Advances in Microbiology
Vol.4 No.2(2014), Article ID:42306,9 pages DOI:10.4236/aim.2014.42018
High Prevalence of Ciprofloxacin Resistance in Community Associated Staphylococcus aureus in a Tertiary Care Indian Hospital
1Society for Innovation and Development, Indian Institute of Science, Bengaluru, India
2St. John’s Hospital and Medical College, Bengaluru, India
3Reference Centre for Staphylococci, Hospices Civils de Lyon, Lyon, France
4Faculte de Medicine Lyon Est, University of Lyon, Inserm U851, Lyon, France
Email: *gayathri.arakere@gmail.com
Copyright © 2014 Bhavya Chakrakodi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Bhavya Chakrakodi et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received November 27, 2013; revised December 25, 2013; accepted January 2, 2014
KEYWORDS
Ciprofloxacin Resistance; Indian ST772 Staphylococcus Aureus; gyrA Mutations
ABSTRACT
We have studied the nature of ciprofloxacin resistance in methicillin sensitive and resistant Staphylococcus aureus among patients in a tertiary care hospital in Bengaluru, South India. All the isolates were highly resistant to ciprofloxacin. Molecular characterization of these samples performed using Staphylococcal Cassette Chromosome typing and multilocus sequence typing showed that 37.5% of total isolates and 59% of MRSA were sequence type (ST)772 and the rest were other STs. This indicates high prevalence of CA-MRSA in this tertiary care hospital serving the Indian community. Mutations responsible for ciprofloxacin resistance among these isolates in DNA gyrase (gyrA and gyrB) and topoisomerase IV (grlA and grlB) were analyzed by PCR amplification of specific fragments and sequencing. We found that for ST772 and five other STs present in this collection, single mutation in the gyrA gene, Ser-84→Leu, was sufficient for the high resistance. In vitro generation of ciprofloxacin resistance in two sensitive ST772 isolates by exposure to increasing antibiotic concentrations also resulted in the same single mutation of gyrA. The factors responsible for high ciprofloxacin resistance are varied and are dependent on the genetic background of the isolates and the environment. This is the first report on the mechanism of ciprofloxacin resistance among the most prevalent Indian CA-MRSA.
1. Introduction
Staphylococcus aureus is an important pathogen in the community and hospital, although 30% of the population is colonized with the organism asymptomatically. Increasing use of systemic antibiotics in the hospitals has resulted in multi drug resistant S. aureus due to selective pressure. The advent of community associated S. aureus into hospitals in the past ten years has resulted in drug resistance in this population too, although the genetic back grounds of community and hospital associated S. aureus are different [1].
Fluoroquinolones (FQs) are broad-spectrum antibiotics effective against gram-positive and -negative organisms, including both methicillin-sensitive and resistant S. aureus (MSSA and MRSA). Ciprofloxacin is the most widely used antimicrobial agent among FQs due to its greater potency. But its use in treatment of S. aureus infections is impaired by the rapid emergence of resistant strains, a feature widely spread among both MRSA and MSSA [2, 3].
FQ drugs act by inhibiting the DNA gyrase (gyrA and B) which relieves DNA super coiling and topoisomerase IV (grlA and B), which separates concatenated DNA strands. Amino acid changes in critical regions of this enzyme-DNA complex reduce the affinity for these drugs. DNA gyrase subunits are common sites of resistant mutations in Gram negative organisms, while topoisomerase IV is most critical in case of Gram positive organisms although there are few exceptions [4]. The stepwise emergence of ciprofloxacin resistance in S. aureus is attributed to first grlA mutations conferring low-level resistance, followed by gyrA mutations leading to high-level resistance [5,6]. Several efflux pump systems also contribute to FQ resistance in S. aureus [7].
Ciprofloxacin is used commonly in Indian hospitals and reports of widespread resistance exist [8,9]. There has been no systematic analysis of ciprofloxacin resistance associated with particular genetic back grounds or diseases [10]. This study involves ciprofloxacin resistant MRSA and MSSA isolates, their genetic backgrounds and mutations important in causing the resistance as we have analyzed the clonal complexes present among Indian S. aureus isolates in an earlier study [11]. ST772 (single locus variant of ST1) is one of the prominent sequence types present among Indian community associated S. aureus, and there is not much information on resistance to this antibiotic and its mechanism [11-13]. This work demonstrates that a high percentage of patients in this tertiary care hospital carry CA-MRSA which is highly resistant to ciprofloxacin and a single mutation in gyrA gene is responsible for high resistance in ST772. Mutations among S. aureus isolates from other genetic back grounds are also examined.
2. Materials and Methods
2.1. Sample Collection
Single sample was collected from each in-patient after admission and cultured for S. aureus within 48 hrs after admission. Forty eight S. aureus isolates were collected over a period of three months (between November 2011 and February 2012) from St. John’s Medical College, a tertiary care teaching hospital, Bengaluru, India. Community associated (CA) S. aureus was defined according to Centers for Disease Control and Prevention (CDC) definition for CA-MRSA when patients did not meet any of the following criteria: 1) history of hospitalization, surgery, or dialysis within one year of the SA culture; 2) presence of an indwelling catheter or a percutaneous device at the time of culture.
Hospital had obtained the informed consent from each patient after explaining the purpose of collection. The mean age of the patient was around 40.
Clinical History: Forty percent of patients had superficial skin infections, 29% profound skin infections, 17% pneumonia, 6.3% bone and joint infections, and 8.3% other infections. Major risk factors for infection were diabetes, immuno suppression and burns.
2.2. Phenotypic Characterization
S. aureus isolates were selected after growth on chromogenic agar medium (chromAgar, bioMérieux, MarcyL’Etoile, France) and identified as described elsewhere [14].
