Advances in Microbiology
Vol.06 No.09(2016), Article ID:69674,9 pages
10.4236/aim.2016.69065
Study of OmpK35 and OmpK36 Expression in Carbapenem Resistant ESBL Producing Clinical Isolates of Klebsiella pneumoniae
Amina Amal Mahmoud Nour El Din, Reem Abdel Hameed Harfoush, Hadir Ahmed Said Okasha, Dina Aly El Sayed Kholeif
Department of Medical Microbiology and Immunology, Faculty of Medicine, Alexandria University, Alexandria, Egypt

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 18 July 2016; accepted 8 August 2016; published 11 August 2016
ABSTRACT
Background: Carbapenem resistant extended spectrum β-lactamase (ESBL) producing Klebsiella pneumoniae (K. pneumoniae) is increasing worldwide. Carbapenem resistance (CR) has been attributed not only to production of carbapenemases but also to permeability barriers due to outer membrane proteins (OmpK35 and OmpK36) disruption. Objective: Phenotypic detection of CR among ESBL producing K. pneumoniae isolates, followed by the evaluation of the role of ompK35 and ompK36 gene expression among carbapenem resistant K. pneumoniae (CR-KP) isolates. Methods: 100 ESBL producing K. pneumoniae isolates were included in this study. Minimum inhibitory concentration (MIC) of imipenem was performed for all isolates by broth microdilution method. For CR-KP isolates, phenotypic detection of K. pneumoniae carbapenemase (KPC), metallo-β- lactamase (MBL) and AmpC enzymes was performed followed by Realtime qRT-PCR to detect and quantify ompK35 and ompK36 gene expression. Results: 42% of our isolates were carbapenem resistant, and all of them were KPC producers either singly or in combination with MBL and/or AmpC production. Reduced expression of both ompK35 and ompK36 was detected in (52.38%) of CR-KP isolates, while reduced expression of ompK36 or ompK35 alone was found in (2.38%) and (33.33%) respectively. Twenty of 42 CR-KP isolates (47.62%), showing reduced ompK35 and ompK36 expression, exhibited high level resistance (HLR) (>32 µg/ml) to imipenem. There was a significant correlation between reduced expression of ompK36 and increase MIC values (p < 0.05). The combined production of MBL or AmpC together with reduced expression of ompK35 and/or ompK36 resulted in significant increase in imipenem MIC (p < 0.05). Conclusion: The combined OmpK35/OmpK36 loss resulted in HLR. However OmpK36 seems to play a major role in those strains. Imipenem MIC was markedly increased among K. pneumoniae showing carbapenemase and/or AmpC production together with loss of OmpK35 and/or OmpK36.
Keywords:
OmpK35, OmpK36, K. pneumoniae, Carbapenem Resistant

1. Introduction
Carbapenems are widely used in the treatment of extended spectrum β-lactamase (ESBL) producing Klebsiella pneumoniae (K. pneumoniae). However, as carbapenems are more frequently utilized, an increasing number of carbapenem resistant K. pneumoniae (CR-KP) has been observed worldwide. The emergence of CR-KP is alarming, as antimicrobial treatment options are limited [1] .
Carbapenem resistance (CR) in K. pneumoniae has been attributed mainly to enzymatic degradation by carbapenemases namely class A Klebsiella pneumoniae carbapenemase (KPC), class B metallo-β-lactamase (MBL) or class D oxacillinases (OXA) [2] . However, other mechanisms for CR in K. pneumoniae exist. These include the combination of outer membrane porin expression disruption and production of β-lactamases that have weak carbapenemase activity such as AmpC among the strains [3] [4] .
Two major porins, namely OmpK35 and OmpK36 have been described in K. pneumonia [5] . Loss of outer membrane proteins (OMPs), including OmpK35 and OmpK36 plays a role in CR in K. pneumoniae strains acting as a permeability barrier. As a result, carbapenems reach low concentrations in the periplasmic space and their activity may be then compromised by enzymes with carbapenemase activity, leading to CR [6] [7] . Moreover, recent studies emphasize the importance of characterization of porin expression in KPC producing K. pneumoniae as this identifies isolates which are most susceptible to the combination of colistin and carbapenems for treatment, and indicating that in the future, molecular characterization of CR-KP isolates might be a practical tool for identifying effective antimicrobial combination regimens [8] [9] .
