Open Journal of Marine Science
Vol.2 No.4(2012), Article ID:23854,11 pages DOI:10.4236/ojms.2012.24021
Screening and Phylogenetic Analysis of Deep-Sea Bacteria Capable of Metabolizing Lignin-Derived Aromatic Compounds
Institute of Biogeosciences, Japan Agency of Marine-Earth Science and Technology (JAMSTEC), Yokosuka, Japan
Email: *ohtay@jamstec.go.jp
Received August 15, 2012; revised September 15, 2012; accepted September 30, 2012
Keywords: Aromatic Compound; Bacteria; Deep Sea; Lignin
ABSTRACT
Lignin is one of the most abundant biomasses in nature. It is composed of aromatic moieties and has great potential for use in the production of chemical alternatives to petroleum products. Because of increasing interest in biocatalysis, the potential for industrial application of microbial metabolism of lignin-derived compounds has gained considerable recent attention. Functional screenings of culturable bacteria isolated from sediments and sunken wood collected from the deep sea revealed the existence of a number of previously unidentified bacteria capable of metabolizing lignin-related aromatic compounds. Of the 510 isolates obtained in the present study, 208 completely or partially metabolized these compounds. The 208 isolates were classified into diverse phyla, including Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria. Among the 208 isolates, 61 unique 16S rRNA gene sequences were detected including previously unidentified marine lineage isolates. The metabolites of the isolates were analysed using liquid chromatography/mass spectrometry (LC/MS) or gas chromatography/mass spectrometry (GC/MS). Most of the representative 61 isolates non-oxidatively decarboxylated the substrates to produce the corresponding aromatic vinyl monomers, which are used as feed stocks for bio-based plastics production. Oxidative metabolism of the lignin-related compounds for assimilation was frequently observed. Our study showed that the deep-sea environment contains an abundance of microorganisms capable of both non-oxidative and oxidative bioconversion of lignin-derived aromatic compounds. The ability for bioconversion of aromatic compounds found in this study will facilitate the development of future biotechnological applications.
1. Introduction
Lignin is the most abundant renewable biomass next to cellulose. It is composed of aromatic moieties and is found in the secondary cell wall of plants, where it is intimately attached to the cellulose microfibrils [1]. More than 70 million tons of various types of lignin preparations are produced as waste material by the paper Industry yearly [2]. The amount of lignin production will increase in the near future as a result of increasing demand for reliable biomass-based renewable energy sources to replace fossil fuels. At present, most of the lignin produced is used for low-value fuels; however, the innate chemistry of lignin has led to increased demand for it in the high-value chemical industries. The efficient production of value-added chemicals from lignin is now a high priority [3,4].
Harnessing the biosynthetic ability of microorganisms is becoming an increasingly important platform for producing value-added chemical products [5]. To date, the most extensively studied lignin-metabolizing microorganisms are the whiteand brown-rot Basidiomycete fungi [6-9]. These fungi initially degrade lignin using an array of oxidative enzymes, such as laccases, lignin peroxidases (LiP’s), and manganese peroxidases (MnP’s). Other synergistic accessory enzymes are known to be involved in lignin degradation. Various low-molecular weight compounds are also produced, which act as diffusible mediators that transfer electrons to substrates from the corresponding enzymes. In contrast to fungal lignin degradation, the involvement of bacteria in lignin metabolism is poorly understood. Some Streptomyces [10] and Nocardia [11] are reportedly capable of degrading lignin; however, the metabolic pathways have not been fully elucidated. Only a limited number of isolates, including Sphingobium sp. SYK-6, have been extensively studied with respect to the metabolism of ligninderived compounds [12]. Using a newly developed spectrophotometric assay, Ahmad et al. recently found that the lignin-metabolizing activity of two bacteria, Rhodococcus jostii RHA1 and Pseudomonas putida mt-2, is comparable to that of some lignin-degrading fungi [13]. These two strains are known to metabolize a range of aromatic compounds that share structural features with lignin. These findings suggest that bacteria play a more prominent role in lignin metabolism than was previously thought [14].
The marine environment, especially at the bottom of the deep sea, is deficient in readily available nutrients due to the lack of primary photosynthetic production. Plant-derived organic materials fallen from terrestrial environments into deep sea supports distinct habitats for both macroand microorganisms [15,16]. Strictly aerobic organisms, i.e., the majority of wood-decaying fungi, are supposedly less active in the marine environment. In addition, some lignin-converting oxidative enzymes are inactivated under saline conditions [17]. Thus, in marine environments, alternative systems are required for the bioconversion of lignin-related compounds.
The marine environment remains relatively unexplored with respect to lignin-metabolizing bacteria, although it is believed that this environment may prove to harbor a variety of such organisms. With the aim of facilitating the development of future biotechnological applications, we therefore conducted functional screening and phylogenetic analyses of lignin-derived aromatic compound metabolizing bacteria cultured from deep-sea sediments and sunken wood.
2. Methods
2.1. Sampling
Deep-sea sediment samples were collected in August 2011 from a depth of 3100 m in the Japan Trench off the Pacific coast of Miyagi Prefecture, northern Japan. A sample of sunken wood, designated as sample A, was recovered in May 2011 from a depth of 260 m in Suruga Bay, off the Pacific coast of Shizuoka prefecture, central Japan. Sample A was identified as a Pinales trunk and was in a highly decomposed state resulting from extensive colonization by Teredothyra matocotana, a teredinid wood-boring bivalve known as the shipworm [18]. A second sample of sunken wood, designated as sample B, a Quercus phillyraeoides trunk, was recovered in January 2012 from a depth of 495 m in the Nansei-Shotō Trench off Miyako Island, Okinawa Prefecture, southern Japan. A 0.1-g portion of each sediment and sunken wood sample was suspended in 2 mL of sterile artificial seawater (Nihon Pharmaceutical, Japan) and shaken briefly on a vortex at room temperature, then used for isolation of bacteria.
2.2. Isolation of Bacteria
The isolation medium consisted of 2% (w/v) milled Japanese beech wood or rice straw, Daigo IMK medium (Nihon Pharmaceutical), and 2% (w/v) DifcoTM bacto agar (BD) in artificial seawater. A suspension of each sample was spread on the solid medium and the plates were incubated at 25˚C for 10 days. A total of 510 colonies representing all observed colony morphologies were randomly selected and isolated in pure culture by repeated streaking and cultivation on DifcoTM Marine agar 2216 (BD).
