Advances in Microbiology
Vol.3 No.3(2013), Article ID:33881,8 pages DOI:10.4236/aim.2013.33041
Revealing Toxin Signatures in Cyanobacteria: Report of Genes Involved in Cylindrospermopsin Synthesis from Saxitoxin-Producing Cylindrospermopsis raciborskii
1Departamento de Microbiología, Instituto de Investigaciones Biológicas Clemente Estable, Montevideo, Uruguay
2CSIC, Universidad de la República, Montevideo, Uruguay
3Sección Limnología, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay
Email: claudia.piccini@gmail.com
Copyright © 2013 Claudia Piccini et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 11, 2013; revised April 11, 2013; accepted May 11, 2013
Keywords: Cylindrospermopsin; Cyr; American Ecotypes; Cyanotoxins; Blooms
ABSTRACT
Cyanotoxins are distinctive molecules in Cyanobacteria whose evolutionary origin, radiation and ecological role are still controversial. The cyanobacterium Cylindrospermopsis raciborskii is alternately capable of producing two types of potent toxins, cylindrospermopsin (CYN) or saxitoxin and analogues (SAX). It has been proposed that this species spread to all continents early in its evolutionary history and biogeographical differences in toxin production are found between populations. Most reports indicate that American strains are able to produce SAX but not CYN, while Australian strains are described to produce CYN but not SAX. Here we describe the presence of three genes belonging to the cylindrospermopsin cluster (cyr), cyrA, cyrB and cyrC, in two SAX producing South American C. raciborskii strains, MVCC14 and MVCC19, which due to their differences in morphology, growth preferences, SAX production and genetic context are defined as different ecotypes. No CYN production was detected in either strain (by ELISA) after growth under nitrogen replete or nitrogen-free nutrient conditions. Phylogenetic analyses of cyrA, cyrB and cyrC partial sequences from both strains showed high similarity (>99%) with CYN genes belonging to C. raciborskii strains from Australia and Germany and to Aphanizomenon strains. This is the first report of the presence of cyr genes in strains known to produce only SAX.
1. Introduction
Secondary metabolites in microorganisms are particularly interesting in the case of cyanotoxins [1] because they can reach high concentrations in cyanobacterial blooms posing a serious threat to human and animal health [2]. Cyanobacterial toxins are produced by strains of different species. The evolutionary spreading of these molecules respond to horizontal or vertical genetic transfer mechanisms depending on the kind of toxin and the ability to produce toxins has been lost repeatedly in different lineages of cyanobacteria [3]. The biological role of cyanotoxins in the ecosystem is still under debate [4] and current hypotheses include predation defense [5], nitrogen storage [6-8], protein-modulating role [9] and chemical signaling [10,11].
There are different kind of cyanotoxins with differences in their chemistry, biosynthesis, regulation and mode of action. Cylindrospermopsin (CYN) (C15H21N5O7S) is a water-soluble alkaloid that has cytotoxic effects on animals and humans. This molecule is present in several cyanobacterial species such as Umezakia natans [12], Aphanizomenon ovalisporum and Apha. flos-aquae [13- 15], Raphidiopsis curvata [16], Anabaena lapponica and Ana. bergii [1,17], Lyngbya wollei [18] and Cylindrospermopsis raciborskii [19]. One key factor to explain the distribution of toxins among species and geographical regions is to understand their biological role in the organisms. Although the role of CYN is not yet established, a recent study suggests that CYN in Apha. ovalisporum is involved in phosphorus acquisition by inducing the secretion of alkaline phosphatase by other phytoplankters [11].
Among the toxic bloom-forming cyanobacteria, Cylindrospermopsis raciborskii (Order Nostocales) has been focus of considerable attention due to its apparent invasive movement from tropical towards temperate lakes in Europe and the Americas [20-24]. In this species, CYN biosynthesis, regulation and export are encoded by the 43 kb-long cyr cluster, which comprises 15 open reading frames (ORFs) [25]. The first three genes in CYN biosynthetic pathway encode for an amidinotransferase (cyrA), a nonribosomal peptide synthetase/polyketide synthasa hybrid (NRPS, cyrB) and a polyketide synthase (PKS, cyrC) [19,26], which correspond to the orthologues aoaA, aoaB and aoaC from Apha. ovalisporum [25]. The first enzyme needed to start CYN synthesis is amidinotransferase, via an amidinotransfer onto glycine. According to Kellmann, et al. [27], the amidinotransferase occurrence in a cyanotoxin gene cluster would be unique to CYN-producing cyanobacteria. The guanidinoacetate formed in the first step is then recruited by the NRPS (cyrB gene product) and passed on to the PKS (cyrC gene product).