The aim of this study was to phenotypically detect CR among ESBL producing K. pneumoniae, followed by the evaluation of the role of ompK35 and ompK36 gene expression among CR-KP isolates.
2. Materials and Methods
2.1. Bacterial Isolation
A total of 100 K. pneumoniae isolates were collected from clinical specimens delivered to the Microbiology lab of Alexandria main university hospital (AMUH). The isolates were identified using standard biochemical reactions [10] and verified using API 20 E system (bioMerieux, Marcy l’Etoile, France). Phenotypic detection of ESBLs was performed as guided by CLSI (2014) [11] . The organism was considered ESBL producer when the inhibitory zone of ceftazidime was ≤22 mm, ceftriaxone was ≤25 mm, aztreonam ≤27 mm and when there was a difference of ≥5 mm between the zones of ceftazidime and ceftazidime-clavulinic acid discs [11] .
2.2. Detection of CR
Minimum inhibitory concentration (MIC) of imipenem was performed for all isolates by broth microdilution method, and the results were interpreted according to the CLSI breakpoints (sensitive ≤1 μg/mL, intermediate =2 μg/mL and resistant ≥4 μg/mL) [11] [12] .
2.3. Phenotypic Detection of Carbapenemases
Modified Hodge test (MHT) was performed according to CLSI guidelines [11] . Class A and B carbapenemases were further identified using carbapenemase inhibition tests (the combined disk method). For class A, KPC enzyme production was suspected when there is ≥5 mm increase of inhibition zone diameter around imipenem/ phenylboronic acid disk (10 μg/400μg) than imipenem disk (10 μg) [13] . For class B, MBL detection is based on the synergy between MBL inhibitor (EDTA) and imipenem. This was achieved when there was a ≥7 mm increase of inhibition zone diameter around imipenem/EDTA (10 μg/750μg) than imipenem disk (10 μg) [14] .
2.4. Phenotypic Detection of AmpC Enzyme
Done according to Rand et al. [15] by Modified Hodge test using cefoxitin. Briefly, a cefoxitin-susceptible 0.5 McFarland adjusted E. coli indicator strain (ATCC 25922) was inoculated on Mueller Hinton agar plate and the cefoxitin disk was placed in the center of the plate. The test organism (K. pneumoniae) was streaked from the edge of the cefoxitin disk to nearly the edge of the plate. If the test organism expresses AmpC, it hydrolyzes the cefoxitin and shows growth of the indicator E. coli along the intersection of the streak and the zone of inhibition from the cefoxitin disk.
2.5. Analysis of ompK35 and ompK36 Gene Expression
Total RNA was obtained from 1 ml (about 108 cells/ml) of mid-logarithmic growth-phase bacterial cultures with ISOLATE II RNA Mini kit (Bioline Ltd, UK) [16] . Reverse transcription and Real-time PCR were performed with the SensiFAST™ SYBR No-ROX One-Step Kit (Bioline Ltd, UK) using StepOne™ Real Time PCR (Applied Biosystems, Foster, CA, USA). The qPCR reactions were performed in a 20 μL final volume containing the selected primers for ompK35, ompK36 and rpoB (housekeeping gene) (Table 1). The reaction conditions consisted of initial denaturation at 95˚C for 10 min, followed by 40 cycles of 15 s at 95˚C, 11s at 54˚C and 22 s at 72˚C [8] . The expression levels were expressed in cycle threshold (CT) units. Expression of each gene was normalized to that of the housekeeping gene rpoB. The relative expression of ompK35 and ompK36 genes was compared to that of a control isolate that is known to express both porins (K. pneumoniae ATCC 13883). To guarantee the specificity of the amplification products, a melting curve analysis was performed. No amplification of unspecific products was observed.
2.6. Ethics
This study was approved by the Ethical Review Committee, Faculty of Medicine, Alexandria University, Egypt.
2.7. Statistical Analysis
Data were fed to the computer using IBM (NY, USA) SPSS software package version 20.0. Qualitative data were described using number and percent. Quantitative data were described using mean and standard deviation. For normally distributed data, comparison between two independent populations was done using Student’s t-test. Correlations between two quantitative variables were assessed using Pearson coefficient. Significance test results are quoted as two-tailed probabilities. Significance of the obtained results was judged at the 5% level.