2.3. Antibiotic Susceptibility and Minimal Inhibitory Concentration (MIC) Determination
Antibiotic susceptibility testing was performed for erythromycin, gentamicin, amikacin, tetracycline, chloramphenicol and ciprofloxacin by Kirby-Bauer disc diffusion according to the guidelines recommended by the Clinical Laboratory and Standards Institute on MuellerHinton agar plates (HIMEDIA, Mumbai, India) at 37˚C. MIC for Ciprofloxacin was determined by the broth dilution method in Mueller-Hinton broth (HIMEDIA) after 24 h of incubation at 37˚C in micro titer plates [15].
2.4. Chromosomal DNA Isolation
DNA was isolated from 18 hour grown cultures in BHI broth according to previously published procedures [16].
2.5. Molecular Characterization
Staphylococcal cassette chromosome mec (SCCmec) typing was carried out by determining the type of mec and ccr complexes and identification of SCC mec types III, IV and V by an updated multiplex PCR using published procedures and primers [17-19]. Detection of the Panton Valentine Leukocidin (pvl) gene and accessory gene regulator (agr) typing was carried out by PCR and multiplex PCR respectively [20,21].
2.6. Staphylococcal Protein A (spa) Typing and Multi Locus Sequence Typing (MLST)
Spa typing was performed according to the procedure of Shopsin et al. and MLST by Enright et al. [22,23]. Pulsed Field Gel Electrophoresis: PFGE was performed using conditions described previously after restriction digestion using SmaI enzyme (Fermentas, India) and CHEFDRIII (BioRad Laboratories [India] Private Limited, Bengaluru, India) device [16].
2.7. Detection of Mutations
Four independent PCRs were carried out to amplify DNA from the quinolone determining region (QRDR) of the gyrA, gyrB, grlA and grlB using the primer sequences published for the four genes [10]. PCR products were purified (QiaQuick PCR purification kit; QIAGEN GmbH, Hilden, Germany) and commercially sequenced (Macrogen, Inc, Seoul, South Korea). Wild type sequences and mutations were identified by comparison with the published sequences of ciprofloxacin sensitive S. aureus strains ATCC 12600 and RN4220 for gyrA, gyrB [24] and grlA, grlB [25] respectively.
2.8. Generation of in Vitro Ciprofloxacin Resistance in Ciprofloxacin Sensitive ST772 Isolates
Two ciprofloxacin sensitive ST772 isolates with MIC <2.0 µgml−1 were chosen and passaged in Mueller Hinton broth containing increasing concentrations of ciprofloxacin (0.5, 5.0, 50.0 and 500 µg−1) growing at each concentration for 48 hrs before increasing to the next. DNA was made at each step, PCRs were performed for gyrA, and PCR purified products were sequenced.
3. Results and Discussion
3.1. Community and Health Care-Associated S. aureus among Admitted Patients
Data collected from interviews and culture results of 48 patients, indicate that about 71% of the admitted patients are colonized with CA and 29% with HA S. aureus according to the criteria given earlier. This is indicative of high rate of colonization among Indian community with S. aureus which could be making an entry in to the hospital. But dividing samples as coming from the community or hospital based on culture results within or after 48 hrs of admission to the hospital under Indian conditions is problematic due to difficulty in gathering accurate data on stay in hospitals, antibiotics taken etc. Most patients would be visiting multiple hospitals and are perhaps not aware of nature of medicines taken. An additional limitation to arrive at conclusions about CA or HA status is the small sample size in this study.
3.2. Molecular Characterization
Among 48 S. aureus isolates studied, 43 were ciprofloxacin resistant and 5 were sensitive out of which 40% of the isolates were from superficial skin, 29% from profound skin infections, 17% from pneumonia, 6.3% from bone osteomyelitis/osteitis, and rest from others (eye, septicemia, UTI etc.: 8.3%). Table 1 presents the antibiotic sensitivity of the isolates to erythromycin, gentamicin, amikacin, tetracycline, chloramphenicol and ciprofloxacin. Majority of isolates were resistant to erythromycin while the other antibiotics were resistant to varying degrees. Out of 19 isolates from superficial skin infections, 17 were ciprofloxacin resistant and 2 were sensitive. Out of 14 isolates from profound skin infections, 12 were ciprofloxacin resistant and 2 sensitive. Among pneumonia and osteomyelitis isolates, all were ciprofloxacin resistant. The minimum inhibitory concentrations (MIC) of all ciprofloxacin resistant isolates were >512 µg·ml−1 and sensitive isolates were <2 µg·ml−1. Molecular characterization of the 48 isolates is presented in Table 2. Among these 48 isolates collected within the span of 3 months, 27 MRSA isolates belonged to the following sequence types (ST): ST772 (59.3%), ST239 (26%), ST22 (11%), and ST1208 (4%). Except for two ST772 isolates (1 MSSA and MRSA each) which were ciprofloxacin sensitive, all the rest were resistant. There was wide diversity of STs among the 21 MSSAs and they belonged to ST772 (9.5%), 30 (28.6%), 291 (9.5%), 5 (19.0%), 7 (9.5%), 9 (4.8%), 672 (4.8%), 88 (4.8%), 779 (4.8%) and 1 (4.8%) of which 17 were ciprofloxacin resistant. Single isolate of S. aureus ST779 (first reported in Ireland) was ciprofloxacin sensitive and has not been reported in India till now. Among the ciprofloxacin resistant isolates belonging to various STs, ST772 constitute 37.5% of total isolates (18/48) and 59.2% (16/27) ciprofloxacin resistant MRSA isolates. Majority of infections caused by ST772 were skin and soft tissue infections (83% SSTI) while ST30 and ST5 were responsible for 50% and ST239 for 43% SSTI. Figure 1 depicts pulsed Field Gel Electrophoresis (PFGE) patterns and dendrogram of all ciprofloxacin resistant isolates, their genetic back ground and their relatedness.