2.3. Metabolic Screening of Bacterial Isolates and Metabolite Analysis
Metabolic screening of the bacterial isolates was performed using DifcoTM Marine broth 2216 (BD) as a basal medium. Eight compounds with the same basic molecular structure of the lignin biosynthesis precursors pcoumaryl alcohol (1 in Figure 1), coniferyl alcohol (2 in Figure 1), and sinapyl alcohol (3 in Figure 1), were selected as lignin model compounds. The substrates were purchased from Sigma-Aldrich Japan, and were added to the medium at a final concentration of 2 mM: p-coumaric acid (4 in Figure 1), ferulic acid (5 in Figure 1), sinapic acid (6 in Figure 1), vanilin (10 in Figure 1), syringaldehyde (11 in Figure 1), protocathecuic acid (12 in Figure 1), p-hydroxybenzaldehyde (13 in Figure 1), and veratryl alcohol (14 in Figure 1). Bacteria were pre-cultured in Marine broth 2216 for 2 days. Each well of a deep-well plate containing 1 mL of medium was inoculated with 50 μL of bacterial suspension. The plates were then incubated at 25˚C with gentle shaking. After 5 days of incubation, the culture supernatants were collected by centrifugation for 5 min at 4800 rpm. A 0.1 mL aliquot of each sample supernatant was mixed with 0.9 mL of methanol and then centrifuged. The resulting supernatant was collected and analyzed using liquid chromatography (LC). Samples were analyzed by LC using an HSS T3 C18 reverse phase column (1.8 μm particle size, 100 × 2.1 mm, Waters) coupled to a Waters Accuity H-class UPLCTM system operated at a flow rate of 0.4 mL/min. The eluate was monitored at 270 nm. The gradient was as follows: 5% - 95% methanol/2 mM sodium acetate and 0.05% formic acid from 0 to 6 min, 95% MeOH/2 mM sodium acetate and 0.05% formic acid from 6 to 7 min. The amount of substrate added to the cultivation medium was calculated based on the area of the resulting chromatographic peaks. The metabolic activity of an isolate was considered positive if the residual amount of a sub-

Figure 1. Molecular structures of the lignin-related aromatic compounds examined in the present study.
strate was less than 50% of that in control medium samples incubated under the same conditions without bacteria. When chromatographs of test samples revealed new peaks not detected in analyses of control culture supernatants, these samples were analyzed further using liquid chromatography/mass spectrometry (LC/MS) or gas chromatography/mass spectrometry (GC/MS). LC/MS data were generated using a Waters Xevo G2TM quadrupole time-of-flight mass spectrometer operated in negative ion electrospray ionization (ESI) mode. The inlet system was the same as that used for the LC system described above. Data were acquired over the mass range 100 - 1000 Da with a 0.45-s scan time using a desolvation temperature of 400˚C, source temperature of 120˚C, and cone voltage of 30 V. GC/MS data were generated using a quadrupole mass spectrometer (TRACE DSQ, Thermo Fisher Scientific) equipped with an Agilent DB5TM column (30 m × 0.25 mm). The column temperature was held at 60˚C for 5 min and then increased to 80˚C, after which it was raised to 225˚C at a rate of either 1.5 or 3˚C/min. The temperatures of injection and detection were 100˚C and 225˚C, respectively. Helium gas served as the mobile phase, and the flow rate was 1.1 mL/min. Bacterial culture and metabolite analysis described in this section were repeated twice in two independent experiments.
2.4. DNA Isolation and Sequence Analysis