3. Results
In the present study, the highest isolation of ESBL producing K. pneumoniae isolates was from urine specimens (33%), followed by sputum and bronchoalveolar lavage (BAL) specimens (31%), then pus specimens (28%). The lowest isolation percentage was from blood and aspirated fluids (7% and 1%) respectively.
In this study, CR was detected in 42% of the 100 ESBL producing K. pneumoniae isolates where 12 isolates (12%) exhibited low level resistance (LLR) (≥4 - 32 µg/ml) and 30 isolates (30%) exhibited high level resistance (HLR) (>32 - 512 µg/ml).
Out of the 42 CR-KP strains, 20 (47.62%) were isolated from ICUs, 12 (28.57%) from surgical wards and 10 isolates (23.81%) from internal medicine wards. The highest isolation was observed among sputum and BAL specimens, where 16/42 isolates (38.1%) were identified, followed by urine and pus specimens (12/42, 28.57%) each, and blood culture specimens (2/42, 4.76%).
Table 1. Nucleotide sequence of primers used for real time quantitative PCR analysis of mRNA expression of ompK35 and ompK36 genes [8] .
By the phenotypic detection methods, all 42 isolates were KPC producers whether singly in 9/42 isolates or in combination with other enzymes: [KPC/AmpC] and [KPC/AmpC/MBL] in 13/42 isolates for each, and [KPC/ MBL] in 7/42 isolates.
The analysis of expression levels of ompK35 and ompK36 genes by real time qRT-PCR showed that among the 42 isolates, only 5 isolates (11.91%) showed normal expression of both porins (OmpK35 and OmpK36), one isolate (2.38%) showed reduced expression of ompK36 only, 14 (33.33%) showed reduced expression of ompK35 only while reduced expression of both ompK35 and ompK36 occurred in a significant number of isolates (22/42, 52.38%) (p = 0.011).
When comparing the MIC results with the expression levels of porins, among the 30 HLR isolates, 20 isolates exhibited reduced expression of both ompK35 and ompK36 and 9 isolates exhibited reduced expression of ompk35 only and this was statistically significant (p < 0.05).
Meanwhile, among the 12 LLR isolates, 5 isolates showed normal expression of both porins, 5 isolates showed reduced expression of ompk35 alone while 2 isolates only showed reduced expression of both ompK35 and ompK36.
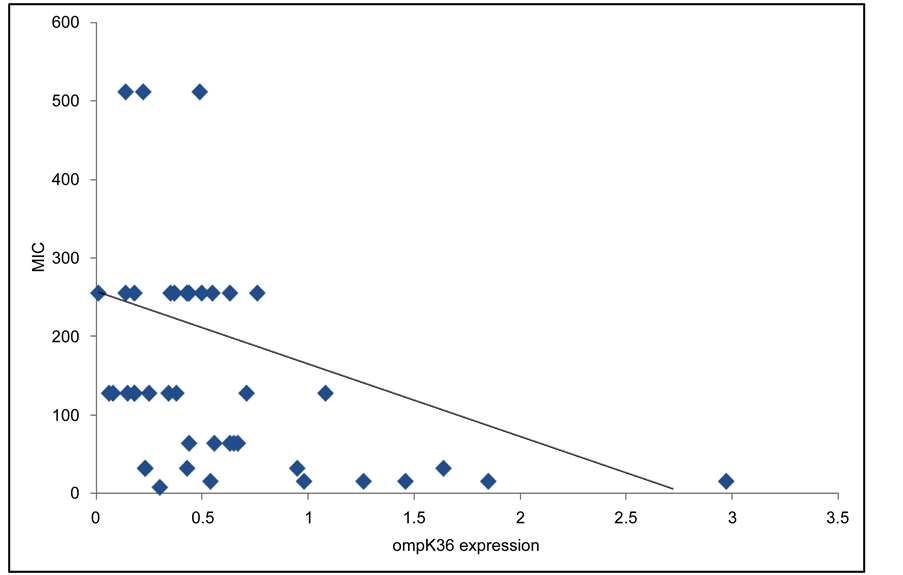
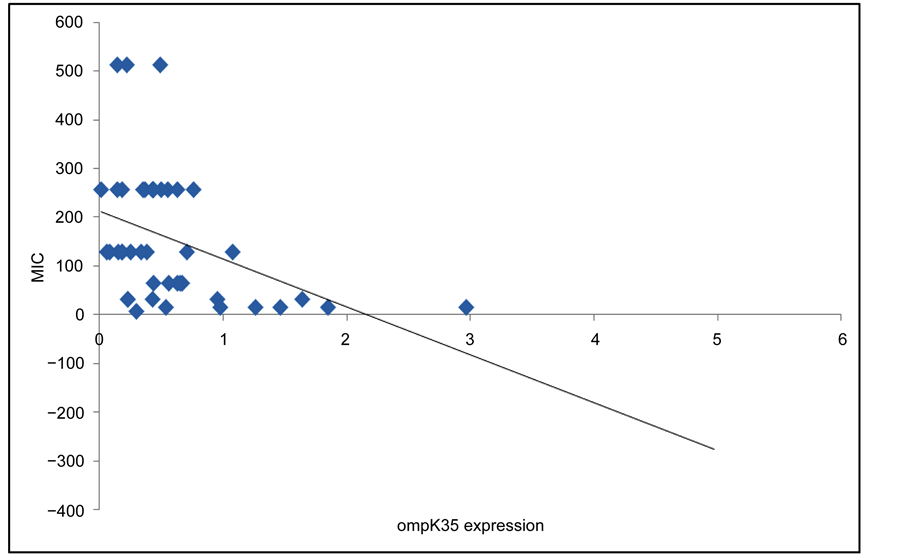
To study the contribution of reduced expression of these porins to the increase of imipenem MIC values, the correlation between ompK35/ompK36 genes expression and MIC values was analyzed. Our data demonstrated a significant correlation between reduced expression of ompK36 and increased MIC values, while this correlation could not be established between reduced expression of ompK35 and high MIC values (Figure 1 and Figure 2).
Our data demonstrated that the combined effect of MBL production together with reduced expression of ompK35 and ompK36 resulted in significant increase in imipenem MIC. Furthermore, the combined effect of AmpC production together with reduced expression of ompK35 resulted in significant increase in imipenem MIC (Table 2).