It has been described that C. raciborskii strains produce alternatively one of two kinds of toxins, cylindrospermopsin (CYN, hepatotoxin) or saxitoxin and its analogues (SAX, neurotoxins) [28,29], however there are no reports of strains producing both toxins simultaneously. Furthermore, the relationship between phylogeny and toxicity in C. raciborskii has not yet been elucidated. CYN-related toxicity in Australian strains has been shown to be unrelated to phylogenetic affiliation, and genes involved in CYN biosynthesis were only present in hepatotoxic strains, suggesting the absence of mutated or inactivated biosynthetic genes in non-toxic strains [30]. In addition, hepatotoxic or CYN-producing strains have been isolated only from Europe, Australia and Asia [31,32], while in the Americas or in Africa have not yet been found [29,31,33]. In addition, strains isolated from the Americas have been shown to produce only SAX [29,33,34]. However, a recent study detected CYN in natural samples from blooms when Aphanizomenon and Cylindrospermopsis were detected (Bittencourt-Oliveira et al. 2011), suggesting that some Brazilian Cylindrospermopsis strains could produce CYN.
Generally, the sequence similarity between toxin-encoding genes is higher than that found for the 16S rRNA. Stucken, et al. [35] have recently described the CYN gene cluster from the C. raciborskii strain CS-505 isolated from Australia, which showed perfect synteny and high sequence conservation with the CYN cluster of C. raciborskii AWT205 described by Mihali, et al. [25]. This finding together with the atypical GC content and the evidence of transposase activity would suggest that CYNproducing C. raciborskii strains recently acquired the cyr cluster by horizontal gene transfer, being Australia the dispersion centre [27,30,36].
In a previous study, we have described two C. raciborskii isolates from Uruguay (MVCC14 and MVCC19), which were phylogenetically distinct (based on ITS and nifH gene sequences) and showed significantly different morphology and growth behavior under different light intensities and phosphate supply [34]. In that study, when analyzing nifH sequences, we could identify subclusters inside the American populations, suggesting an early spread and diversification of the species within the continent. Therefore, we proposed that phenotypic and genetic data support the hypothesis about the existence of different C. raciborskii ecotypes whose success is subject to the local environmental conditions. Both ecotypes showed differences in SAX content (under identical conditions MVCC19 had significantly higher SAX concentration than MVCC14) and were unable to produce CYN under sufficient nutrient conditions [34]. In this study, we describe for the first time the presence of putative CYN biosynthesis encoding genes, cyrA, cyrB and cyrC, in saxitoxin-producing C. raciborskii strains that were described as being different ecotypes. We also performed phylogenetic analyses of their nucleotide sequences and checked for the production of CYN by quantitative Enzyme Linked Immunosorbent Assays (ELISA).
2. Materials and Methods
2.1. Strains and Culture Conditions
Cultures of C. raciborskii strains MVCC14, MVCC19 (Uruguay) and CYP011K (Australia) were grown in BG11 culture medium, modified to have 25% of the CuSO4 of the original recipe after Stanier, et al. [37]. Two growth conditions were assayed: with nitrogen (sodium nitrate) (+N) and nitrogen-free (−N), with no addition of sodium nitrate to BG11 medium. Cultures were grown at 26˚C and 80 µmol photons m−1·s−1 provided by daylight fluorescent tubes with 16:8-h light:dark photoperiod. Static cultures were maintained in 500 mL borosilicate culture bottles that were continuously bubbled with saturated humid air, pre-bubbled in ultrapure sterilized water and pre-filtered by 0.45 µm. According to previous works, MVCC14 and MVCC19 produce SAX but not CYN and the strain CYP011K is a CYN-producer that was used as positive control [34].
2.2. ELISA Test
Cultures of the three strains (MVCC14, MVCC19 and CYP011K) growing in +N and –N culture medium were harvested when in exponential growth phase for toxin detection and kept frozen until analysis. Quantitative analysis of cylindrospermopsin was performed by ELISA (Abraxis, LLC, Warminster, PA, USA) with a detection limit of 0.05 µg·L−1. Five milliliters of cultures were frozen and thawed three times to break the filaments and obtain the toxin. ELISA analyses were performed in accordance with the instructions of the manufacturer. The final CYN concentration was transformed to pg cell−1 after cell counting as described in Piccini, et al. [34].
2.3. DNA Extraction and PCR
Genomic DNA from C. raciborskii strains was extracted according to the protocol described in Piccini, et al. [34]. The concentration and purity of DNA were determined spectrophotometrically at 260 and 280 nm (Nanodrop). DNA from MVCC14, MVCC19 and CYP011K was subjected to PCR using primers designed in this study to amplify partial cyrA gene sequences of C. raciborskii, as well the primers for cyrA, cyrB and cyrC described in Kellmann, et al. [27] (Table 1). The primers were designed based on the sequence from C. raciborskii AWT205 (Australia, Accession number EU140798) and checked against the whole GenBank database using Primer-BLAST (NCBI). All amplifications were performed in a volume of 25 µL containing 1 × PCR buffer (Invitrogen), 1 Unit of Taq (Invitrogen), 0.4 mM of each of the four dNTPs (Invitrogen), 3 mM MgCl2, 10 pM (each) primer, 1.75 µL of bovine seroalbumin (BSA, 30 mg·mL−1) and 2 µL of DNA. PCR water was used as a negative control. PCR conditions consisted of an initial 5 min step at 94˚C, followed by 30 cycles of 1 min at 94 ˚C, 30 sec. at 55˚C and 1 min at 72˚C. PCR products were sent to Macrogen Inc. (Korea) for purification and sequencing using the PCR primers (Table 1).
