Open Journal of Medical Microbiology
Vol.3 No.3(2013), Article ID:37060,5 pages DOI:10.4236/ojmm.2013.33030
Plasmid Profiling and Curing Analysis of Fluoroquinolone Multidrug Resistant Pseudomonas aeruginosa in Benin City, Nigeria
1Department of Hematology, College of Health Sciences, Igbinedion University, Okada, Nigeria
2Lahor Research and Medical Centre, 121 Old Benin—Agbor Road, Benin City, Nigeria
3Department of Medical Laboratory Science, Nnamdi Azikiwe University, Awka, Nigeria
4Department of Medical Laboratory Science, College of Health Sciences Niger Delta University, Wilberforce Island, Amassoma, Bayelsa State
5Department of Microbiology, Faculty of Life Sciences, University of Benin, Benin City, Nigeria
6Department of Medical Microbiology, College of Health Sciences, Igbinedion University Okada, Okada, Nigeria
7Faculty of Pharmacy, Delta State University, Abraka, Nigeria
Email: *fredleo2547@yahoo.com
Copyright © 2013 F. A. Ehiaghe et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 27, 2013; revised May 27, 2013; accepted June 4, 2013
Keywords: Pseudomonas aeruginosa; Fluoroquinolone; Plasmids Analysis; Benin City
ABSTRACT
Aim: To determine the Plasmid DNA profile of the multidrug resistant strains of Pseudomonas aeruginosa in the clinical isolates. Materials and Methods: Of the 150 clinical samples (Ear swab, Urine, Wound swab, Sputa and Semen) received at Lahor Research Laboratory and Medical center in Benin City, between January 2010 and December 2012, 36 (24%) yielded significant growth of P. aeruginosa. Samples were cultured on MacConkey and Blood agar. Clinical isolates were identified using standard method. Antibiotics susceptibility test employing agar disc diffusion method was used. Clinical isolates were subjected to Plasmid DNA profiling and curing test was carried out at Lahor Molecular Laboratory. This was followed by a post plasmid curing susceptibility test. Agarose gel electrophoresis was carried out to separate the Plasmid DNA using standard method. Bands were visualized using UV illuminator. Results: Wound swabs had the highest numbers of clinical isolates of P. aeruginosa (55.6%) followed by Urine, Semen, Sputa and Ear swab (19.4%, 11.0%, 8.3%, and 5.6%) respectively. Before the isolates were cured of their plasmid, 39% of the P. aeruginosa strains were found to be resistant to Ciprofloxacin (CPX), 47%, Ofloxacin (OFX), 44% Pefloxacin (PEF) and 56% Sparfloxacin (SPX). After plasmid curing, the new antibiogram of the isolates showed that some clinical isolates that hitherto were resistant to a given Fluoroquinolone became susceptible, 36% to CPX, 12% to OFX, 12.5% to PEF and 15% to SPX. Agarose gel electrophoresis carried out on the Plasmid DNA revealed that there was detectable Plasmid DNA in 13.9% of the clinical isolates analyzed. Conclusion: There is an alarming increase of clinical infections caused by multidrug resistant strains of P. aeruginosa.13.9% of the multidrug resistance strains of P. aeruginosa in Benin City were Plasmid mediated. Treatment should be based on current Laboratory Susceptibility Test results of the isolates.
1. Introduction
Pseudomonas aeruginosa is a Gram-negative aerobic, coccobacillus bacterium with unipolar motility. In mixed cultures, it can be isolated as clear colonies on MacConkey agar (as it does not ferment lactose) which will test positive for oxidase [1]. It is often preliminarily identified by its pearlescent appearance and grape like odor invitro. Definitive clinical identification of P. aeruginosa often includes identifying the production of pyocyanin and fluorescein as well as its ability to grow at 42˚C. P. aeruginosa is capable of growth in diesel and jet fuel, where it is known as a hydrocarbon-using microorganism [2].