3.3. Relationship between Ciprofloxacin and Methicillin Resistance
Several previous studies have indicated that a high percentage of MRSA are resistant to ciprofloxacin compared to MSSA [26]. This seems to be true of clinical infections of various organs including the eye although resistance has been reported to be on the rise even among the MSSA [27,28]. Among the isolates we have studied, high ciprofloxacin resistance (>512 µg·ml−1) is prevalent
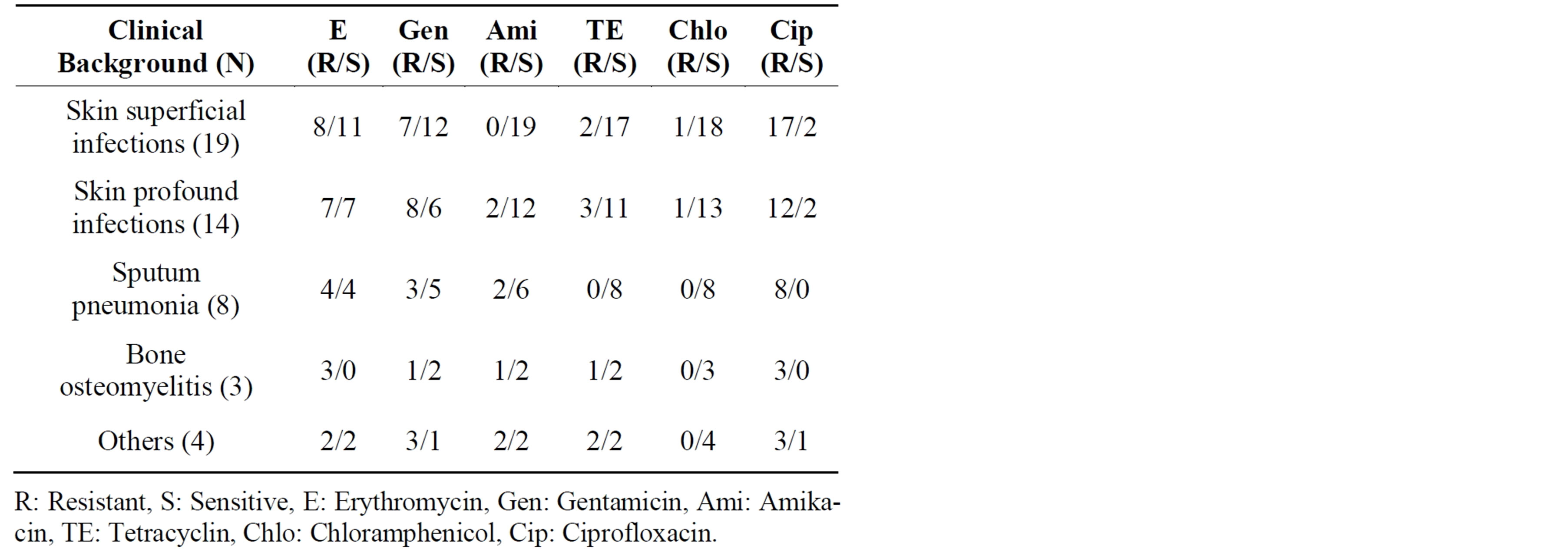
Table 1. Clinical background and antibiotic sensitivity of 48 S. aureus isolates.
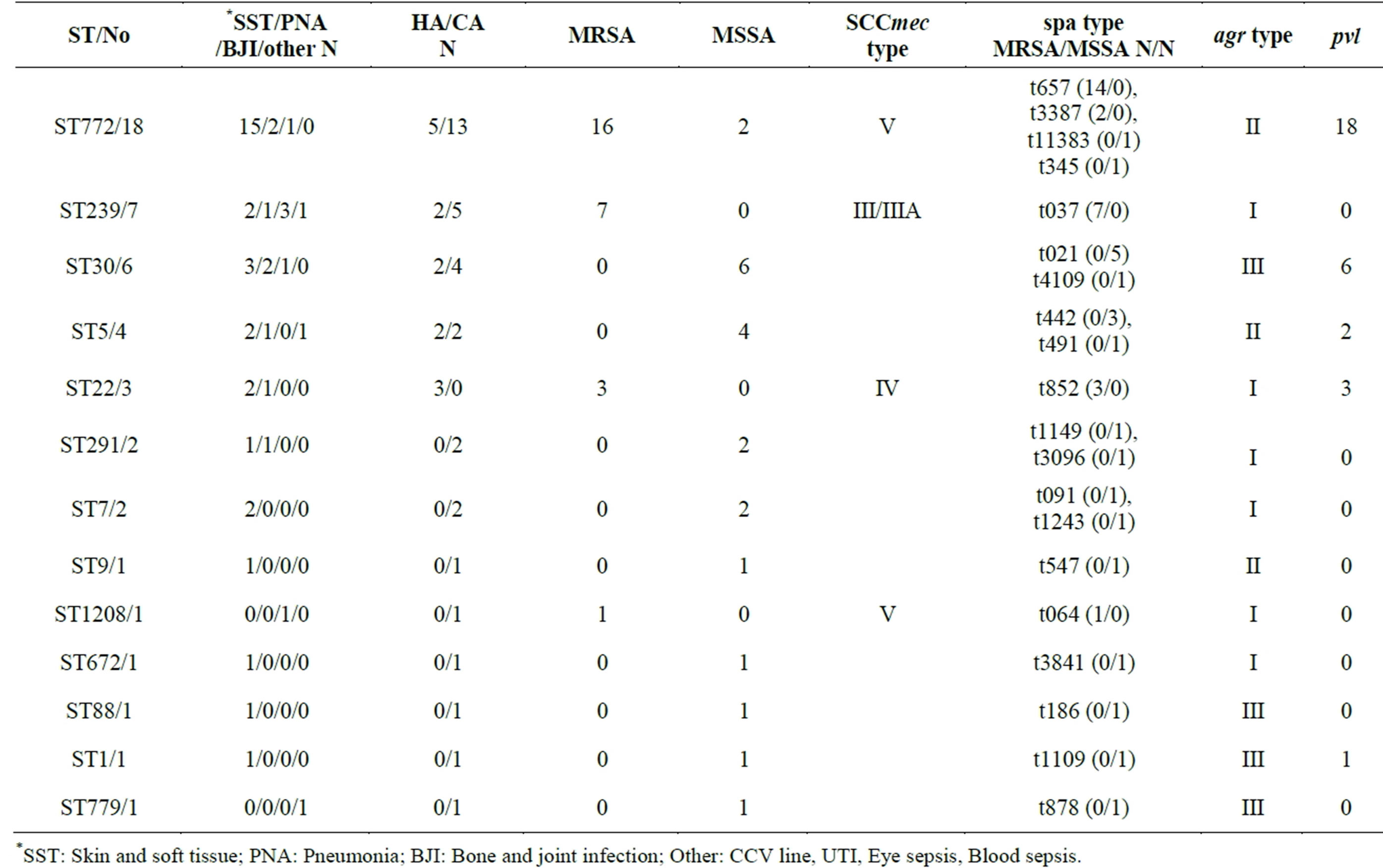
Table 2. Molecular characterization of ciprofloxacin sensitive and resistant MRSA and MSSA isolates.