2.4. Phylogenetic Analyses
DNA sequences were assembled and edited using DNA Baser Sequence Assembly software (Heracle BioSoft). The multiple sequence alignment tools from ClustalW [38] was used to align the sequences. Alignments were checked manually and maximum-likelihood (ML) phylogenetic analyses were conducted using MEGA version 5 [39]. The number of nucleotides included in the phylogenetic analyses was 459 for cyrA, 285 for cyrB and 300 for cyrC.
3. Results and Discussion
The assessment of CYN-production by the studied strains using an ELISA assay gave results below the detection limit of the technique (0.05 µg·L−1) for MVCC14 and MVCC19 grown in –N and +N culture medium. On the other hand, CYN concentration of the strain used as positive control, CYP011K, was 5.66 pg·cell−1 and 0.44 pg·cell−1, under –N and +N growing conditions, respectively. The results found in the CYN-producer strain CYP011K are in agreement with previous findings that showed that CYN concentrations are highest when C. raciborskii grow in the absence of nitrogen [40]. However, other environmental constraints, like low temperature, are also proposed to promote the CYN production [41]. Thus, further studies have to be performed to determine if under different environmental conditions the assayed strains are able to express CYN.
When the presence of cyrA was assessed by PCR amplifications using the primers described in Kellmann, et al. [27] amplicons where not detected neither in MVCC14 nor in MVCC19. However, when DNA amplifications were performed using the newly designed primers, CYLTOX-f and CYLTOX-r, amplicons of the expected size (823 bp) were detected from both strains. Nucleotide sequences corresponding to the obtained amplicons (accession numbers JN014840 and JN014843 for MVCC14 and MVCC19, respectively) shared 99% - 100% similarity with cyrA genes from other C. raciborskii strains (Blast, NCBI). Amplicons obtained from
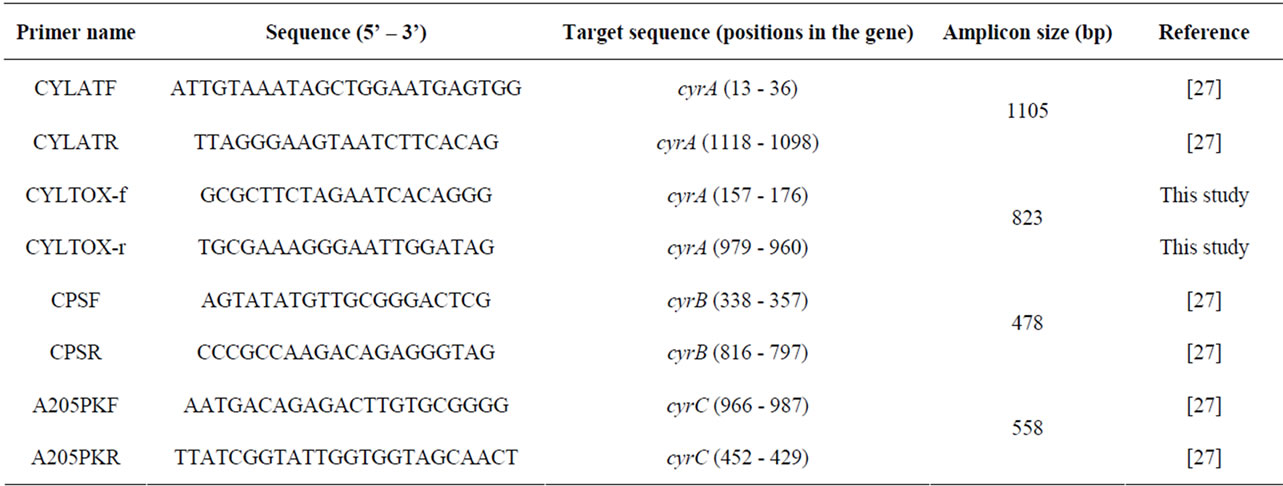
Table 1. Primers used in this study. Expected size of amplicons and positions on the cyrA, cyrB and cyrC genes according to GenBank sequences are shown.
cyrB primers were 478 bp long and the gene sequences (accession numbers JX141619 and JX141620 for MVCC14 and MVCC19, respectively) showed similarities ≥99% with cyrB sequences from Australian and Brazilian C. raciborskii strains, and from Raphidiopsis and Aphanizomenon isolates. In the case of cyrC the size of the amplicon was 558 bp and the obtained sequences were highly similar (99% identity) to those obtained from CYN-producing Australian and Brazilian strains (accession numbers JX141621 and JX141622 for MVCC14 and MVCC19, respectively).