Total genomic DNA was extracted from bacterial isolates using a NucleoSpinTM plant II DNA Isolation Kit (Takara Bio), according to the manufacturer’s instructions. The isolated DNA was used as the template for PCR reactions with the 16S rDNA universal primers 27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1525r (5’-AAAGGAGGTGATCCAGCC-3’). Each PCR reaction (25 μL) consisted of 1× PCR buffer with MgCl2, 200 μM dNTPs, 0.6 μM 27f, 0.6 μM 1525r, 1.4 U of LA Taq DNA Polymerase (Takara Bio), and 1.2 μL of DNA. Cycling conditions were as follows: 97˚C for 2 min, 30 cycles of 97˚C for 30 s, 60˚C for 1 min, and 72˚C for 90 s, followed by 72˚C for 5 min. The PCR products were sequenced using an ABI 3730 XL DNA Analyzer (Applied Biosystems). The 208 sequences obtained by PCR were manually edited and corrected prior to BLAST (blastn) searching against the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/blast). The closest relatives were identified based on the best blastn match in the non-redundant database.
2.5. Accession Numbers
Nucleotide sequences determined in this study were submitted to the Genbank database under the accession numbers AB733523-AB733583 (61 entries). The accession number for each isolate is listed in Table 1.
3. Results
3.1. Isolate Identification
A total of 510 bacterial colonies were isolated in pure culture following initial growth on the simple medium containing milled wood or rice straw as the nutrient source and subsequent repeated transfer and cultivation on fresh medium. Metabolism of lignin-related aromatic compounds was assessed by incubating each isolate in liquid medium containing each of 8 lignin-related compounds and then measuring the changes in the level of the target compound using LC. A total of 208 isolates metabolized one or more compounds. Based on the 16S rRNA gene sequences, the 208 isolates were classified into diverse phyla, including Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria. The majority of isolates (131) were classified into the Bacilli class of the phylum Firmicutes. These isolates were found in the sediment samples and sunken wood sample A, which
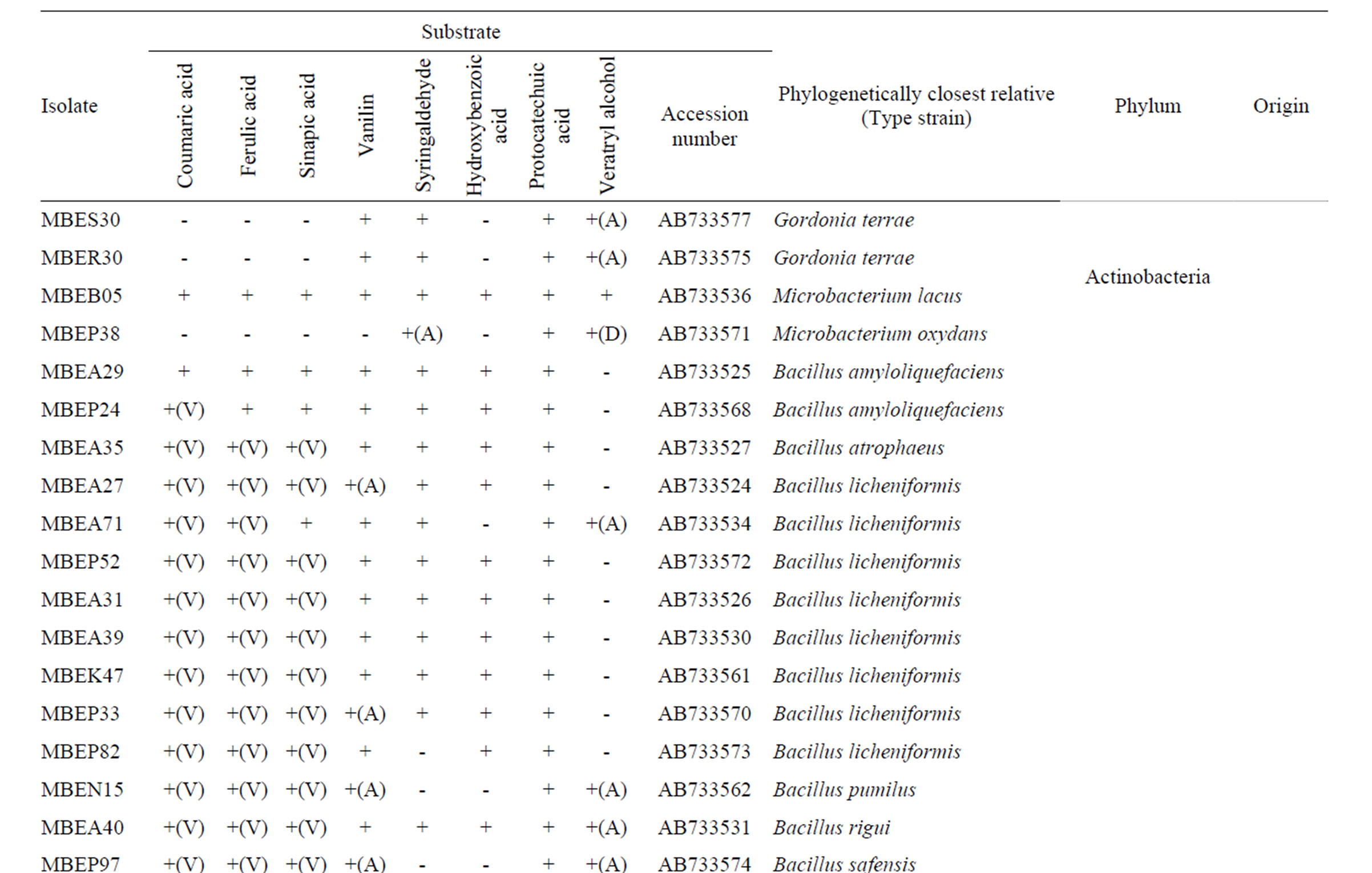
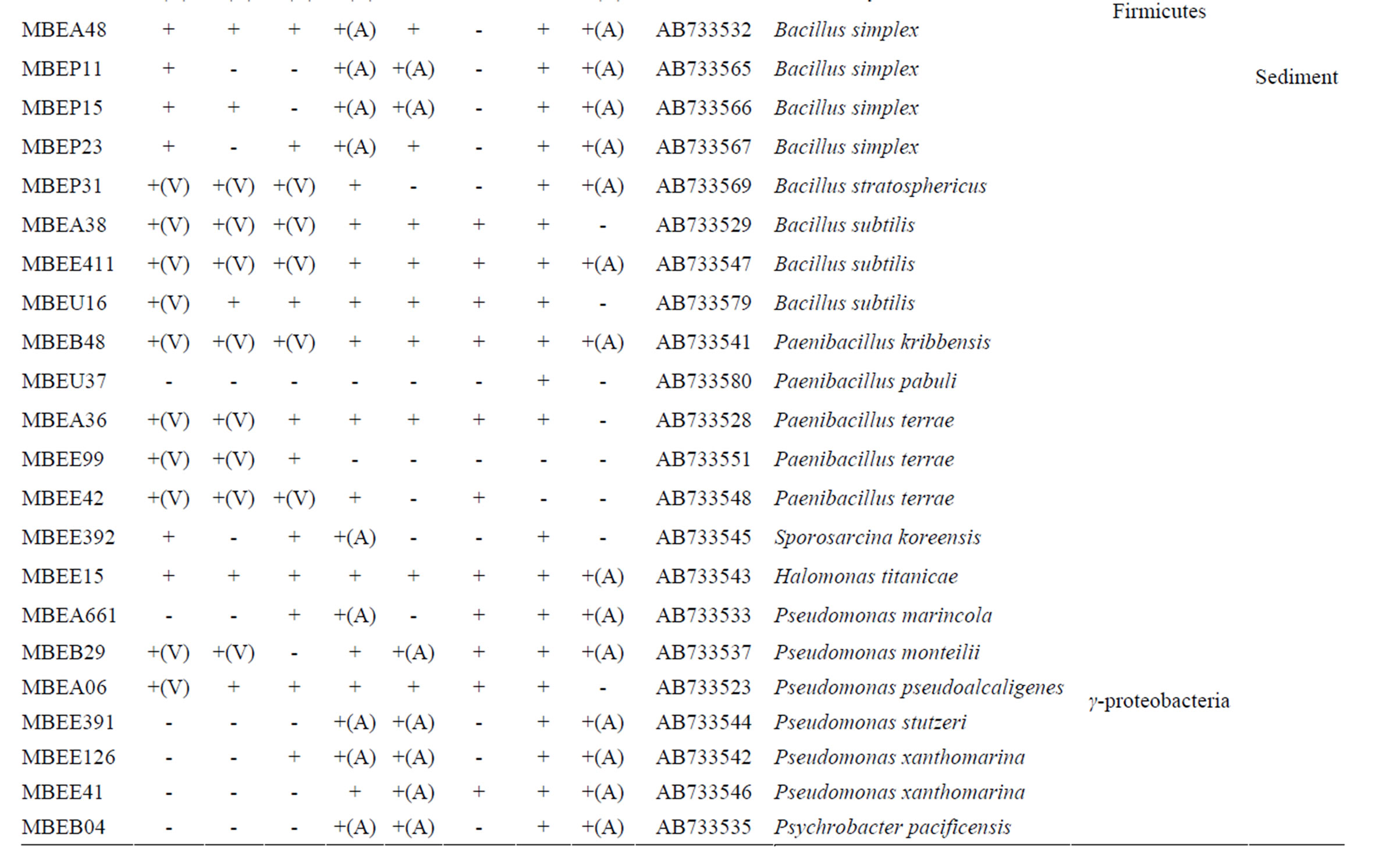

Table 1. Phylogenetic associations and metabolism of lignin-related substrates by deep-sea bacteria.
was extensively colonized by shipworms. In contrast, members of the α- and γ-Proteobaceria classes of the phylum Proteobacteria dominated among the isolates identified in sunken wood sample B. The microbial communities in the different sunken wood samples were apparently distinct from each other. Of all the DNA sequences determined, 61 were found to be unique (Table 1). Blastn searching against the non-redundant nucleotide sequence database revealed that 59 isolates possessed novel 16S rRNA gene sequences dissimilar to any other sequences currently deposited in the database. The phylogenetic positions of the representative 61 isolates and relation between them and closely related strains based on the 16S rRNA gene sequences were shown in Figure 2. Some isolates were found to be of marine lineage. For example, Sulfitebacter, which was isolated from sunken wood sample B, is a member of the Rosebacter clade, a marine lineage of the class α-Proteobacteria. These bacteria are known to play a key role in marine carbon and sulfur cycles [19], and some species reportedly degrade aromatic compounds [20]. Novosphingobium is also a member of the α-Proteobacteria class. Some members of the Novosphingobium genus reportedly degrade polycyclic aromatic hydrocarbons [21]. A Novosphingobium species has been shown to degrade phenol and catechol under saline conditions [22].