4. Discussion
CR is a major threat to the antimicrobial treatment of infections with multidrug resistant organisms. One report cites that these resistant strains can contribute to death in up to 50% of patients who become infected. The optimal treatment of these infections is not well established and the clinical outcome data remain sparse [17] .
Figure 1. correlation between ompK36 expression and MIC values. r = −0.446 (p < 0.05).
Figure 2. correlation between ompK35 expression and MIC values. r = −0.162 (p = 0.304).
Table 2. The combined effect of enzymes production and reduced porins expression on imipenem MIC.
t = Student’s t-test; *Significant p values at the level of <0.05.
The present study demonstrated that (42%) of ESBL producing K. pneumoniae isolates were CR-KP. Similar and variable results were detected by different studies [18] - [20] . These discrepant results might be attributed to the different antibiotic susceptibility pattern of K. pneumoniae isolated all over the countries and the diverse antibiotic policies that may aid in selection of certain antibiotic resistant pathogen than another. The high prevalence of CR-KP in the current study may be attributed to the excessive empirical use of carbapenems in our hospital for infections with multidrug resistant pathogens.
In this study, the highest percentage of CR-KP were isolated from ICUs, similar to a study by Carrër et al. [21] and Arnold et al. [22] . This can be attributed to the nature of patients admitted to this section; many of our patients were immunocompromised, subjected to invasive procedures and devices, complicated or end-stage cases referred from other hospital wards, where they had prolonged and numerous antibiotic therapies, with long periods of hospital stay.
Carbapenem resistance in K. pneumoniae has been attributed also to enzymatic degradation. In the current study, all 42 isolates were KPC producers. Our result was concordant with previous studies that revealed high percentage of KPC production among their isolates [23] - [25] . This high percentage can be explained by the great potential for spread of this enzyme due to its location on plasmids in CR-KP strains [18] .
The combined production of KPC/MBL/AmpC was observed among our isolates. Co-production of [KPC and MBL] was detected among 7/42 isolates (16.67%) and [KPC, MBL and AmpC] among 13/42 isolates (30.95%).
Several Egyptian studies described the combined carbapenemase production [18] [26] . Hospital infections caused by K. pneumoniae co-producing KPCs and MBLs have also been described in other regions of the world [27] .
OmpK35 and OmpK36 are the major outer membrane porins of K. pneumoniae. In this study, the expression of both porins genes has been estimated to evaluate their role in CR. The results showed a reduced expression of both ompK35 and ompK36 in a significant number of isolates (52.38%) while the rest of the isolates had either reduced expression of ompK36 only (2.38%) or ompK35 only (33.33%) or normal expression of both porins in (11.91%).