Phylogeny of cyrA gene sequences obtained from the analyzed strains and from GenBank database showed a cluster that was clearly separated from the cluster of sequences belonging to Aphanizomenon sp. and confirmed the identity of the genes detected in MVCC14 and MVCC19 (Figure 1(a)). These findings also suggest that the reports about the lack of the gene cluster based on the absence of cyrA in several C. raciborskii strains could be the result of a methodological limitation [27,42], and consequently lacking gene cluster strains should be perhaps re-analyzed.
Phylogenetic analyses based on the gene sequences obtained in this study showed that Uruguayan strains grouped together to C. raciborskii strains from Australia and Europe (Figures 1(b) and (c)). In order to increase the number of sequences in the phylogenetic analyses of cyr genes we included sequences belonging to a strain of Aphanizomenon. This genus is related to Cylindrospermopsis (Nostocaceae) and strains of Aphanizomenon have been reported as CYN producers [13,15]. The cyrC sequences obtained from the Uruguayan C. raciborskii strains grouped in a cluster that included sequences from Australia, Brazil and Poland (Figure 1(c)).
When evaluating the molecular phylogeny of cyrA, cyrB and cyrC in strains of C. raciborskii from different geographical regions, Kellmann, et al. [27] found these genes exclusively in CYN-producing Australian strains, with the exception of a non toxic Hungarian strain having cyrB but lacking cyrA, cyrC and unable to produce the toxin. Similarly, Rasmussen, et al. [43] detected the cyrB gene in a C. raciborskii strain from Australia that lacked CYN production. In a screening involving non-toxic C. raciborskii strains isolated from Lake Guiers (Senegal), Berger, et al. [44] did not detect the presence of cyrB or cyrC. Similarly, Fathalli, et al. [42] found that Tunisian strains of C. raciborskii did not show neither amidinotransferase, NRPS and PKS genes nor the SAX gene sxtI encoding for a carbamoyltransferase. Putting together, these data suggest that evolutionary mechanisms leading to toxin production by C. raciborskii are quite complex and should be analyzed in a wider biogeographical context.
Genomic studies by Stucken, et al. [35] showed that
 (a)
(a) (b)
(b)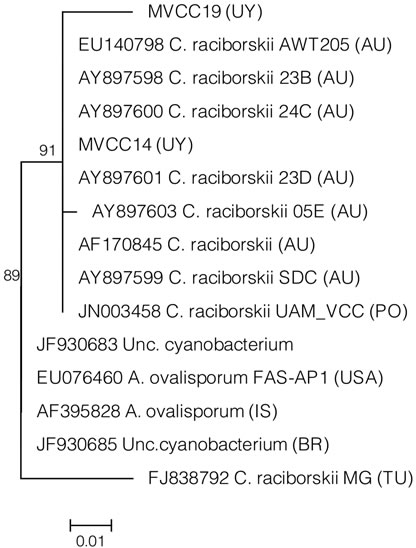 (c)
(c)
Figure 1. Maximum likelihood phylogenetic trees obtained for cyrA (a); cyrB (b) and cyrC (c) gene sequences from C. raciborskii MVCC14 and MVCC19 strains and sequences from GenBank database. Bootstrap values as percentages are given at the nodes (1000 bootstrap resampling). GenBank accession numbers of sequences are shown. Origin of the strains is shown between brackets, AU, Australia; BR, Brazil, GE, Germany; HUN, Hungary; PO, Poland; TU, Tunisia; USA, United States of America; UY, Uruguay; IS, Israel.
cyrA is located between transposase genes (cyrL, cyrM and CRC_01709) in the CYN-producing C. raciborskii strain CS-505. Also, the flanking regions of cyr cluster contained hyp genes at both ends, with molecular chaperones involved in hydrogenase maturation, suggesting that the toxin gene cluster was likely inserted as a unit in this genome region, interrupting the HYP gene cluster and generating a genetic rearrangement [25,35]. The finding of cyrA, cyrB and cyrC genes in SAX-producing C. raciborskii strains unable to synthesize CYN suggests the existence of a non-functional cyr gene cluster. In a recent study Ballot, et al. [45] detected cyrA, cyrB and cyrC in non-CYN producing Apha ovalisporum isolates, suggesting that production of CYN may have been a widespread ability in species of different geographical regions and open the question about which evolutionary mechanisms are involved in the lack of this ability.
According to our previous findings, phylogeny based on ITS, nifH and 16S rRNA showed that MVCC14 and MVCC19 ecotypes isolated from Uruguayan lakes are closely related to other American isolates, particularly from Brazil [34]. Phylogenetic analyses of cyr sequences obtained from the Uruguayan strains showed that they are highly similar to those from Australian CYN-producing, Cylindrospermopsis strains. Moreover, sequences from the three analyzed genes showed to be also closely related to cyr sequences from Brazilian populations. Therefore, there are two possible scenarios for explaining these findings. The first one is in agreement with the hypothesis of early acquisition of CYN-encoding genes in the evolution of this species [27,30,36] before its spreading, with a loss of genes by selection mechanisms. The second scenario would be the acquisition of the cyr cluster by horizontal gene transfer after the species has spread to the American continent. Further research using C. raciborskii strains from the entire American continent and addressing not only genetic but also ecophysiological traits are needed.