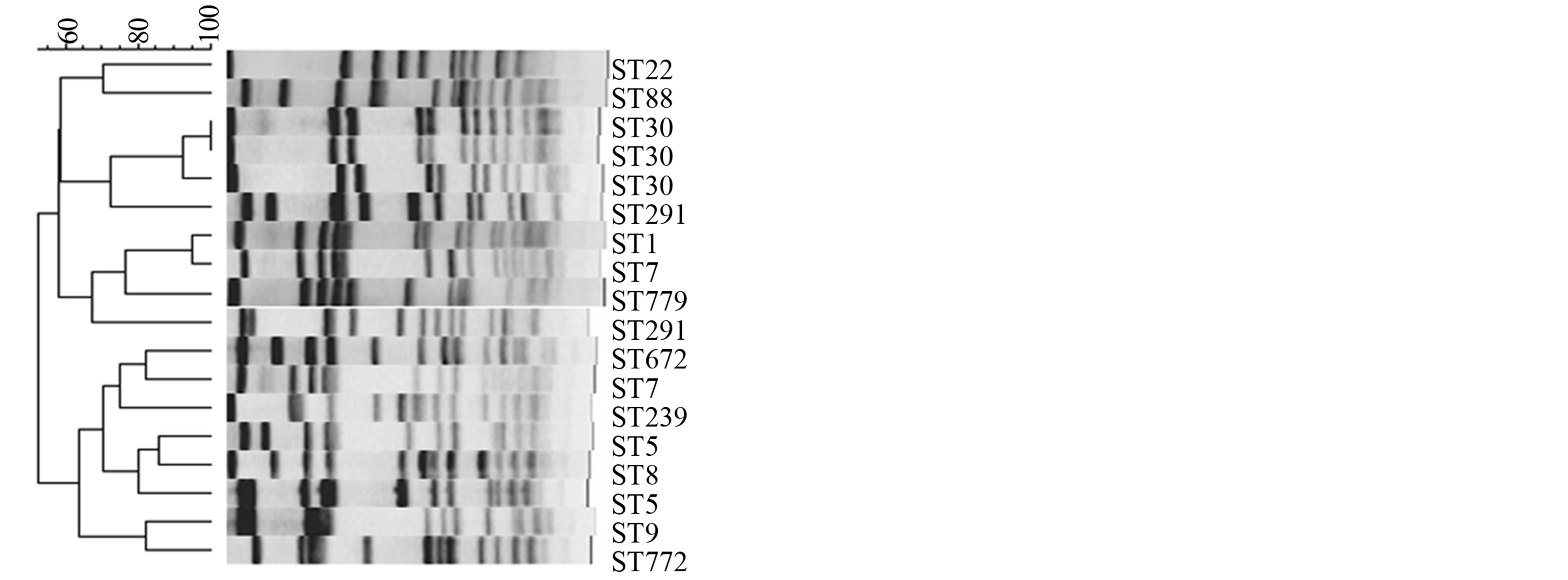
Figure 1. PFGE patterns and dendrogram of ciprofloxacin resistant and sensitive isolates belonging to different genetic backgrounds.
among MRSA and MSSA, the number of MSSA isolates studied being lower. High resistance is observed across all isolates with different genetic and clinical backgrounds. Although it is difficult to categorize S. aureus as community or hospital associated among Indian isolates, from the molecular characterization data, it is clear that ciprofloxacin resistance is more prevalent in MRSA carrying SCCmec elements IV and V (ST772, 22, 1208) than III (ST239) indicating S. aureus of community rather than nosocomial origin. Ciprofloxacin resistant MSSA isolates belong to more diverse genetic backgrounds of ST30, 5, 772, 291 etc., also pointing to community origin of the organism. From our data collected in 2003-4 on molecular characterization of S. aureus collected from this and few other Indian hospitals, it was apparent that majority of isolates were MRSA belonging to ST239 carrying SCC mec elements III or IIIa [16]. A percentage distribution of 41, 34 and 25 for presence of SCCmec elements V, IV and III respectively, has been published for S. aureus collected in a tertiary care hospital in Mumbai, India, from 2006-9 [12]. Our samples collected three months ago have significantly higher proportion of ST772 isolates in this tertiary care hospital.