These results were similar to previous studies by Hashemi et al. [28] , Ma et al. [29] , Ruiz [30] and Findlay et al. [31] who also reported defects in OmpK35 and/or OmpK36 production in CR-KP isolates.
As regards the carbapenem resistance level in correlation to porins expression, our findings revealed that loss of both porins contributed to HLR and clearly agree with previous studies by Davies et al. [32] , Kitchel et al. [33] and Clancy et al. [9] .
To elucidate the contribution of porins in CR, studies by Tsai et al. [34] and Doumith et al. [4] noted that a double deletion of ompK35 and ompK36 genes (ΔompK35/36) led to a marked increase in the MIC of meropenem and ertapenem respectively and the increased MIC could be restored to the original levels following complementation by plasmids carrying ompK35 and ompK36 genes.
Furthermore, we demonstrated a significant correlation between reduced expression of ompK36 and increased MIC values of imipenem while we failed to do so for of ompK35. These observations coincide with the findings previously published by Palasubramaniam et al. [35] , Landman et al. [36] and Netikul et al. [37] . Several genetic studies also highlighted the role of OmpK36 in CR; according to Tsai et al. [34] , the deletion of ompK36 gene resulted in greater drug resistance compared to the deletion of ompK35 thus, the emergence of clinical antibiotic resistance due to OmpK36 loss was predicted. Meanwhile, Clancy et al. [9] noted that a certain mutation in ompK36 gene was an independent predictor for HLR to meropenem.
In the present study, CR was attributed to the combined effect of enzymes production together with loss of OmpK35 and/or OmpK36. From previous reports, we know that KPC production combined with porins loss can result in higher carbapenem MIC [33] [36] .
Our data also demonstrated that the combined production of MBL together with reduced expression of ompK35 and ompK36 resulted in significant increase in imipenem MIC. These findings were compatible with those obtained by Sho et al. [38] and Chen et al. [39] . Moreover, the production of AmpC together with reduced expression of ompK35 resulted in significant increase in imipenem MIC in accordance with previous studies by Palasubramaniam et al. [35] and Shin et al. [7] .
The current study and those cited emphasize the importance of characterization of porin expression in CR-KP, as referred by Hong et al. [8] and Clancy et al. [9] who documented that screening for porin expression identifies CR-KP which are most susceptible to colistin/carbapenems combination in attempt for treatment.
5. Conclusion
The high percentage of CR calls for more rigid application of infection control measures and establishing strict antibiotic policies mainly in ICUs. Our data revealed an interplay between enzymes production and reduced expression of ompK35 and/or ompK36 that affected the CR level and hence the options of treatment available including combination therapies. It is mandatory to correctly identify patients for whom combination antimicrobial therapies are most likely to be effective based on the molecular biology of specific isolates, types of infection to be treated, underlying diseases and other host factors in order to both optimize the outcomes and minimize the potential for toxicity.
Cite this paper
Amina Amal Mahmoud Nour El Din,Reem Abdel Hameed Harfoush,Hadir Ahmed Said Okasha,Dina Aly El Sayed Kholeif, (2016) Study of OmpK35 and OmpK36 Expression in Carbapenem Resistant ESBL Producing Clinical Isolates of Klebsiella pneumoniae. Advances in Microbiology,06,662-670. doi: 10.4236/aim.2016.69065
References
- 1. Leavitt, A.I., Chmelnitsky, R., Colodner, I., Ofek, Y., Carmeli, A. and Venezia, S.N. (2009) Ertapenem Resistance among Extended-Spectrum-Beta-Lactamase-Producing Klebsiella Pneumoniae Isolates. Journal of Clinical Microbiology, 47, 969-974.
http://dx.doi.org/10.1128/JCM.00651-08 - 2. Gupta, N., Limbago, B.M., Patel, J.B. and Kallen, A.J. (2011) Carbapenem-Resistant Enterobacteriaceae: Epidemiology and Prevention. Clinical Infectious Diseases, 53, 60-67.
http://dx.doi.org/10.1093/cid/cir202 - 3. Nordmann, P., Cuzon, G. and Naas, T. (2009) The Real Threat of Klebsiella pneumoniae Carbapenemase-Producing Bacteria. Lancet Infectious Diseases, 9, 228-236.