The genome of P. aeruginosa is relatively large (6 - 7 mb) and encodes around 6000 open reading frames (ORFs), depending on the strain. There are 5021 genes that are conserved across all 5 (five) genomes analyzed with at least 70% sequence identity [3]. One of the most worrisome characteristics of P. aeruginosa is its antibiotic susceptibility, which is attributable to a concerted action of multidrug efflux pump with chromosomally encoded antibiotic resistance genes (mexAB, mexXY) and the low permeability of the bacterial cellular envelopes. P. aeruginosa can easily develop acquired resistance either by mutation in chromosomally encoded genes or by the horizontal Plasmid DNA transfer of antibiotic resistance determinants [4].
Plasmid DNA are extra chromosomal element of finite size, usually stably inherited within a bacterial cell line and potentially capable of transfer between strains, species or genera [5]. They were first discovered in enteric bacteria and from the late 1950s onwards were increaseingly associated with antibiotics resistance. Its main function is to carry antibiotics resistant genes, which are responsible for the increase pathogenicity of most bacteria [5,6].
The aim of this investigation is to determine the antimicrobial susceptibility of clinical isolates P. aeruginosa to four Fluoroquinolone and to determine the percentage of multidrug resistance of P. aeruginosa strains that are plasmid mediated.
2. Materials and Methods
2.1. Samples Collection
150 samples were collected from Lahor Research Laboratory and medical center, 121 Old Benin-Agbor Road, Ikpoba Hill, Benin City, Edo state, Nigeria. Of which include specimen of ear swab, wound swab, mid stream urine, sputum and semen. Samples were collected between January 2010 to December 2012. Ethical approval and patient consent was obtained before sample collection.
2.2. Cultural Methods
With a grease pencil, the bottom of MacConkey agar and Blood agar Plates were divided into three parts; the samples were cultured on the media and incubated at 37˚C for 24 to 48 hours depending on the nature of the sample. Of the 150 samples collected, 36(24%) yielded significant growth of P. aeruginosa and were stored on nutrient agar slant of MarCartney bottle.
2.3. Identification of Isolates
The isolates were identified by standard techniques [7]. This included colonial appearance of the organism on the media, morphological characteristics such as size, form, elevation, opacity, colour, odour and edge. Gram’s technique was used to classify the organism into Gram negative coccobacillus; they were further subjected to specific biochemical tests such as citrate utilization, indole production, urease production, oxidase test, sugar fermentation (sucrose, salicin, lactose, maltose and mannitol) test along side with motility test.
2.4. Antibiotics Susceptibility Testing
Susceptibility tests were performed on each P. aeruginosa isolated using the agar disc diffusion method described by [8]. Antibiotic disc of Ciprofloxacin (10 ug), Ofloxacin (10 ug), Pefloxacin (10 ug) and Sparfloxacin (10 ug) were used. Zones of inhibition less than 13 mm were regarded as resistant while those measuring 15 mm and above as susceptible. The multi resistant isolate that were resistant to all the four Antibiotics were taking for Plasmid DNA curing.
2.5. Plasmids Curing
Isolates that were resistant to the Fluoroquinolone were subjected to standard plasmid curing method [9].
• Overnight broth culture was inoculated into 4.5 ml nutrient broth.
• 0.5 ml of Sodium deodecyl sulphate was added and Incubated for 48 hours at 37˚C.
• 0.5 ml of the broth was added into a freshly prepared 4.5 ml nutrient broth, it was incubate for another 24 hours at 37˚C.
• After which post plasmid Antibiotic susceptibility was carried out.
On each of the clinical isolates of P.aeruginosa as described [8].
The reason for this second susceptibility test is to check if the curing agent has removed the resident plasmid present in the resistant isolates.
Preparations of Reagents
Reagents Composition Reagents
1. Buffer 1A (Cell Resuspension 50 mM Tris-Cl
Buffer)
pH 8.0
10 mM EDTA
100 ug/ml
DNase free RNase
2. Buffer 2B (Neutralization 3.0 mM Potassium
solution) acetate, pH 5
3. Buffer 3C (Equilibration 6 M guanidine
Buffer) hydrochloride
4. Cell Lysis Buffer 200 mM NaOH
1% Sodium deodecyl
sulphate
0.8% Agarose Gel
preparation
5. 100 ml of Tris Boric EDTA 108 g Tris
(TBE Buffer) 55 g Boric acid
40 ml of 0.5 M EDTA,
pH 8
8 g of Agarose powder
2.6. Agarose Gel Electrophoresis
Plasmid isolation was performed using the alkaline lysis method described by [10]. After isolation of plasmid DNA, a horizontal agarose gel electrophoresis was carried out with the method described by [11].