3.2. Metabolism of 3-Phenyl 2-Propenoic Acids
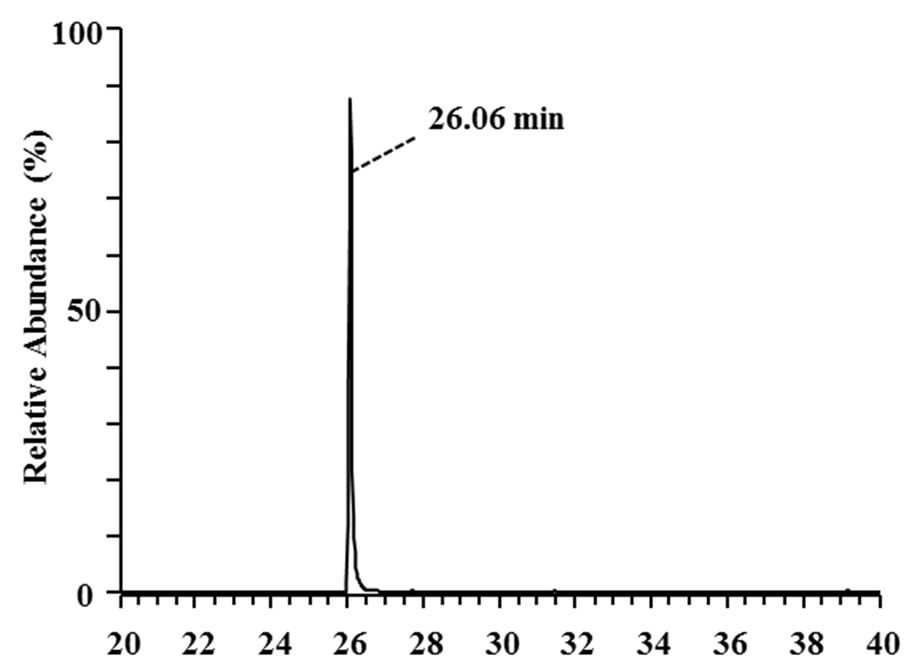
Three 3-phenyl 2-propenoic acids (p-coumaric acid, ferulic acid, and sinapic acid) were used as substrates to analyze the major metabolites produced in the culture supernatants of the isolates. Metabolites in the culture supernatants were identified using GC/MS. The most abundant metabolites of p-coumaric and ferulic acid are shown in Figure 3. The peaks with retention times of 26.06 and 30.83 min (Figures 3(a) and (c)) corresponded to 4-vinyl phenol (7 in Figure 1) and 4-vinyl guaiacol (8 in Figure 1), respectively, judging from a database search against the GC mass spectral references compiled by the National Institute of Standards and Technology using the MS spectra in Figures 3(b) and (d) as the queries. The data revealed that strain MBEP82 produces vinyl decarboxylation derivatives from p-coumaric and ferulic acid. A total of 29 strains, including 23 from the genus Bacillus, 4 from the genus Paenibacillus, and 2 from the genus Pseudomonas, produced 4-vinyl phenol from p-coumaric acid. A total of 25 strains produced 4- vinyl guaiacol from ferulic acid, and 20 strains produced 4-vinyl syringol (9 in Figure 1) from sinapic acid. These aromatic vinyl compounds are produced by enzymatic
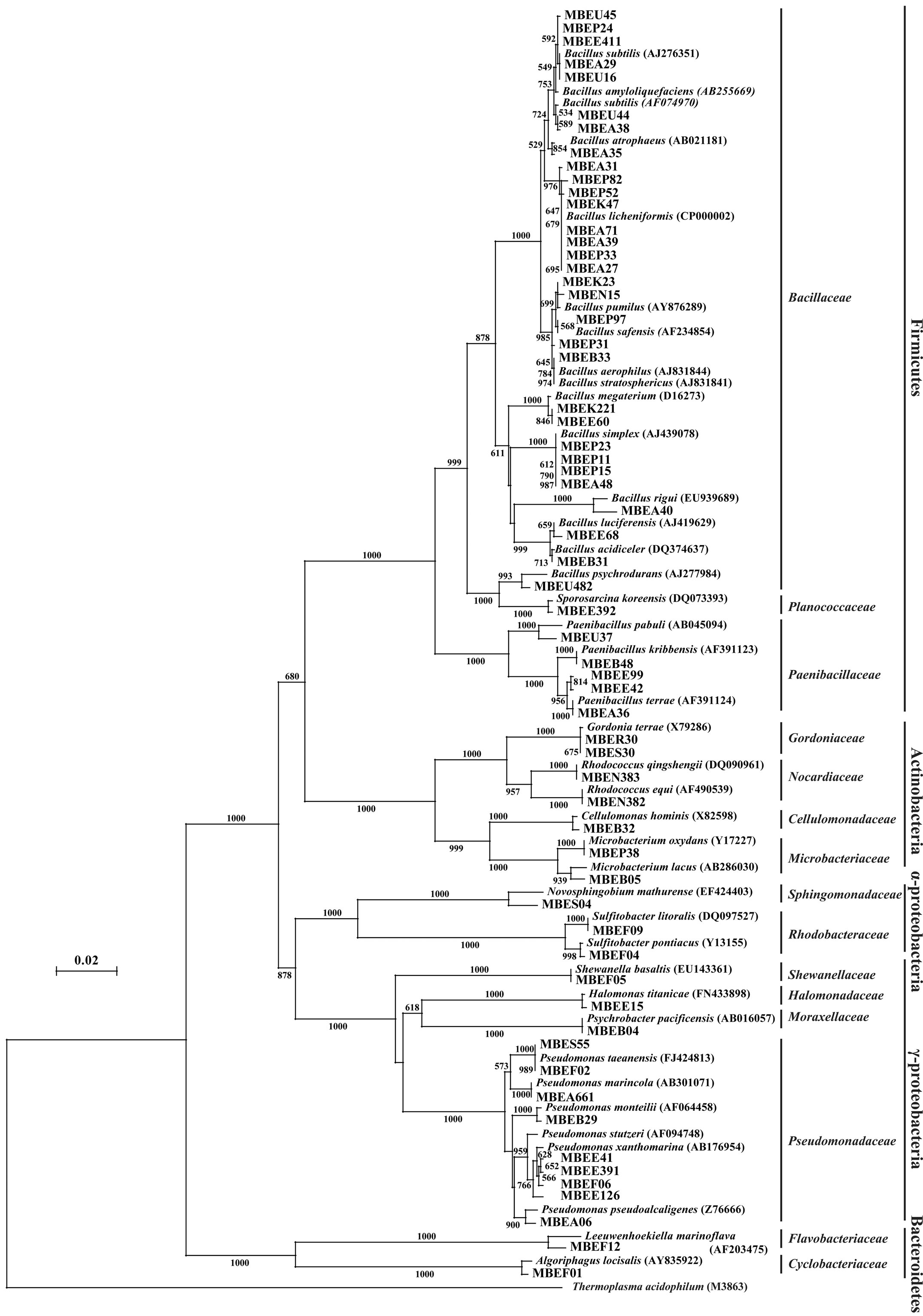
Figure 2. Neighbor-joining tree showing the phylogenetic relationships and closest related strains of the deep-sea bacterial isolates examined in the present study. Data are based on 16S rRNA gene sequences. Bootstrap values were derived from 1000 replicates. The scale bar indicates 2% sequence divergence.