To date, available information indicates that C. raciborskii strains have different toxicity patterns related to their biogeography, with most Australian [41,46] and some Asian strains [47] producing CYN, South American strains (Brazil and Uruguay) producing SAX [29,34] and non toxic African or European strains [31,32]. Yılmaz, et al. [14] tested Florida isolates of this species and found that they do not have the ability to produce CYN. Putting together, these findings would indicate a high diversification of this species within this continent; which agrees with the evidence of subclusters within the nifH American cluster of C. raciborskii described by Piccini, et al. [34].
4. Conclusion
Here we report for the first time the presence of genes belonging to the CYN gene cluster, cyrA, cyrB and cyrC in two C. raciborskii ecotypes that produce SAX. Current dispersal hypothesis for C. raciborskii suggests that the ability to produce CYN originated in Australia and subsequently radiated to Europe and Asia but not to the Americas [27]. In addition, transcontinental radiation of the species might have occurred at an early stage of evolution, as evidenced by three clear clusters in ITS-based phylogenies (America, Europe, Africa-Australia) [33,48]. It has been reported that CYN genes are evolutionary more conserved than genes encoding for other toxins [36]. The presence of cyr genes highly similar to those belonging to current CYN-producers in C. raciborskii strains that cannot produce the toxin could indicate either that this cluster may have been present in American populations in the past and are being lost or that these genes have been recently acquired by horizontal transfer.
5. Acknowledgements
We thank Dermot Antoniades for performing the English correction and a critical revision of the manuscript, A. Humpage (SA Water and University of Adelaide Medical School, Adelaide, Australia) who kindly provided the CYP011K strain and Beatriz Brena for performing the ELISA tests. This research was funded by ANII grant FCE_2007-353, ANII PR FCE_2009_1_2330 and PEDECIBA (Uruguay).
REFERENCES
- L. Pearson, T. Mihali, M. Moffitt, R. Kellmann and B. Neilan, “On the Chemistry, Toxicology and Genetics of the Cyanobacterial Toxins, Microcystin, Nodularin, Saxitoxin and Cylindrospermopsin,” Marine Drugs, Vol. 8, No. 5, 2010, pp. 1650-1680. doi:10.3390/md8051650
- I. Chorus and J. Bartram, “Toxic Cyanobacteria in Water. A Guide to Their Public Health Consequences, Monitoring and Management,” Chapman & Hall, London, 1999. doi:10.4324/9780203478073
- A. Rantala, D. P. Fewer, M. Hisbergues, L. Rouhiainen, J. Vaitomaa, T. Börner and K. Sivonen, “Phylogenetic Evidence for the Early Evolution of Microcystin Synthesis,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 101, No. 2, 2004, pp. 568- 573. doi:10.1073/pnas.0304489101
- B. A. Neilan, L. A. Pearson, J. Muenchhoff, M. C. Moffitt and E. Dittmann, “Environmental Conditions That Influence Toxin Biosynthesis in Cyanobacteria,” Environmental Microbiology, Vol. 15, No. 5, 2012, pp. 1239-1253.
- A. E. Wilson, O. Sarnelle and A. R. Tillmanns, “Effects of Cyanobacterial Toxicity and Morphology on the Population Growth of Freshwater Zooplankton: Meta-Analyses of Laboratory Experiments,” Limnology and Oceanography, Vol. 51, No. 4, 2006, pp. 1915-1924. doi:10.4319/lo.2006.51.4.1915
- F. Pomati, M. C. Moffitt, R. Cavaliere, and B. A. Neilan, “Evidence for Differences in the Metabolism of Saxitoxin and C1+2 Toxins in the Freshwater Cyanobacterium Cylindrospermopsis raciborskii T3,” Biochimica et Biophysica Acta (BBA)—General Subjects, Vol. 1674, No. 1, 2004, pp. 60-67. doi:10.1016/j.bbagen.2004.05.006
- A. Cembella and U. John, “Ecological Studies,” In: E. Granéli and J. T. Turner, Eds., Molecular Physiology of Toxin Production and Growth Regulation in Harmful Algae, Springer, Berlin, 2006, pp. 215-227.