The plasmid profiling was carried out by sub-culturing those isolate onto nutrient agar plate and incubated for 24 hour at 37˚C. The isolate was harvested into Eppendorf tube. 200 µl of buffer 1A (Cell Resuspension Buffer) was added and vortexed for proper mixing. 200 µl of Cell lysing solution was added. The tubes were Inverted 20 times at room temperature. Add 300 µl of ice-cold buffer 2B (Neutralization solution), vortexed and keep on ice for 30 minutes, centrifuged at 3000 × g for 15 minutes. To supernatant, 700 µl of chloroform was added and centrifuged at 3000 × g for 10 minutes. To 500 µl aqueous layer, 1 ml of absolute ethanol was added and kept in ice for 1 hour. This was centrifuged at 3000 × g for 30 mins and the pellet was washed in 70% ethanol. 100 µl of buffer 3C (Equilibration Buffer) was added to the dried pellet.
After the DNA have been extracted from the cells, 0.8% agarose gel with wells was prepared, in which 2 ul of the loading dye was mixed with 10 ul of the 100 base pair molecular markers and transferred into the first well. 2 ul of the loading dye was mixed with the sample and it was transferred into the other wells. Agarose gel Electrophoresis was carried out on the plasmid DNA at 90 v for 60 minutes. It was viewed under UV Trans-illuminetion for the band harboring plasmid.
3. Results
The results obtained are shown in Tables 1-3 and Plate 1.
Table1: Shows P. aeruginosa from various clinical sources. Isolates were highest from wound swabs.
Table 2: Shows the Antibiogram pre Plasmid curing of P. aeruginosa isolates. Whereas isolates showed the highest resistance to Sparfloxacin (56%), they exhibited the lowest resistance to Ciprofloxacin (39%).
Table 3: Shows the Antibiogram post Plasmid Curing of multidrug resistant strains of P. aeruginosa to four Fluoroquinolone. Of the 20 strains that were resistant to Sparfloxacin, 3 were plasmid mediated while none was plasmid mediated in the 14 strains that were resistant to Ciprofloxacin.
Plate 1: Shows the Results of the Plasmids DNA profiling analyzed in a 0.8% Agarose gel Electrophoresis. Five samples 16, 18, 26, 28 and 29 were plasmid positive.
4. Discussion
Our study revealed that wound swab had the highest number of clinical isolates of P. aeruginosa (55.6%) followed by urine sample (19.4%), Semen (11%) and Sputa sample (8.3%). These may be due to the fact that surgical wound sample site are exposed to many ubiquitous environmental pathogen which include P. aureginosa found in unsterile surface, water and soil. These findings have been reported by [12]. P. aeruginosa has a prevalence of 29.6% among pathogens isolated from surgical wound site. The infection appears to be common in hospital with relaxed hygienic measure. This is also in accordance with these findings. It is the most common pathogen causing wound infection especially in developing countries. It is an epitome of opportunistic nosocomial pathogen, which causes a wide spectrum of infection and lead to substantial morbidity in immune compromised patients [13-15].