3.4. Mutations
As our isolates belonged to a diverse group of STs, with different genetic back grounds, we studied the mutations in gyrA, gyrB, and grlA, grlB genes that have been shown to play a prominent role in resistance. PCRs were done for the four products using published primers and purified PCR products were sequenced to check for mutations in the four genes as shown in Table 3. A common mutation in gyrA serine 84 to leucine was found in S. aureus isolates of all genetic back grounds. STs 772, 30, 291, 5, 7, and 672 had only this mutation in gyrA while



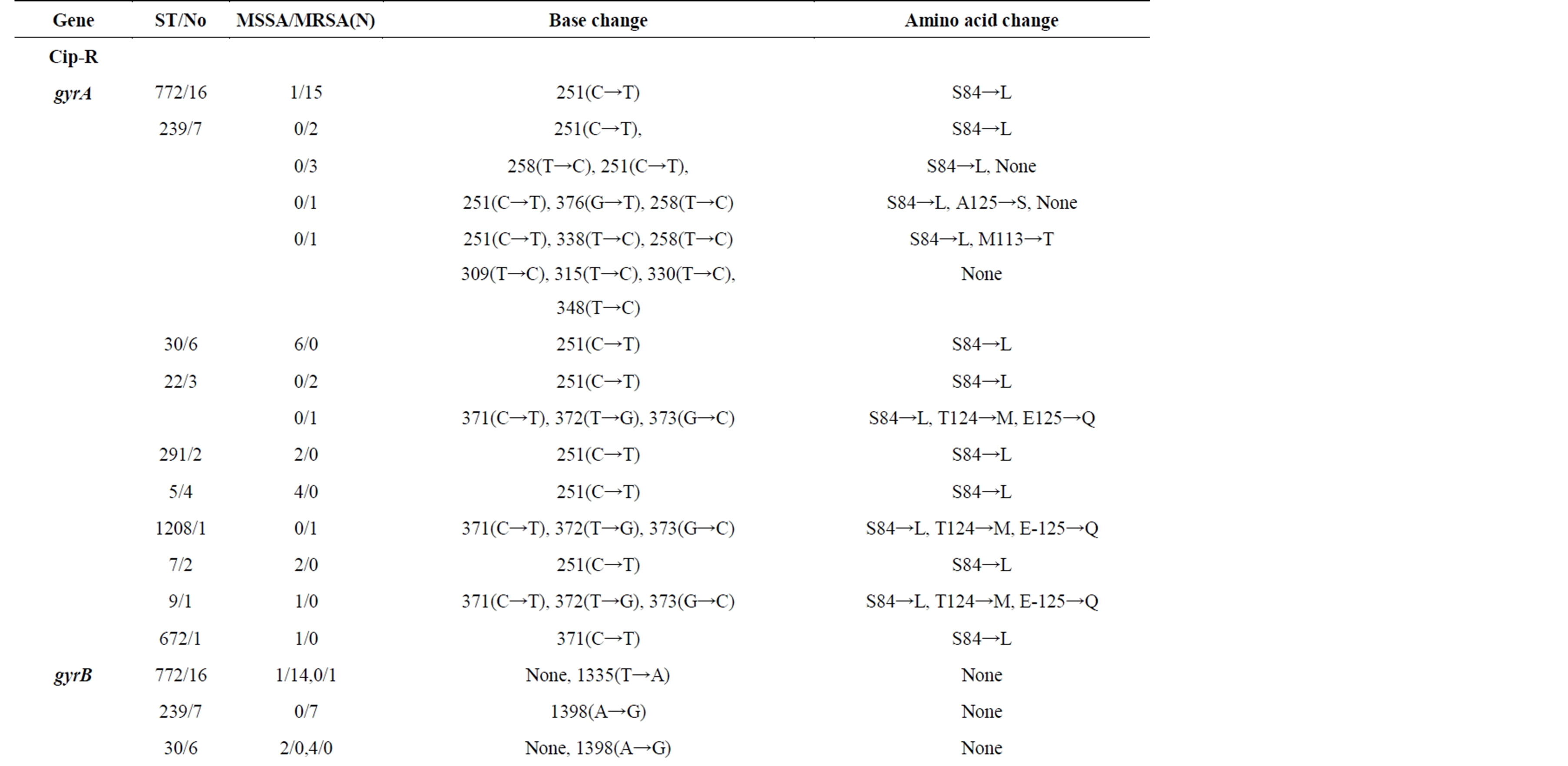
Table 3. Mutations in gyrA, gyrB, grlA and grlB genes among S. aureus Isolates of Different Genetic Backgrounds.
few isolates belonging to STs 239 (2/7), 22(1/3), 1208(1/1) and 9(1/1) had other mutations in addition to serine 84 to leucine mutation. There were no mutations in protein sequences in gyrB while there were few base changes in some STs. GrlA mutations are known to play an important role in ciprofloxacin resistance among MRSA. ST772 and ST1208 S. aureus isolates had mutation in 239(C→A) and change of serine to tyrosine while other STs had mutation in 239(C→T) and change of serine to phenylalanine. But ciprofloxacin sensitive ST772 (MSSA and MRSA) also contained the same mutations in grlA and hence this mutation does not seem important at least in ST772. As we did not have ciprofloxacin sensitive and resistant isolates in other STs to analyze the changes in these genes, we are not certain about the role of this mutation. There were several base changes in grlB with very few amino acid changes. Ciprofloxacin sensitive as well as resistant ST772 isolates did not have any change in bases or amino acids in grlB (Figure 2).