 (a)
(a)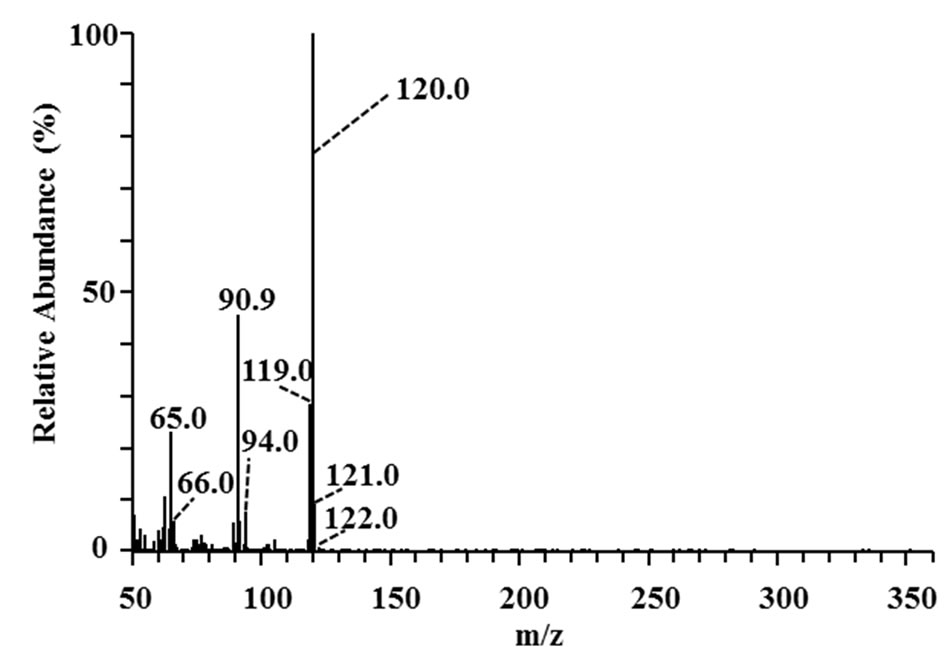 (b)
(b)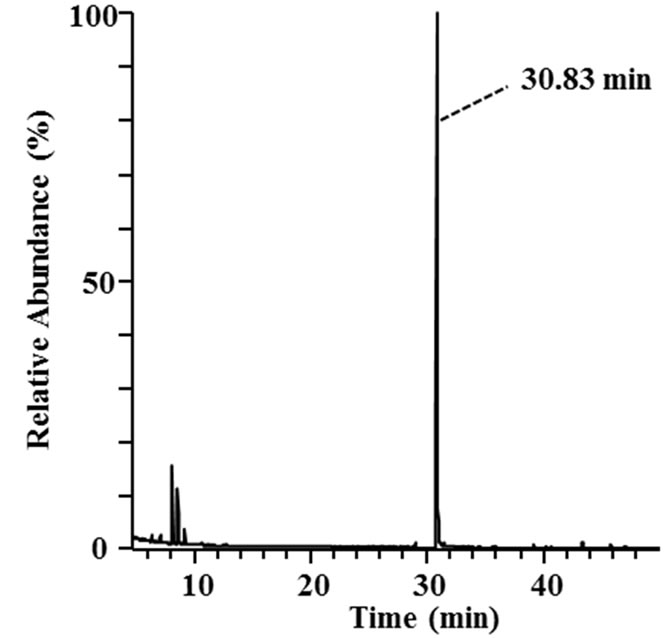 (c)
(c)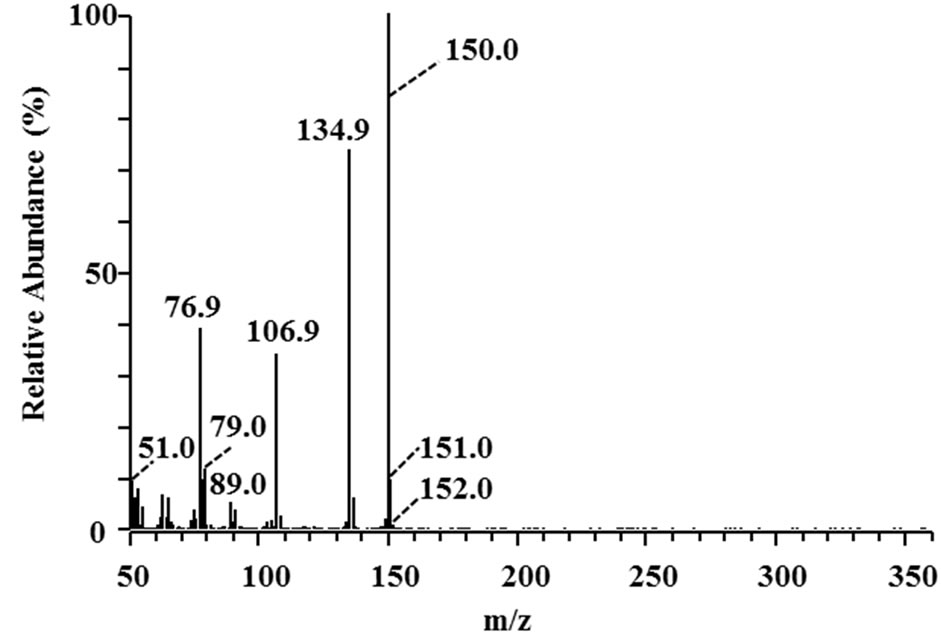 (d)
(d)
Figure 3. GC/MS analysis of metabolites of p-coumaric and ferulic acid produced by strain MBEP82. (a) Total ion chromatogram of the supernatant of culture broth containing p-coumaric acid; (b) MS spectrum of the peak at 26.06 min in (a); (c) Total ion chromatogram of the supernatant of culture broth containing ferulic acid; (d) MS spectrum of the peak at 30.83 min in (c).
non-oxidative decarboxylation of 3-phenyl 2-propenoic acids [23]. Another metabolite of ferulic acid was identified by LC/MS analysis in the culture broth supernatant of Shewanella strain (MBEF05) (Figure 4). Ferulic acid was detected in the MS chromatograms of the culture medium with retention times of 4.04 min (Figure 4(a)) and 4.07 min (Figure 4(b)). The major ESI negative mode ion associated with these peaks was detected at m/z 193.05 (Figures 4(c) and (d)), which corresponds to the one-proton loss molecular ion of ferulic acid (calculated monoisotopic mass = 194.06 Da). A new metabolite with a retention time of 3.96 min was detected after cultivation with strain MBEF05 (Figure 4(b)). The major ion associated with this peak was detected at m/z 195.07 (Figure 4(e)). The elemental composition of this com0 pound was determined as C10H12O4, indicating that the metabolite was produced by hydration of ferulic acid (elemental composition: C10H10O4). Strain MBEF05 is closely related to Shewanella basaltis and S. oneidensis. S. oneidensis is a psychrophilic species pre-dominantly found in deep-sea anaerobic habitats, and reportedly reduces heavy metals through outer membrane cytochromes [24]. No residual 3-phenyl 2-propenoic acid or any intermediate metabolite derived from the 3-phenyl 2-propenoic acid substrates were detected in the supernatants of four isolates (MBEE05, MBEA29, MBEE15, MBES04) which respectively belong to the genera Microbacterium, Bacillus, Halomonas, and Novosphingobium, suggesting rapid and complete cellular uptake of these substrates and/or their metabolites. These isolates are considered nutritional generalists that are capable of assimilating a wide variety of phenolic compounds in the marine environment.