It was observed that at Antibiogram pre curing stage of P. aeruginosa, the results show that 39% of the microorganism was found to be resistant to Ciprofloxacin, 47% to Ofloxacin, 44% to Pefloxacin and 59% to Sparfloxacin. This finding has been reported by other Authors. A high number of acquired resistance genes coding for aminoghyloside modifying enzymes and β-Lactamases have been noted in P. aeruginosa [16]. Extended spectrum
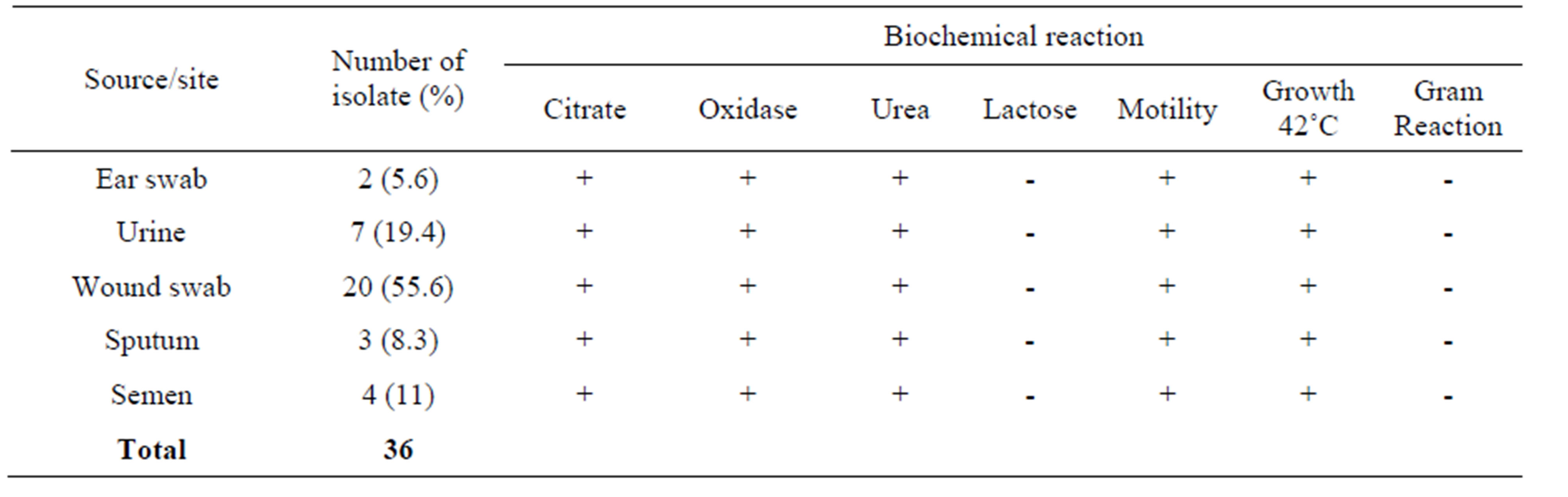
Table 1. P. aeruginosa isolated from various sources.

Table 2. Antibiogram pre curing of P. aeruginosa to four fluoroquinolone.
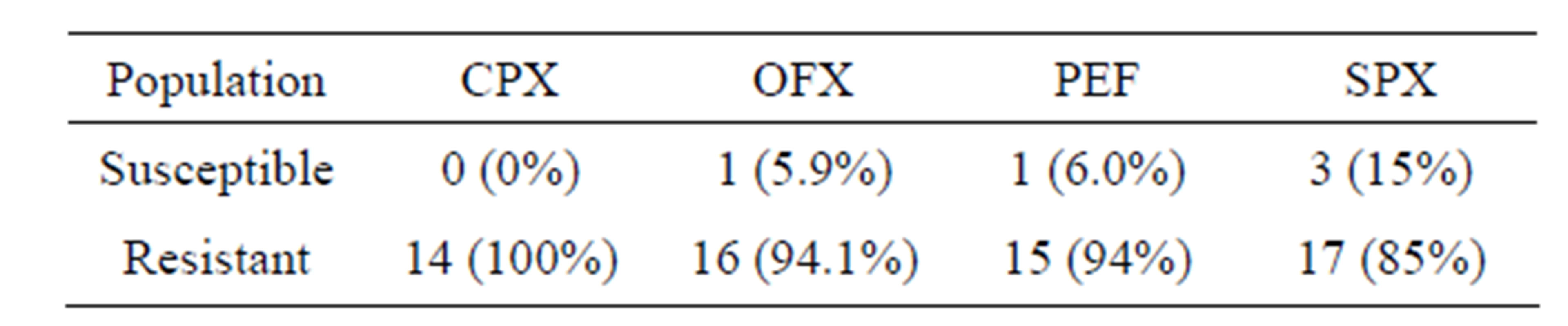
Table 3. Antibiogram post plasmid curing of multidrug resistant strains of Pseudomonas aeruginosa to four fluoroquinolone.

Plate 1 . Results of the plasmid DNA profile analysed in a 0.8% agarose gel, L is a DNA ladder with size range of 0.1 - 5.1 kbp, samples 16, 18, 26, 28 and 29 have a plasmid band greater than 1.5 kbp.
β-Lactamases have been increasingly reported about this micro organism [17]. Metallo β-Lactamases have also emerged in P. aeruginosa [18].