- K. Soto-Liebe, A. A. Murillo, B. Krock, K. Stucken, J. J. Fuentes-Valdés, N. Trefault, A. Cembella, and M. Vásquez, “Reassessment of the Toxin Profile of Cylindrospermopsis raciborskii T3 and Function of Putative Sulfotransferases in Synthesis of Sulfated and Sulfonated PSP Toxins,” Toxicon, Vol. 56, No. 8, 2010, pp. 1350- 1361. doi:10.1016/j.toxicon.2010.07.022
- Y. Zilliges, J.-C. Kehr, S. Meissner, K. Ishida, S. Mikkat, M. Hagemann, A. Kaplan, T. Börner and E. Dittmann, “The Cyanobacterial Hepatotoxin Microcystin Binds to Proteins and Increases the Fitness of Microcystis under Oxidative Stress Conditions,” PLoS ONE, Vol. 6, No. 3, 2011, Article ID: e17615. doi:10.1371/journal.pone.0017615
- D. Schatz, Y. Keren, A. Vardi, A. Sukenik, S. Carmeli, T. Börner, E. Dittmann and A. Kaplan, “Towards Clarification of the Biological Role of Microcystins, a Family of Cyanobacterial Toxins,” Environmental Microbiology, Vol. 9, No. 4, 2007, pp. 965-970. doi:10.1111/j.1462-2920.2006.01218.x
- Y. Bar-Yosef, A. Sukenik, O. Hadas, Y. Viner-Mozzini and A. Kaplan, “Enslavement in the Water Body by Toxic Aphanizomenon ovalisporum, Inducing Alkaline Phosphatase in Phytoplanktons,” Current Biology, Vol. 20, No. 17, 2010, pp. 1557-1561. doi:10.1016/j.cub.2010.07.032
- K.-I. Harada, I. Ohtani, K. Iwamoto, M. Suzuki, M. F. Watanabe, M. Watanabe and K. Terao, “Isolation of Cylindrospermopsin from a Cyanobacterium Umezakia natans and Its Screening Method,” Toxicon, Vol. 32, No. 1, 1994, pp. 73-84. doi:10.1016/0041-0101(94)90023-X
- R. Banker, S. Carmeli, O. Hadas, B. Teltsch, R. Porat and A. Sukenik, “Identification of Cylindrospermopsin in Aphanizomenon ovalisporum (Cyanophyceae) Isolated from Lake Kinneret, Israel,” Journal of Phycology, Vol. 33, No. 4, 1997, pp. 613-616. doi:10.1111/j.0022-3646.1997.00613.x
- M. Yılmaz, E. J. Phlips, N. J. Szabo and S. Badylak, “A Comparative Study of Florida Strains of Cylindrospermopsis and Aphanizomenon for Cylindrospermopsin Production,” Toxicon, Vol. 51, No. 1, 2008, pp. 130-139. doi:10.1016/j.toxicon.2007.08.013
- K. Preußel, A. Stüken, C. Wiedner, I. Chorus and J. Fastner, “First Report on Cylindrospermopsin Producing Aphanizomenon flos-aquae (Cyanobacteria) Isolated from Two German Lakes,” Toxicon, Vol. 47, No. 2, 2006, pp. 156-162. doi:10.1016/j.toxicon.2005.10.013
- R. Li, W. W. Carmichael, S. Brittain, G. K. Eaglesham, G. R. Shaw, Y. Liu and M. M. Watanabe, “First Report of the Cyanotoxins Cylindrospermopsin and Deoxycylindrospermopsin from Raphidiopsis curvata (Cyanobacteria),” Journal of Phycology, Vol. 37, No. 6, 2001, pp. 1121-1126. doi:10.1046/j.1529-8817.2001.01075.x
- L. Spoof, K. A. Berg, J. Rapala, K. Lahti, L. Lepistö, J. S. Metcalf, G. A. Codd and J. Meriluoto, “First Observation of Cylindrospermopsin in Anabaena lapponica Isolated from the Boreal Environment (Finland),” Environmental Toxicology, Vol. 21, No. 6, 2006, pp. 552-560. doi:10.1002/tox.20216
- M. Seifert, G. McGregor, G. Eaglesham, W. Wickramasinghe and G. Shaw, “First Evidence for the Production of Cylindrospermopsin and Deoxy-Cylindrospermopsin by the Freshwater Benthic Cyanobacterium, Lyngbya wollei (Farlow ex Gomont) Speziale and Dyck,” Harmful Algae, Vol. 6, No. 1, 2007, pp. 73-80. doi:10.1016/j.hal.2006.07.001
- M. A. Schembri, B. A. Neilan and C. P. Saint, “Identification of Genes Implicated in Toxin Production in the Cyanobacterium Cylindrospermopsis raciborskii,” Environmental Toxicology, Vol. 16, No. 5, 2001, pp. 413-421. doi:10.1002/tox.1051
- J. Padisák, “Cylindrospermopsis raciborskii (Woloszyska) Seenayya et Subba Raju, an Expanding, Highly Adaptive Cyanobacterium: Worldwide Distribution and Review of Its Ecology,” Archiv für Hydrobiologie Supplement, Vol. 107, 1997, pp. 563-593.