3.5. Number of Mutations
In the last twenty years, S. aureus isolates from different parts of the globe have been tested for mutations causing ciprofloxacin resistance [3]. Schmitz et al. had tested isolates from seven different countries and had found that all isolates that did not have grlA mutation were ciprofloxacin susceptible and all isolates which had grlA mutation of Ser-80→Phe and gyrA mutation of Ser-84→Leu or Glu-88→Lys had an MIC of > 4 µg·ml−1 [10]. Ng et al. had provided evidence that grlA was the primary target of fluoroquinolones in S. aureus and not gyrA [6]. There was a large study in Japan with 451 isolates looking at gyrA mutations where Ser-84→Leu or Ser-84→ Val showed the highest level of resistance, and Glu-88→ Lys the second highest level of resistance to FQs. The resistance of strains with double mutations was higher than ones with single mutations [29]. The same studies have shown increasing MIC to ciprofloxacin with multiple
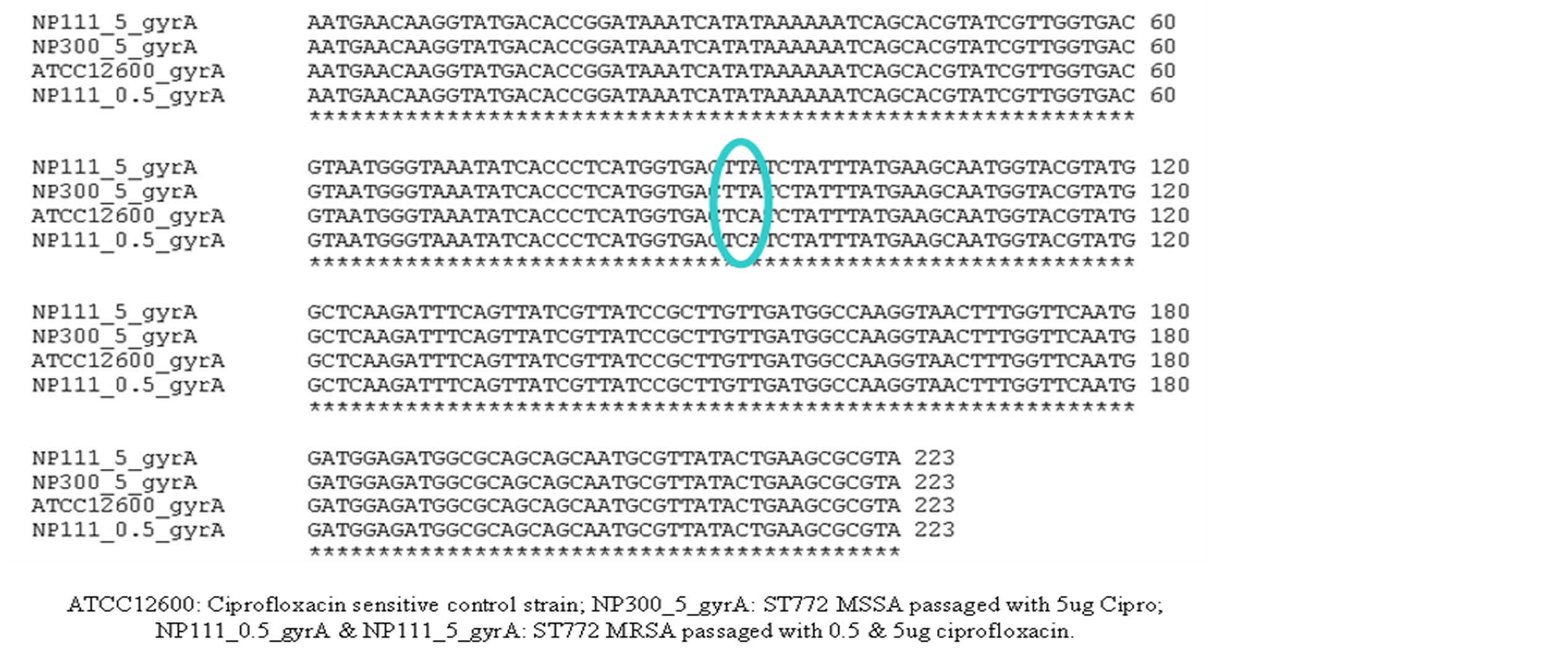
Figure 2. ClustalW alignment of gyrA gene sequences of ciprofloxacin passaged ST772 MSSA and MRSA with ciprofloxacin sensitive control ATCC12600.
mutations. The MICs reported in most studies were not as high as we are seeing in our isolates. Only one mutation Ser-84→Leu seems sufficient for six STs in our collection to become highly resistant to ciprofloxacin. As ST772 is 37.5% of total number of isolates and 59% of MRSA in our collection, a single amino acid change at Ser-84→Leu seems the only important factor for resistance in this major Indian ST of importance in the Indian hospitals and community.
3.6. In Vitro Generation of Ciprofloxacin Resistance in ST772 Isolates
As all our ciprofloxacin resistant isolates had high MIC (>512 µg·ml−1), we chose two ciprofloxacin sensitive ST772 isolates, one MSSA and one MRSA, to check mutations in gyrA gene at different concentrations of ciprofloxacin. We generated ciprofloxacin resistance by step wise exposure and examined the change in the gyrA gene at every step. Except for passage with 0.5 µg·ml−1 ciprofloxacin, all other higher concentrations up to 500 µg·ml−1 had the same Ser-84→Leu mutation with no other change in the gyrA gene sequence corroborating the data obtained from ST772 isolates from the collection. We have not checked with other STs as samples were few and we did not have ciprofloxacin sensitive S. aureus isolates.
4. Conclusion
Most important mutations that have been reported in the literature for ciprofloxacin resistance in S. aureus involve these four genes and our studies indicate that the majority of Indian STs present require only a single mutation in gyrA to acquire high resistance to ciprofloxacin. Few isolates from other STs, 239, 22, 1208, and 9 had multiple mutations and the roles played by these in acquiring the resistance have not been looked into. We have not studied roles played by other factors like efflux proteins in ciprofloxacin resistance in our isolates. As a single mutation is leading to high antibiotic resistance in Indian STs, the genomic background and the environment in which isolates have to survive (high usage of antibiotics in India) might play a very important role. Single mutation in gyrA gene resulting in high resistance to ciprofloxacin may also explain the success of ST772 as a pathogen similar to ST22 isolates with SNPs resulting in ciprofloxacin resistance [30].