Once colonization and infection are established, P. aeruginosa becomes one of the worst pathogens of human and it is known to possess intrinsic multi-drug resistance capabilities [19,20]. It has a large genome of about 6.3 million base pairs (bp), house 8 virulence genes, the large genome size increase the probability of possible mutation sites and thus gives reasons for its virulence versatile, its growing multi-drug resistance and the high mortality rate associated with its infection [21].
It was also observed that at Antibiogram pre-curing stage of P. aeruginosa, results revealed that 61.1% of the isolates were sensitive to CPX, 52.8% to OFX, 56.0% to PEF and 44.4% Sparfloxacin. It might be due to that some clinical isolates of P. aeruginosa do not hyper expressed the efflux system genes that confer resistance to Antibiotics. This is in line with these finding. The hyper expression of MexAB-OprM efflux pump mainly found in P. aeruginosa are not present in some clinical isolates of P. aeruginosa, hence they are susceptible to most Antibiotics [22].
At Antibiogram post curing stage of P. aeruginosa, our result shows that some clinical isolates that were resistant to the same Fluoroquinolone at Antibiogram precuring stage became susceptible at the Antibiogram post curing stage. 5.9% to OFX, 6.8% to PEF and 15% to SPX. These might be due to the fact that the plasmid borne multidrug resistance gene had been denatured or removed by the sodium deodecyl sulphate used as the curing agent. This is in agreement with these findings. P. aeruginosa is a classic opportunistic pathogen especially because of its innate resistance to many antibiotics and its acquired resistance due to plasmid mediation [23].
The plasmids analysis revealed that there were detectable plasmid DNA genes in sample 16, 18, 26, 28 and 29, which represents 13.9% of the multidrug resistance strains of the clinical isolates analyzed. The plasmids DNA genes isolated in the 0.8% Agarose gel Electrophoresis might be the bacterial contents that confer plasmid resistance on the clinical isolates of P. aeruginosa. This is in accordance with these findings. The plasmids profile analysis examines the total bacterial plasmid content and is separated by electrophoresis. This method is used to assess the spread of antibiotics plasmid DNA resistance determinates [23].
5. Conclusion and Recommendation
There is an alarming increase of clinical infection caused by multidrug resistant strains of P. aeruginosa. Plasmid analysis revealed that 13.9% of the multidrug resistant strains of P. aeruginosa were plasmid mediated in Benin City. Treatment should be based on current laboratory susceptibility test results.
REFERENCES
- K. J. Ryan and C. G. Ray, “Sheris Medical Microbiology,” 4th Edition, McGraw Hill, New York, 2004.
- A. Balcht and R. Smith, “Pseudomonas aeruginosa: Infection and Treatment,” Informa Healthcare, 1994, pp. 83-84.
- K. Mathee, G. Narasimhan, C. Valdes, X. Qiu, J. M. Metewish, M. Koehrsen, A. Rokas and C. N. Yandava, “Dynamics of Pseudomonas aeruginosa Genome Evolution,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 105, No. 8, 2008, pp. 3100-3105. doi:10.1073/pnas.0711982105
- K. Poole, “Efflux Mediated Multiresistance in Gram-Negative Bacteria,” Clinical Microbiology and Infection, Vol. 10, No. 1, 2004, pp. 12-26. doi:10.1111/j.1469-0691.2004.00763.x
- V. Alan, M. Jesus, and W. J. Rober, “The Role of Plasmid in Phytopathogenic Bacteria: Mobile Arsenals,” Journal of General Virology, Vol. 147, No. 4, 2001, pp. 4763- 4780.
- N. Datta and V. M. Hughes, “Plasmid of the Same Inc. Group in Enterobacteria before and after the Medical Use of Antibiotics,” Nature, Vol. 306, No. 5943, 1983, pp. 616-613. doi:10.1038/306616a0
- S. T. Cowan and S. Steel, “Manual for the Identification of Medical Bacteria,” 3rd Edition, Cambridge University Press, Cambridge, 1974.
- Clinical Laboratory Standard Institute, “Performance Standard for Antimicrobial Disk Susceptibility Tested: Approved Standard,” 2006.