3.3. Metabolism of Phenolic Aldehydes
Vanillin (10 in Figure 1) and syringaldehyde (11 in Figure 1) served as test compounds to assess the metabolism of phenolic aldehyde substrates. The vast majority of the representative isolates listed in Table 1 (58 of 61) were able to metabolize vanillin. In most cases, vanillin was completely consumed, indicating that the isolates assimilated it as their energy source. Vanillic acids accumulated in the supernatants of 21 of the 58 vanillin-metabolizing strains, indicative of oxidation of the substrate (denoted with an (A) in Table 1). Compared to the number of vanillin metabolizers, fewer isolates (42 of 61) metabolized syringaldehyde. The molecular structures of these compounds are quite similar, the only difference is that syringaldehyde carries two methoxy moieties on the aromatic ring while vanillin carries only one. Many soil bacteria can assimilate both vanillin and syringaldehyde via the β-ketoadipate pathway [25]. A different pathway existing upstream of β-ketoadipate pathway is required for the assimilation of each compound. Demethylation of
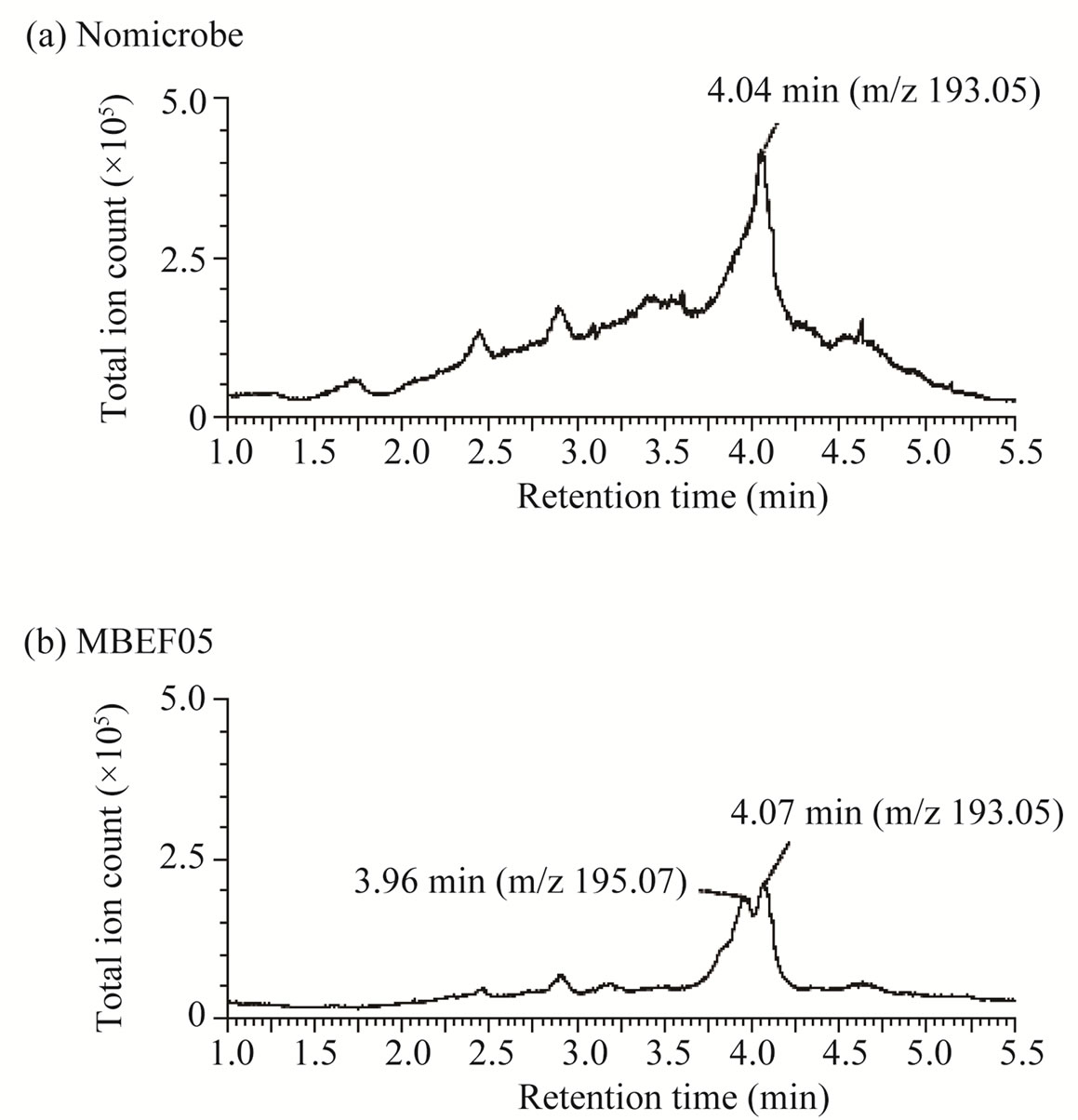

Figure 4. LC/MS analysis of metabolites of ferulic acid produced by strain MBEF05. (a) Total ion chromatogram of culture medium containing ferulic acid without microbes; (b) MS spectrum of the peak at 4.04 min in (a); (c) Total ion chromatogram of the post-culture supernatant of culture broth containing ferulic acid; (d) MS spectrum of the peak at 4.07 min in (b); (e) MS spectrum of the peak at 3.96 min in (b).
syringaldehyde is an energy consuming step that requires tetrahydrofolate as a source of reducing power [26], and therefore demethylation may not occur when other substrates are available for use as energy sources. The difference in the number of isolates capable of metabolizing vanillin and syringaldehyde can be explained by the requirement of each of pathways for each metabolism. Vanillin-metabolizing strains, indicative of oxidation of the substrate (denoted with an (A) in Table 1). Compared to the number of vanillin metabolizers, fewer isolates (42 of 61) metabolized syringaldehyde. The molecular structures of these compounds are quite similar, the only difference is that syringaldehyde carries two methoxy moieties on the aromatic ring while vanillin carries only one. Many soil bacteria can assimilate both vanillin and syringaldehyde via the β-ketoadipate pathway [25]. A different pathway existing upstream of β-ketoadipate pathway is required for the assimilation of each compound. Demethylation of syringaldehyde is an energy consuming step that requires tetrahydrofolate as a source of reducing power [26], and therefore demethylation may not occur when other substrates are available for use as energy sources. The difference in the number of isolates capable of metabolizing vanillin and syringaldehyde can be explained by the requirement of each of pathways for each metabolism.
3.4. Metabolism of Phenolic Acids
p-Hydroxybenzoic acid and protocatechuic acid were used as the phenolic acid substrates in this study. Approximately half of the representative isolates listed in Table 1 (30 of 61) metabolized p-hydroxybenzoic acid. This compound was relatively recalcitrant compared to the other substrates tested. Generally, aromatic ring cleavage proceeds via ring hydroxylation followed by funneling of the intermediate organic acids into the β- ketoadipate pathway [25]. More methoxylated aromatic rings tend to be susceptible to hydroxylation. Since phydroxybenzoic acid has no methoxy moiety, it is more recalcitrant than similar compounds containing methoxy moieties.
Almost all of the isolates (58 of 61) completely assimilated protocatechuate within the incubation period. Protocatechuate is a central intermediate in the metabolic pathways that lead to the degradation of a variety of not only natural aromatic monomers, but also various xenobiotics, such as halogenated, heterocyclic, and polycyclic aromatic hydrocarbons.
3.5. Oxidation of Veratryl Alcohol
Veratryl alcohol is a mediator of lignin peroxidases in fungal extracellular lignin biodegradation and modification systems [7]. Veratryl alcohol is oxidized by veratryl alcohol oxidase and plays an important role in electron transfer to the high redox potential substrate. Veratryl alcohol oxidase is referred to as an accessory enzyme in the lignin degradation system; therefore, we examined the ability of the isolates to oxidize veratryl alcohol. A total of 38 isolates produced oxidized compounds of the corresponding carboxylic acid or aldehyde (denoted with an (A) or (D), respectively, in Table 1), whereas a Microbacterium strain completely assimilated veratryl alcohol.