- A. D. Chapman and C. L. Schelske, “Recent Appearance of Cylindrospemopsis (Cyanobacteria) in Five Hypereutrophic Florida Lakes,” Journal of Phycology, Vol. 33, No. 2, 1997, pp. 191-195. doi:10.1111/j.0022-3646.1997.00191.x
- A. Stüken, J. Rücker, T. Endrulat, K. Preussel, M. Hemm, B. Nixdorf, U. Karsten and C. Wiedner, “Distribution of Three Alien Cyanobacterial Species (Nostocales) in Northeast Germany: Cylindrospermopsis raciborskii, Anabaena bergii and Aphanizomenon aphanizomenoides,” Phycologia, Vol. 45, No. 6, 2006, pp. 696-703. doi:10.2216/05-58.1
- S. Bonilla, L. Aubriot, M. C. S. Soares, M. GonzálezPiana, A. Fabre, V. L. M. Huszar, M. Lürling, D. Antoniades, J. Padisák and C. Kruk, “What Drives the Distribution of the Bloom-Forming Cyanobacteria Planktothrix agardhii and Cylindrospermopsis raciborskii?” FEMS Microbiology Ecology, Vol. 79, No. 3, 2012, pp. 594-607. doi:10.1111/j.1574-6941.2011.01242.x
- A. Sukenik, O. Hadas, A. Kaplan and A. Quesada, “Invasion of Nostocales (Cyanobacteria) to Subtropical and Temperate Freshwater Lakes Physiological, Regional and Global Driving Forces,” Frontiers in Microbiology, Vol. 3, 2012, pp. 2-9.
- T. K. Mihali, R. Kellmann, J. Muenchhoff, K. D. Barrow and B. A. Neilan, “Characterization of the Gene Cluster Responsible for Cylindrospermopsin Biosynthesis,” Applied and Environmental Microbiology, Vol. 74, No. 3, 2008, pp. 716-722. doi:10.1128/AEM.01988-07
- G. Shalev-Alon, A. Sukenik, O. Livnah, R. Schwarz and A. Kaplan, “A Novel Gene Encoding Amidinotransferase in the Cylindrospermopsin Producing Cyanobacterium Aphanizomenon ovalisporum,” FEMS Microbiology Letters, Vol. 209, No. 1, 2002, pp. 87-91. doi:10.1111/j.1574-6968.2002.tb11114.x
- R. Kellmann, T. Mills and B. Neilan, “Functional Modeling and Phylogenetic Distribution of Putative Cylindrospermopsin Biosynthesis Enzymes,” Journal of Molecular Evolution, Vol. 62, No. 3, 2006, pp. 267-280. doi:10.1007/s00239-005-0030-6
- D. J. Griffiths and M. L. Saker, “The Palm Island Mystery Disease 20 Years on: A Review of Research on the Cyanotoxin Cylindrospermopsin,” Environmental Toxicology, Vol. 18, No. 2, 2003, pp. 78-93. doi:10.1002/tox.10103
- N. Lagos, H. Onodera, P. A. Zagatto, D. Andrinolo, S. M. F. Q. Azevedo and Y. Oshima, “The First Evidence of Paralytic Shellfish Toxins in the Freshwater Cyanobacterium Cylindrospermopsis raciborskii, Isolated from Brazil,” Toxicon, Vol. 37, No. 10, 1999, pp. 1359-1373. doi:10.1016/S0041-0101(99)00080-X
- K. Stucken, A. A. Murillo, K. Soto-Liebe, J. J. FuentesValdés, M. A. Méndez and M. Vásquez, “Toxicity Phenotype Does Not Correlate with Phylogeny of Cylindrospermopsis raciborskii Strains,” Systematic and Applied Microbiology, Vol. 32, No. 1, 2009, pp. 37-48. doi:10.1016/j.syapm.2008.10.002
- M. L. Saker, I. C. G. Nogueira and V. M. Vasconcelos, “Distribution and Toxicity of Cylindrospermopsis raciborskii (Cyanobacteria) in Portuguese Freshwaters,” Limnetica, Vol. 22, No. 3-4, 2003, pp. 129-136
- J. Fastner, R. Heinze, A. R. Humpage, U. Mischke, G. K. Eaglesham and I. Chorus, “Cylindrospermopsin Occurrence in Two German Lakes and Preliminary Assessment of Toxicity and Toxin Production of Cylindrospermopsis raciborskii (Cyanobacteria) Isolates,” Toxicon, Vol. 42, No. 3, 2003, pp. 313-321. doi:10.1016/S0041-0101(03)00150-8
- S. Haande, T. Rohrlack, A. Ballot, K. Røberg, R. Skulberg, M. Beck and C. Wiedner, “Genetic Characterisation of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) Isolates from Africa and Europe,” Harmful Algae, Vol. 7, No. 5, 2008, pp. 692-701. doi:10.1016/j.hal.2008.02.010
- C. Piccini, L. Aubriot, A. Fabre, V. Amaral, M. González-Piana, A. Giani, C. C. Figueredo, L. Vidal, C. Kruk and S. Bonilla, “Genetic and Eco-Physiological Differences of South American Cylindrospermopsis raciborskii Isolates Support the Hypothesis of Multiple Ecotypes,” Harmful Algae, Vol. 10, No. 6, 2011, pp. 644-653. doi:10.1016/j.hal.2011.04.016