http://dx.doi.org/10.1016/S1473-3099(09)70054-4 - 4. Doumith, M., Ellington, M.J., Livermore, D.M. and Woodford, N. (2009) Molecular Mechanisms Disrupting Porin Expression in Ertapenem Resistant Klebsiella and Enterobacter spp. Clinical Isolates from the UK. Journal of Antimicrobial Chemotherapy, 63, 659-667.
http://dx.doi.org/10.1093/jac/dkp029 - 5. Yigit, H., Anderson, J.G., Biddle, J.W., Steward, C.D., Rasheed, J.K., Valera, L.L., et al. (2002) Carbapenem Resistance in a Clinical Isolate of Enterobacter aerogenes Is Associated with Decreased Expression of OmpF and OmpC Porin Analogs. Antimicrobial Agents and Chemotherapy, 46, 3817-3822.
http://dx.doi.org/10.1128/AAC.46.12.3817-3822.2002 - 6. Martinez, L.M. (2008) Extended-Spectrum β-Lactamases and the Permeability Barrier. Clinical Microbiology and Infection, 14, 82-89.
http://dx.doi.org/10.1111/j.1469-0691.2007.01860.x - 7. Shin, S.Y., Bae, I.K., Kim, J., Jeong, S.H., Yong, D., Kim, J.M. and Lee, K. (2012) Resistance to Carbapenems in Sequence Type 11 Klebsiella pneumoniae Is Related to DHA-1 and Loss of OmpK35 and/or OmpK36. Journal of Medical Microbiology, 61, 239-245.
http://dx.doi.org/10.1099/jmm.0.037036-0 - 8. Hong, J.H., Clancy, C.J., Cheng, S., Shields, R.K., Chen, L., Doi, Y., et al. (2013) Characterization of Porin Expression in Klebsiella pneumoniae Carbapenemase (KPC)-Producing K. pneumoniae Identifies Isolates Most Susceptible to the Combination of Colistin and Carbapenems. Antimicrobial Agents and Chemotherapy, 57, 2147-2153.
http://dx.doi.org/10.1128/AAC.02411-12 - 9. Clancy, C.J., Chen, L., Hong, J.H., Cheng, S., Hao, B., Shields, R.K., et al. (2013) Mutations of the OmpK36 Porin Gene and Promoter Impact Responses of Sequence Type 258, KPC-2-Producing Klebsiella pneumoniae Strains to Doripenem and Doripenem-Colistin. Antimicrobial Agents and Chemotherapy, 57, 5258-5265.
http://dx.doi.org/10.1128/AAC.01069-13 - 10. Chander, Y., Ramakrishnam, M.A. and Goyal, S.M. (2011) Differentiation of Klebsiella pneumoniae and Klebsiella oxytoca by Multiplex Polymerase Chain Reaction. International Journal of Applied Research in Veterinary Medicine, 9, 138-142.
- 11. Clinical and Laboratory Standards Institute (2014) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24, Vol. 34, No.1.
- 12. Clinical and Laboratory Standards Institute (2006) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. 7th Edition, CLSI Document M7-A7, Vol. 26, No. 2.
- 13. Tsakris, A., Kristo, I., Poulou, A., Themeli-Digalaki, K., Ikonomidis, A., Petropoulou, D., et al. (2009) Evaluation of Boronic Acid Disk Tests for Differentiating KPC-Possessing Klebsiella pneumoniae Isolates in the Clinical Laboratory. Journal of Clinical Microbiology, 47, 362-367.
http://dx.doi.org/10.1128/JCM.01922-08 - 14. Pandya, N.P., Prajapati, S.B., Mehta, S.J., Kikani, K.M. and Joshi, P.J. (2011) Evaluation of Various Methods for Detection of Metallo-β-Lactamase (MBL) Production in Gram Negative Bacilli. International Journal of Biological and Medical Research, 2, 775-777.