Acknowledgements
We thank Dr. Nastasia EL-ZEENNI for collecting the S. aureus isolates and detailed clinical history from patients admitted to St. John’s Hospital. We would also like to thank Helene Meugnier, University of Lyon, France, for creating the PFGE dendrogram from Bionumerics software. GA has received funding from Department of Biotechnology, Govt of India (BT/PR9649/MED/12/356/ 2007), BT/IN/New Indigo/16/GA/2010 and Swedish International development agency. BC is a project assistant in (BT/PR9649/MED/12/356/2007), SP is a JRF in BT/ IN/New Indigo/16/GA/2010. Part of this study was presented as a poster in the ISSSI 2012 conference held in Lyon, France.
REFERENCES
- H. F. Chambers and F. R. DeLeo, “Waves of Resistance: Staphylococcus aureus in the Antibiotic Era,” Nature Reviews Microbiology, Vol. 7, No. 9, 2009, pp. 629-641. http://dx.doi.org/10.1038/nrmicro2200
- F. D. Lowy, “Antimicrobial Resistance: The Example of Staphylococcus aureus,” Journal of Clinical Investigation, Vol. 111, No. 9, 2003, pp. 1265-1273.
- G. A. Jacoby, “Mechanisms of Resistance to Quinolones,” Clinical Infectious Diseases, Vol. 41, No. S2, 2005, pp. S120-S126. http://dx.doi.org/10.1086/428052
- D. C. Hooper, “Emerging Mechanisms of Fluoroquinolone Resistance,” Emerging Infectious Diseases, Vol. 7, No. 2, 2001, pp. 337-341. http://dx.doi.org/10.3201/eid0702.010239
- L. Fererro, B. Cameron and J. Crouzet, “Analysis of gyrA and grlA Mutations in Stepwise-Selected Ciprofloxacin Resistant Mutant of Staphylococcus aureus,” Antimicrobial Agents and Chemotherapy, Vol. 39, No. 7, 1995, pp. 1554-1558. http://dx.doi.org/10.1128/AAC.39.7.1554
- E. Y. Ng, M. Trucksis and D. C. Hooper, “Quinolone Resistance Mutations in Topoisomerase IV: Relationship to the flqA Locus and Genetic Evidence that Topoisomerase IV Is the Primary Target and DNA Gyrase Is the Secondary Target of Fluoroquinolones in Staphylococcus aureus,” Antimicrobial Agents and Chemotherapy, Vol. 40, No. 8, 1996, pp. 1881-1888.
- S. S. Costa, C. Falcão, M. Viveiros, D. Machado, M. Martins J. Melo-Cristino, et al., “Exploring the Contribution of Efflux on the Resistance to Fluoroquinolones in Clinical Isolates of Staphylococcus Aureus,” BMC Microbiology, Vol. 11, No. 10, 2011, pp. 241-252. http://dx.doi.org/10.1186/1471-2180-11-241
- R. Sharma, C. L. Sharma and B. Kapoor, “Antibacterial Resistance: Current Problems and Possible Solutions,” Indian Journal of Medical Science, Vol. 59, No. 3, 2005, pp. 120-129.
- R. S. Phakade, G. Nataraj, S. K. Kuyare, U. S. Khopkar and P. R. Mehta, “Is Methicillin Resistant Staphylococcus aureus Involved in Community Acquired Skin and Soft Tissue Infections? Experience from a Tertiary Care Centre in Mumbai,” JPGM, Vol. 58, No. 1, 2012, pp. 3-7.
- F. J. Schmitz, M. E. Jones, B. Hofmann, B. Hansan, S. Scheuring, M. Luckefahr, et al., “Characterization of grlA, grlB, gyrA, and gyrB Mutations in 116 Unrelated Isolates of Staphylococcus aureus and Effects of Mutations on Ciprofloxacin MIC,” Antimicrobial Agents and Chemotherapy, Vol. 42, No. 5, 1998, pp. 1249-1252.
- S. Shambat, S. Nadig, S. Prabhakara, M. Bes, J. Etienne and G. Arakere, “Clonal Complexes and Virulence Factors of Staphylococcus aureus from Various Cities in India,” BMC Microbiology, Vol. 12, No. 5, 2012, pp. 64-73. http://dx.doi.org/10.1186/1471-2180-12-64
- N. D’Souza, C. Rodrigues and A. Mehta, “Molecular Characterization of Methicillin-Resistant Staphylococcus aureus with Emergence of Epidemic Clones of Sequence Type (ST) 22 and ST 772 in Mumbai, India,” Journal of Clinical Microbiology, Vol. 48, No. 5, 2010, pp. 1806- 1811. http://dx.doi.org/10.1128/JCM.01867-09
- S. Nadig, N. Velusamy, P. Lalitha, S. Kar, S. Sharma and G. Arakere, “Staphylococcus aureus Eye Infections in Two Indian Hospitals: Emergence of ST772 as a Major Clone,” Clinical Optholmology, Vol. 6, No. 1, 2012, pp. 165-173.
- D. Baird, “Staphylococcus: Cluster Forming Gram Positive Cocci,” In: J. G. Collee, A. G. Fraser, B. P. Marmion and A. Simmons, Eds., Mackie and McCartney Practical Medical Microbiology, Vol. 2, 1996, pp. 245-261.
- Clinical and Laboratory Standards Institute, “Performance Standards for Antimicrobial Susceptibility Testing,” 19th Informational Supplement. M100-S19, Clinical and Laboratory Standards Institute, Wayne, 2009.