4. Discussion
Our screening demonstrated that a wide variety of marine bacteria are capable of metabolizing aromatic compounds. A previous study showed that marine enrichment cultures using lignin as a substrate contain an abundance of Bacillus species [27]. The population of isolates identified in marine sediments in this study was similar to that in the previous report. However, the populations of isolates in the present study were quite different than those isolated from decayed wood in terrestrial ecosystems [28,29]. Wood-decaying fungi in terrestrial environments, primarily Basidiomycetes, initiate the decay process by loosening the hard structure of wood through the excretion of an array of oxidative enzymes. Natural wood in advanced stages of decay is highly acidic due to the actions of Basidiomycete-released enzymes. Phylogenetic analyses have shown that populations of culturable bacteria found in decayed wood are enriched in acid-tolerant members of the Proteobacteria and Acidobacteria, with a lower abundance of Verrucomicrobia or Bacteroidetes species. The possible presence of bacteria belonging to other phyla, including Frimicutes, Verrucomicrobia, Planctomycetes, Actinobacteria, and TM7, which is a phylogenetic group of unculturable bacteria, was suggested only by the results of non-culture PCRbased methods. In the marine environment, where the concentration of organic matter is usually low, fallen wood represents a significant proportion of the organic input. In marine ecosystems, the tightly organized structure of lignin and cellulose components in woody plant materials is primarily disrupted by molluscs and other macrofauna [15,16,30]. These wood-boring animals scatter the fine-grained particles in the surrounding area, rendering the nutrients accessible to other scavengers. Falls of organic material such as wood often lead to the development of active ecosystems capable of supporting locally dense populations of a variety of organisms.
The strains isolated in this study metabolized ligninderived aromatic compounds through both non-oxidative and oxidative reactions. A previous study described a range of bacteria and fungi capable of decarboxylating p-coumaric acid and ferulic acid [31]. Surprisingly, only some of the responsible enzymes have been studied in detail. Pseudomonas fluorescens ferulic acid decarboxylase functions at neutral pH at temperatures of 27˚C - 30˚C [32]. The enzyme prefers ferulic acid to p-coumaric acid as a substrate. A p-coumaric acid decarboxylase isolated from Bacillus pumillus and Lactobacillus plantarum functions at acidic pHs and temperatures below 37˚C [23,33]. This enzyme decarboxylates p-coumaric acid more efficiently than it decarboxylates ferulic acid. These enzymes have recently become the focus of ecofriendly industrial applications. Aromatic vinyl monomers produced by the decarboxylation of 3-phenyl 2- propenoic acids is now being used as a feed stock for bio-based plastics production [34]. Because it links the polysaccharide components in plant cell walls, ferulic acid is abundant in nature, and can be recovered from agricultural wastes such as bran or straw from rice or wheat using thermochemical or biological reactions [31]. Ferulic acid is now used as a starting material for the biological synthesis of a range of useful aromatic compounds in the chemical, pharmaceutical, and food Industries [35].
Vanillin is also a very important compound used in the chemical industry. Vanillin can be recovered by oxidation of lignin waste produced by the pulp industry. Interest in the bioconversion of vanillin is drawing increased attention for its use in the production of value-added chemicals.
The majority of aromatic-metabolizing bacteria discovered to date were originally identified based on their ability to metabolize xenobiotic compounds. Xenobiotic compounds produced by the petrochemical industry are used as ingredients in fuels and as raw materials for the chemical synthesis of pharmaceuticals, pesticides, polymers, and other products. Aromatic-degrading bacteria have often been detected at xenobiotic-contaminated sites. The degradation pathways bacteria use to metabolize xenobiotics commonly involve catecholic intermediates. Dioxigenases, which catalyze ring cleavage of catecholic compounds, play a key role in determining the specificity of these pathways [36]. While such pathways have been identified in a number of bacterial genera inhabiting in soil, including Acinetobacter, Alcaligenes, Azotobacter, Bacillus, Pseudomonas, Rhodococcus, and Streptomyces, it remains unknown whether organisms using these pathways are common in marine communities. The diversity of a ring-cleaving dioxygenase gene, pcaH, in a salt marsh bacterial community was assessed by Buchan et al. [37]. Their study described the wide distribution of aromatic compound degradation genes in intertidal communities, especially among bacteria in the Rosebacter group, suggesting that the ability to metabolize aromatic compounds is widespread in marine environments. Recently, pyrosequence analyses conducted by Wang et al. [38] provided evidence that genes involved in aromatic compound metabolism are concentrated in the Atlantis II Deep brine pool in the Red Sea, suggesting that in situ consumption of such compounds occurs in the deep-sea environment.
Previous studies have shown that deep-sea microorganisms are highly diverse, and that psychrophiles, thermophiles, halophiles, and barophiles can be found in deep-sea environments. The microorganisms found in the deep sea are likely to be great sources of useful enzymes. In this study, we found that a wide variety of microorganisms, including those of marine lineage, are capable of metabolizing lignin-related aromatic compounds, indicating that the deep-sea environment holds great potential as a source of organisms that may be used in the non-oxidative and oxidative bioconversion of aromatic compounds.
5. Acknowledgements
The authors are grateful to Dr. Toshiro Saruwatari (Atmosphere and Ocean Research Institute, University of Tokyo), Dr. Yoshihisa Shirayama (JAMSTEC) and the captains and crews of the commercial trawler Hinodemaru (Heda, Shizuoka, Japan) and research vessel Natsushima (JAMSTEC) for the collection of sunken wood samples. We thank Mr. Ukichi Abe, Ms. Harui Abe, and Mr. Hiroyuki Takano for generously providing the plant materials. This research was supported in part by the Japan Society for the Promotion of Science (JSPS) Funding Program for Next Generation World-Leading Researchers (no. GS031) to Y.O. and by a Grant-in-Aid for JSPS fellows (no. 23 7855) to T.H.
REFERENCES
- T. K. Kirk and R. L. Farrell, “Enzymatic Combustion— The Microbial-Degradation of Lignin,” Annual Review of Microbiology, Vol. 41, No. 1, 1987, pp. 465-505. doi:10.1146/annurev.mi.41.100187.002341
- M. N. S. Kumar, A. K. Mohanty, L. Erickson and M. Misra, “Lignin and Its Applications with Polymers,” Journal of Biobased Materials and Bioenergy, Vol. 3, No. 1, 2009, pp. 1-24. doi:10.1166/jbmb.2009.1001
- D. Stewart, “Lignin as a Base Material for Materials Applications: Chemistry, Application and Economics,” Industrial Crops and Products, Vol. 27, No. 2, 2008, pp. 202-207.
- F. G. Calvo-Flores and J. A. Dobado, “Lignin as Renewable Raw Material,” Chemsuschem, Vol. 3, No. 11, 2010, pp. 1227-1235. doi:10.1002/cssc.201000157
- J. Du, Z. Y. Shao and H. M. Zhao, “Engineering Microbial Factories for Synthesis of Value-Added Products,” Journal of Industrial Microbiology & Biotechnology, Vol. 38, No. 10, 2011, pp. 873-890. doi:10.1002/cssc.201000157
- M. H. Gold and M. Alic, “Molecular-Biology of the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium,” Microbiological Reviews, Vol. 57, No. 3, 1993, pp. 605-622.