- 15. Rand, K.H., Turner, B., Seifert, H., Hansen, C., Johnson, J.A. and Zimmer, A. (2011) Clinical Laboratory Detection of AmpC β-Lactamase. Does It Affect Patient Outcome? American Journal of Clinical Pathology, 135, 572-576.
http://dx.doi.org/10.1309/AJCP7VD0NMAMQCWA - 16. Yasufuku, T., Shigemura, K., Shirakawa, T., Matsumoto, M., Nakano, Y., Tanaka, K., et al. (2011) Correlation of Overexpression of Efflux Pump Genes with Antibiotic Resistance in Escherichia coli Strains Clinically Isolated from Urinary Tract Infection Patients. Journal of Clinical Microbiology, 49, 189-194.
http://dx.doi.org/10.1128/JCM.00827-10 - 17. Holt, K.E., Wertheim, H., Zadoks, R.N., Baker, S., Whitehouse, C.A., Dance, D., et al. (2015) Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance in Klebsiella pneumoniae, an Urgent Threat to Public Health. Proceedings of the National Academy of Sciences of the United States of America, 112, 3574-3581.
http://dx.doi.org/10.1073/pnas.1501049112 - 18. El-Sweify, M.A., Gomaa, N.I., El-Maraghy, N.N. and Mohamed, H.A. (2015) Phenotypic Detection of Carbapenem Resistance among Klebsiella pneumoniae in Suez Canal University Hospitals, Ismailiya, Egypt. International Journal of Current Microbiology and Applied Sciences, 4, 10-18.
- 19. Bratu, S., Landman, D., Haag, R., Recco, R., Eramo, A., Alam, M., et al. (2005) Rapid Spread of Carbapenem Resistant Klebsiella pneumoniae in New York City: A New Threat to Our Antibiotic Armamentarium. Archives of Internal Medicine, 165, 1430-1435.
http://dx.doi.org/10.1001/archinte.165.12.1430 - 20. Khaleid, M., Ibraheim, Z. and Eman, M. (2010) Surgical Site Infections and Associated Risk Factors in Egyptian Orthopedic Patients. Journal of American Science, 6, 272-280.
- 21. Carrer, A., Poirel, L., Eraksoy, H., Cagatay, A., Badur, S. and Nordmann, P. (2008) Spread of OXA-48-Positive Carbapenem-Resistant K. pneumoniae Isolates in Istanbul, Turkey. JAMA, 52, 2950-2954.
- 22. Arnold, R., Thom, K., Sharma, S., Phillips, M., Johnson, K. and Morgan, D. (2012) Emergence of Klebsiella pneumoniae Carbapenemase (KPC)-Producing Bacteria. Southern Medical Journal, 104, 40-45.
http://dx.doi.org/10.1097/SMJ.0b013e3181fd7d5a - 23. Shabban, M. and Abdel-Rahman, S. (2013) Detection of Klebsiella pneumoniae Carbapenemases among Gram Negative Clinical Isolates. New Egyptian Journal of Microbiology, 36, 73-85.
- 24. Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P. and Yu, Y. (2011) ST11, the Dominant Clone of KPC-Producing Klebsiella pneumoniae in China. Journal of Antimicrobial Chemotherapy, 66, 307-312.
http://dx.doi.org/10.1093/jac/dkq431 - 25. Robledo, I.E., Aquino, E.E. and Va’zquez, G.J. (2011) Detection of the KPC Gene in Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa and Acinetobacter baumannii during a PCR-Based Nosocomial Surveillance Study in Puerto Rico. Antimicrobial Agents and Chemotherapy, 55, 2968-2970.
http://dx.doi.org/10.1128/AAC.01633-10 - 26. Fattouh, M., Nasr El-din, A. and Omar, M.A. (2015) Detection of Klebsiella pneumoniae Carbapenemase (KPC) Producing Gram Negative Superbugs: An Emerging Cause of Multidrug-Resistant Infections in General Surgery Department of Sohag University Hospital, Egypt. International Journal of Current Microbiology and Applied Sciences, 4, 1-15.
- 27. Hrabak, J., Chudackova, E. and Papagiannitsis, C.C. (2014) Detection of Carbapenemases in Enterobacteriaceae: A Challenge for Diagnostic Microbiological Laboratories. Clinical Microbiology and Infection, 20, 839-853.
http://dx.doi.org/10.1111/1469-0691.12678 - 28. Hashemi, A., Fallah, F., Erfanimanesh, S., Hamedani, P., Alimehr, S. and Goudarzi, H. (2014) Detection of β-Lactamases and Outer Membrane Porins among Klebsiella pneumoniae Strains Isolated in Iran. Scientifica, 2014, Article ID: 726179.