- K. Stucken, U. John, A. Cembella, A. A. Murillo, K. Soto-Liebe, J. J. Fuentes-Valdés, M. Friedel, A. M. Plominsky, M. Vásquez and G. Glöckner, “The Smallest Known Genomes of Multicellular and Toxic Cyanobacteria: Comparison, Minimal Gene Sets for Linked Traits and the Evolutionary Implications,” PLoS ONE, Vol. 5, No. 2, 2010. doi:10.1371/journal.pone.0009235
- A. Stüken and K. S. Jakobsen, “The Cylindrospermopsin Gene Cluster of Aphanizomenon sp. Strain 10E6: Organization and Recombination,” Microbiology, Vol. 156, No. 8, 2010, pp. 2438-2451. doi:10.1099/mic.0.036988-0
- R. Stanier, R. Kunisawa, M. Mandel and G. Cohen-Bazire, “Purification and Properties of Unicellular BlueGreen Algae (Order Chroococcales),” Bacteriological Reviews, Vol. 35, No. 1971, pp. 171-205.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, “Clustal W and Clustal X Version 2.0,” Bioinformatics, Vol. 23, No. 21, 2007, pp. 2947- 2948. doi:10.1093/bioinformatics/btm404
- K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, “MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods,” Molecular Biology and Evolution, Vol. 28, No. 10, 2011, pp. 2731- 2739. doi:10.1093/molbev/msr121
- M. L. Saker and B. A. Neilan, “Varied Diazotrophies, Morphologies, and Toxicities of Genetically Similar Isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from Northern Australia,” Applied and Environmental Microbiology, Vol. 67, No. 4, 2001, pp. 1839- 1845. doi:10.1128/AEM.67.4.1839-1845.2001
- M. L. Saker and D. J. Griffiths, “The Effect of Temperature on Growth and Cylindrospermopsin Content of Seven Isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from Water Bodies in Northern Australia,” Phycologia, Vol. 39, No. 4, 2000, pp. 349-354. doi:10.2216/i0031-8884-39-4-349.1
- A. Fathalli, A. Jenhani, C. Moreira, J. Azevedo, M. Welker, M. Romdhane, A. Antunes and V. Vasconcelos, “Genetic Variability of the Invasive Cyanobacteria Cylindrospermopsis raciborskii from Bir M’cherga Reservoir (Tunisia),” Archives of Microbiology, Vol. 193, No. 8, 2011, pp. 595-604. doi:10.1007/s00203-011-0704-y
- J. P. Rasmussen, S. Giglio, P. T. Monis, R. J. Campbell and C. P. Saint, “Development and Field Testing of a RealTime PCR Assay for Cylindrospermopsin-Producing Cyanobacteria,” Journal of Applied Microbiology, Vol. 104, No. 5, 2008, pp. 1503-1515. doi:10.1111/j.1365-2672.2007.03676.x
- C. Berger, N. Ba, M. Gugger, M. Bouvy, F. Rusconi, A. Couté, M. Troussellier and C. Bernard, “Seasonal Dynamics and Toxicity of Cylindrospermopsis raciborskii in Lake Guiers (Senegal, West Africa),” FEMS Microbiology Ecology, Vol. 57, No. 3, 2006, pp. 355-366. doi:10.1111/j.1574-6941.2006.00141.x
- A. Ballot, J. Ramm, T. Rundberget, R. N. Kaplan-Levy, O. Hadas, A. Sukenik and C. Wiedner, “Occurrence of Non-Cylindrospermopsin-Producing Aphanizomenon valisporum and Anabaena bergii in Lake Kinneret (Israel),” Journal of Plankton Research, Vol. 33, No. 11, 2011, pp. 1736-1746. doi:10.1093/plankt/fbr071
- P. R. Hawkins, N. R. Chandrasena, G. J. Jones, A. R. Humpage and I. R. Falconer, “Isolation and Toxicity of Cylindrospermopsis raciborskii from an Ornamental Lake,” Toxicon, Vol. 35, No. 3, 1997, pp. 341-346. doi:10.1016/S0041-0101(96)00185-7
- D. Chonudomkul, W. Yongmanitchai, G. Theeragool, M. Kawachi, F. Kasai, K. Kaya and M. M. Watanabe, “Morphology, Genetic Diversity, Temperature Tolerance and Toxicity of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) Strains from Thailand and Japan,” FEMS Microbiology Ecology, Vol. 48, No. 3, 2004, pp. 345-355. doi:10.1016/j.femsec.2004.02.014
- M. Gugger, R. Molica, B. Le Berre, P. Dufour, C. Bernard, and J.-F. Humbert, “Genetic Diversity of Cylindrospermopsis strains (Cyanobacteria) Isolated from Four Continents,” Applied and Environmental Microbiology, Vol. 71, No. 2, 2005, pp. 1097-1100. doi:10.1128/AEM.71.2.1097-1100.2005