- T. J. Sijhary, M. L. Bermann and L. W. Enquist, “Experiments with Gene Fusions,” Cold Spring Habor Laboratory Press, New York, 1984.
- H. Bimboim and J. Doly, “A Rapid Alkaline Extraction Procedure for Screening Recombinant Plasmid DNA,” Nucleic Acids Research, Vol. 7, No. 6, 1979, pp. 1513- 1523. doi:10.1093/nar/7.6.1513
- J. A. Meyers, D. Sanchez, L. P. Elwell and S. Falkow, “Simple Agarose Gel Electrophoresis Method for the Identification and Characterization of Plasmid Deoxyribonucleic Acid,” Journal of Bacteriology, Vol. 127, No. 3, 1976, pp. 1529-1537.
- K. P. Ranjan, N. Ranjan, K. S. Bansal and P. R. Arora, “Prevalence of P. aeruginosa in Most Operative Wound Infection in a Referral Hospital in Haryana,” Indian Journal of Laboratory Physicians, Vol. 2, No. 2, 2010, pp. 74-77. doi:10.4103/0974-2727.72153
- N. M. R. Bojary and M. Hajia, “Multidrug Resistant P. aeruginosa Strains in Tehran Reference Burn Hospital, Tehran, Iran,” African Journal of Microbiology Research, Vol. 6, No. 7, 2012, pp. 1393-1396.
- M. Hajia, M. Qorbanalizadehgan, M. Rahbar and M. Izadi, “Laboratory Evaluations of Iranian Commercially Provided Antibiotic Disks with Conventional E-Test for Susceptibility Testing in Three Most Isolated Multi-Drug Resistant Organisms,” Internet Journal of Microbiology, Vol. 5, 2008, pp. 23-28.
- M. Rahbar, H. Mehragan and N. Hajiali-Akbori, “Prevalence of Drug Resistance in Non fermenter Gram Negative Bacilli,” Iranian Journal of Pathology, Vol. 5, 2010, pp. 90-96.
- R. A. Bonomo and D. Szabo, “Mechanisms of Multidrug Resistance in Acinetobacter Species and Pseudomonas aeruginosa,” Clinical Infectious Diseases, Vol. 43, No. 2, 2006, pp. 49-56. doi:10.1086/504477
- G. F. Weldhagen, L. Poirel and P. Nordmann, “Ambler Class A Extended Spectrum β-Lactamases in Pseudomonas aeruginosa:novel developments and Clinical Impact,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 8, 2003, pp. 2385-2392. doi:10.1128/AAC.47.8.2385-2392.2003
- T. R. Walsh, M. A. Toleman, L. Poirel and P. Nordmann, “Metallo—b-Lactamases: The Quiet before Storm? Clinical Microbiology Reviews, Vol. 18, No. 2, 2005, pp. 306-325. doi:10.1128/CMR.18.2.306-325.2005
- G. Agarwal, A. Kapil, S. K. Kabra, B. K. Das and N. Dwivedi, “Characterization of Pseudomonas Aeruginosa Isolated from Chronically Infected Children with Cystic Fibrosis in Indian,” BMC Microbiology, Vol. 5, No. 1, 2005, pp. 43-47. doi:10.1186/1471-2180-5-43
- H. Y. Chen, K. Yunn and D. M. LiverMore, “Mechanism of Resistance to Beta-Lactam Antibiotics among Pseudomonas Aeruginosa Isolates Collected,” Medical Microbiology, Vol. 43, No. 4, 1995, pp. 300-309.
- S. Rahmatis, S. Yang, A. L. Davidson and E. L. Zechiedrich, “Control of the Acr AB Multi-Drug Efflux Pump by Quorum Sensing Regulator,” Molecular Microbiology, Vol. 43, No. 3, 2002, pp. 677-850. doi:10.1046/j.1365-2958.2002.02773.x
- M. Shadid and N. Malik, “Plasmid Mediated Amikacin Resistance in Clinical Isolate of Pseudomonas aeruginosa,” Indian Journal of Medical Microbiology, Vol. 22, No. 3, 2004, pp. 182-184.
- R. C. Clowers, “Molecular Structure of Bacterial Plasmids,” Bacteriological Reviews, Vol. 36, No. 3, 1972, pp. 361- 405.
NOTES
*Corresponding author.