http://dx.doi.org/10.1155/2014/726179 - 29. Ma, L., Lu, P.L., Siu, L.K. and Hsieh, M.H. (2013) Molecular Typing and Resistance Mechanisms of Imipenem Non Susceptible Klebsiella pneumoniae in Taiwan: Results from the Taiwan Surveillance of Antibiotic Resistance (TSAR) Study, 2002-2009. Journal of Medical Microbiology, 62, 101-107.
http://dx.doi.org/10.1099/jmm.0.050492-0 - 30. Ruiz, E. (2012) Acquisition of Carbapenem Resistance in Multiresistant Klebsiella pneumoniae Strains Harboring blaCTX-M-15, qnrS1 and aac(6’)-Ib-cr Genes. Journal of Medical Microbiology, 61, 672-677.
http://dx.doi.org/10.1099/jmm.0.038083-0 - 31. Findlay, J., Hamouda, A., Dancer, S.J. and Amyes, S.G.B. (2012) Rapid Acquisition of Decreased Carbapenem Susceptibility in a Strain of Klebsiella pneumoniae Arising during Meropenem Therapy. Clinical Microbiology and Infection, 18, 140-146.
http://dx.doi.org/10.1111/j.1469-0691.2011.03515.x - 32. Davies, T.A., Queenan, A.M., Morrow, B.J., Shang, W., Amsler, K., He, W., et al. (2011) Longitudinal Survey of Carbapenem Resistance and Resistance Mechanisms in Enterobacteriaceae and Non-Fermenters from the USA in 2007-09. Journal of Antimicrobial Chemotherapy, 66, 2298-2307.
http://dx.doi.org/10.1093/jac/dkr290 - 33. Kitchel, B., Rasheed, J.K., Endimiani, A., Hujer, A.M., Anderson, K.F., Bonomo, R.A., et al. (2010) Genetic Factors Associated with Elevated Carbapenem Resistance in KPC-Producing Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy, 54, 4201-4206.
http://dx.doi.org/10.1128/AAC.00008-10 - 34. Tsai, Y.K., Fung, C.P., Lin, J.C., Chen, J.H., Chang, F.Y., Chen, T.L., et al. (2011) Klebsiella pneumoniae Outer Membrane Porins OmpK35 and OmpK36 Play Roles in both Antimicrobial Resistance and Virulence. Antimicrobial Agents and Chemotherapy, 55, 1485-1493.
http://dx.doi.org/10.1128/AAC.01275-10 - 35. Palasubramaniam, S., Karunakaran, R., Gin, G.G., Muniandy, S. and Parasakthi, N. (2007) Imipenem-Resistance in Klebsiella pneumoniae in Malaysia Due to Loss of OmpK36 Outer Membrane Protein Coupled with AmpC Hyperproduction. International Journal of Infectious Diseases, 11, 472-474.
http://dx.doi.org/10.1016/j.ijid.2007.01.005 - 36. Landman, D., Bratu, S. and Quale, J. (2009) Contribution of OmpK36 to Carbapenem Susceptibility in KPC-Producing Klebsiella pneumoniae. Journal of Medical Microbiology, 58, 1303-1308.
http://dx.doi.org/10.1099/jmm.0.012575-0 - 37. Netikul, T. and Kiratisin, P. (2015) Genetic Characterization of Carbapenem-Resistant Enterobacteriaceae and the Spread of Carbapenem-Resistant Klebsiella pneumonia ST340 at a University Hospital in Thailand. PLoS ONE, 10, e0139116.
http://dx.doi.org/10.1371/journal.pone.0139116 - 38. Sho, T., Muratani, T., Hamasuna, R., Yakushiji, H., Fujimoto, N. and Matsumoto, T. (2013) The Mechanism of High-Level Carbapenem Resistance in Klebsiella pneumoniae: Underlying OMP Deficient Strains Represent a Threat of Emerging High-Level Carbapenem-Resistant K. pneumoniae with IMP-1 β-Lactamase Production in Japan. Microbial Drug Resistance, 19, 274-281.
http://dx.doi.org/10.1089/mdr.2012.0248 - 39. Chen, L.R., Zhou, H.W., Cai, J.C., Zhang, R. and Chen, G.X. (2009) Combination of IMP-4 Metallo-Beta-Lactamase Production and Porin Deficiency Causes Carbapenem Resistance in a Klebsiella oxytoca Clinical Isolate. Diagnostic Microbiology and Infectious Disease, 65, 163-167.
http://dx.doi.org/10.1016/j.diagmicrobio.2009.07.002