- G. Arakere, S. Nadig, G. Swedberg, R. Macaden, S. Amarnath and D. Raghunath, “Genotyping of Methicillin Resistant Staphylococcus Aureus Strains from Two Hospitals in Bangalore, South India,” Journal of Clinical Microbiology, Vol. 43, No. 7, 2005, pp. 3198-3202. http://dx.doi.org/10.1128/JCM.43.7.3198-3202.2005
- D. C. Oliveira and H. de Lencastre, “Multiplex PCR Strategy for Rapid Identification of Structural Types and Variants of the Mec Element in Methicillin-Resistant Staphylococcus Aureus,” Antimicrobial Agents and Chemotherapy, Vol. 46, No. 7, 2002, pp. 2155-2161. http://dx.doi.org/10.1128/AAC.46.7.2155-2161.2002
- Y. Kondo, T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, et al., “Combination of Multiplex PCRs for Staphylococcal Chromosome Mec Type Assignment: Rapid Identification System for mec, ccr, and Major Differences in Junk Yard Regions,” Antimicrobial Agents and Chemotherapy, Vol. 51, No. 9, 2007, pp. 264-274. http://dx.doi.org/10.1128/AAC.00165-06
- C. Milheirico, D. C. Oliveira and H. de Lencastre, “Update to the Multiplex PCR Strategy for the Assignment of Mec Element Types in Staphylococcus Aureus,” Antimicrob Agents Chemotherapy, Vol. 51, No. 9, 2007, pp. 3374-3377. http://dx.doi.org/10.1128/AAC.00275-07
- G. Lina, Y. Piemont, F. Godail-Gamot, et al., “Involvement of Panton—Valentine Leukocidin-Producing Staphylococcus Aureus in Primary Skin Infections and Pneumonia,” Clinical Infectious Diseases, Vol. 29, No. 5, 1999, pp. 1128-1132. http://dx.doi.org/10.1086/313461
- P. Gilot, G. Lina, T. Cochard and B. Poutrel, “Analysis of the Genetic Variability of Genes Encoding the RNA IIIActivating Components Agr and TRAPb in a Population of Staphylococcus Aureus Strains Isolated from Cows with Mastitis,” Journal of Clinical Microbiology, Vol. 40, No. 11, 2002, pp. 4060-4067. http://dx.doi.org/10.1128/JsCM.40.11.4060-4067.2002
- B. Shopsin, M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, et al., “Evaluation of Protein A Gene Polymorphic Region DNA Sequencing for Typing of Staphylococcus aureus Strains,” Journal of Clinical Microbiology, Vol. 37, No. 11, 1999, pp. 3556- 3563.
- M. C. Enright, N. P. Day, C. E. Davies, S. J. Peacock and B. G. Spratt, “Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus Aureus,” Journal of Clinical Microbiology, Vol. 38, No. 3, 2000, pp. 1008-1015.
- H. Ito, H. Yoshida, M. Bogaki-Shonnai, T. Niga, H. Hattori and S. Nakamura, “Quinolone Resistance Mutations in the DNA Gyrase gyrA and gyrB Genes of Staphylococcus aureus,” Antimicrobial Agents and Chemotherapy, Vol. 38, No. 9, 1994, pp. 2014-2023. http://dx.doi.org/10.1128/AAC.38.9.2014
- J. I. Yamagishi, T. Kojima, Y. Oyamada, K. Fujimoto, H. Hattori, S. Nakamura, et al., “Alterations in the DNA Topoisomerase IV grlA Gene Responsible for Quinolone Resistance in Staphylococcus aureus,” Antimicrobial Agents and Chemotherapy, Vol. 40, No. 5, 1996, pp. 1157-1163.
- J. A. Lindsay, “Hospital Associated MRSA and Antibiotic Resistance—What Have We Learnt from Genomics?” International Journal of Medical Microbiology, Vol. 303, No. 6-7, 2013, pp. 318-323. http://dx.doi.org/10.1016/j.ijmm.2013.02.005
- H. M. Blumberg, D. Rimland, D. J. Carroll, P. Terry and K. I. Wachsmuth, “Rapid Development of Ciprofloxacin Resistance in Methicillin-Susceptible and Resistant Staphylococcus aureus,” Journal of Infectious Diseases, Vol. 163, No. 6, 1991, pp. 1279-1285. http://dx.doi.org/10.1093/infdis/163.6.1279
- F. B. Marangon, D. Miller, M. S. Muallem, A. C. Romano and E. C. Alfonso, “Ciprofloxacin and Levofloxacin Resistance among Methicillin-Sensitive Staphylococcus Aureus Isolates from Keratitis and Conjunctivitis,” American Journal of Ophthalmology, Vol. 137, No. 3, 2004, pp. 453-458. http://dx.doi.org/10.1016/j.ajo.2003.10.026
- T. Takenouchi, C. Ishii, M. Sugawara, Y. Tokue and S. Ohya, “Incidence of Various gyrA Mutations in 451 Staphylococcus aureus Strains Isolated in Japan and Their Susceptibilities to 10 Fluoroquinolones,” Antimicrobial Agents and Chemotherapy, Vol. 39, No. 7, 1995, pp. 1414- 1418. http://dx.doi.org/10.1128/AAC.39.7.1414
- M. T. G. Holden, L. Hsu, K. Kurt, L. A. Weinert, A. E. Mather, S. R. Harris, et al., “A Genomic Portrait of the Emergence, Evolution, and Global Spread of a Methicillin-Resistant Staphylococcus aureus Pandemic,” Genome Research, Vol. 23, No. 4, 2013, pp. 653-664. http://dx.doi.org/10.1101/gr.147710.112
NOTES
*Corresponding author.

