCCAATTGGGTTTGCTGTGTTCATGGTCCACTTGGCAACAATCCCCATCACTGGTACCTGCCCGGGCGGCCGCTCGAA
NOTES
*This project was financially supported by the National Natural Science Foundation of China (30871653), the National Science and Technology Support Plan Subject of China (2007BAD48B04-001), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry of China and the Key Science & Technology of Hainan, China.
#Works now at Yunnan Academy of Agricultural Sciences.
†Corresponding author.