- P. Kersten and D. Cullen, “Extracellular Oxidative Systems of the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium,” Fungal Genetics and Biology, Vol. 44, No. 2, 2007, pp. 77-87. doi:10.1002/cssc.201000157
- D. Martinez, L. F. Larrondo, N. Putnam, M. D. S. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, et al., “Genome Sequence of the Lignocellulose Degrading Fungus Phanerochaete chrysosporium strain RP78,” Nature Biotechnology, Vol. 22, No. 6, 2004, pp. 695-700. doi:10.1038/nbt967
- D. Martinez, J. Challacombe, I. Morgenstern, D. Hibbett, M. Schmoll, C. P. Kubicek, et al., “Genome, Transcriptome, and Secretome Analysis of Wood Decay Fungus Postia Placenta Supports Unique Mechanisms of Lignocellulose Conversion,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 106, No. 6, 2009, pp. 1954-1959.
- D. L. Crawford, A. L. Pometto and R. L. Crawford, “Lignin Degradation by Streptomyces viridosporus—Isolation and Characterization of a New Polymeric Lignin Degradation Intermediate,” Applied and Environmental Microbiology, Vol. 45, No. 3, 1983, pp. 898-904.
- J. Trojanowski, K. Haider and V. Sundman, “Decomposition of C-14-Labeled Lignin and Phenols by a Nocardia sp.,” Archives of Microbiology, Vol. 114, No. 2, 1977, pp. 149-153. doi:10.1007/BF00410776
- E. Masai, Y. Katayama and M. Fukuda, “Genetic and Biochemical Investigations on Bacterial Catabolic Pathways for Lignin-Derived Aromatic Compounds,” Bioscience Biotechnology and Biochemistry, Vol. 71, No. 1, 2007, pp. 1-15. doi:10.1271/bbb.60437
- M. Ahmad, C. R. Taylor, D. Pink, K. Burton, D. Eastwood, G. D. Bending and T. D. H. Bugg, “Development of Novel Assays for Lignin Degradation: Comparative Analysis of Bacterial and Fungal Lignin Degraders,” Molecular Biosystems, Vol. 6, No. 5, 2010, pp. 815-821. doi:10.1039/b908966g
- T. D. H. Bugg, M. Ahmad, E. M. Hardiman and R. Singh, “The Emerging Role for Bacteria in Lignin Degradation and Bio-product Formation,” Current Opinion in Biotechnology, Vol. 22, No. 3, 2011, pp. 394-400. doi:10.1016/j.copbio.2010.10.009
- E. B. Garethjones, R. D. Turner, S. E. J. Furtado and H. Kuhne, “Marine Biodeteriogenic Organisms. 1. Lignicolous Fungi and Bacteria and Wood Boring Mollusca and Crustacea,” International Biodeterioration Bulletin, Vol. 12, No. 4, 1976, pp. 120-134.
- M. Pailleret, T. Haga, P. Petit, C. Prive-Gill, N. Saedlou, F. Gaill and M. Zbinden, “Sunken Wood from the Vanuatu Islands: Identification of Wood Substrates and Preliminary Description of Associated Fauna,” Marine Ecology—An Evolutionary Perspective, Vol. 28, No. 1, 2007, pp. 233-241.
- S. Rohrmann and H. P. Molitoris, “Screening for WoodDegrading Enzymes in Marine Fungi,” Canadian Journal of Botany-Revue Canadienne de Botanique, Vol. 70, No. 10, 1992, pp. 2116-2123. doi:10.1139/b92-263
- W. Appeltans, P. Bouchet, G. A. Boxshall, C. De Broyer, N. J. de Voogd, D. P. Gordon, B. W. Hoeksema, T. Horton, M. Kennedy, J. Mees, G. C. B. Poore, G. Read, S. Stöhr, T. C. Walter and M. J. Costello, “World Register of Marine Species,” 2012. http://www.marinespecies.org
- I. Wagner-Dobler and H. Biebl, “Environmental Biology of the Marine Roseobacter lineage,” Annual Review of Microbiology, Annual Reviews, Vol. 60, 2006, 2006, pp. 255-280.
- M. A. Moran, R. Belas, M. A. Schell, J. M. Gonzalez, F. Sun, S. Sun, B. J. Binder, A. Buchan, et al., “Ecological Genomics of Marine Roseobacters,” Applied and Environmental Microbiology, Vol. 73, No. 14, 2007, pp. 4559- 4569. doi:10.1128/AEM.02580-06
- O. Pinyakong, H. Habe and T. Omori, “The Unique Aromatic Catabolic Genes in Sphingomonads Degrading Polycyclic Aromatic Hydrocarbons (PAHs),” Journal of General and Applied Microbiology, Vol. 49, No. 1, 2003, pp. 1-19. doi:10.2323/jgam.49.1
- M. T. Garcia, A. Ventosa and E. Mellado, “Catabolic Versatility of Aromatic Compound-degrading Halophilic Bacteria,” FEMS Microbiology Ecology, Vol. 54, No. 1, 2005, pp. 97-109. doi:10.1016/j.femsec.2005.03.009
- H. Rodriguez, J. Landete, J. Curiel, B. Rivas, J. Mancheno and R. Munoz, “Characterization of the p-Coumaric Acid Decarboxylase from Lactobacillus plantarum CECT 748T,” Journal of Agricultural and Food Chemistry, Vol. 56, No. 9, 2008, pp. 3068-3072.
- J. F. Heidelberg, I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, et al., “Genome Sequence of the Dissimilatory Metal Ion-Reducing Bacterium Shewanella oneidensis,” Nature Biotechnology, Vol. 20, No. 11, 2002, pp. 1118-1123. doi:10.1038/nbt749
- C. S. Harwood and R. E. Parales, “The Ketoadipate Pathway and the Biology of Self-Identity,” Annual Review of Microbiology, Vol. 50, 1996, pp. 553-590. doi:10.1146/annurev.micro.50.1.553
- E. Masai, M. Sasaki, Y. Minakawa, T. Abe, T. Sonoki, K. Miyauchi, Y. Katayama and M. Fukuda, “A Novel Tetrahydrofolate-Dependent O-Demethylase Gene Is Essential for Growth of Sphingomonas paucimobilis SYK-6 with Syringate,” Journal of Bacteriology, Vol. 186, No. 9, 2004, pp. 2757-2765. doi:10.1128/JB.186.9.2757-2765.2004
- J. M. Gonzalez, W. B. Whitman, R. E. Hodson and M. A. Moran, “Identifying Numerically Abundant Culturable Bcteria from Complex Communities: An Example from a Lignin Enrichment Culture,” Applied and Environmental Microbiology, Vol. 62, No. 12, 1996, pp. 4433-4440.
- B. Y. Tian, Q. G. Huang, Y. Xu, C. X. Wang, R. R. Lv and J. Z. Huang, “Microbial Community Structure and Diversity in a Native Forest Wood-Decomposed HollowStump Ecosystem,” World Journal of Microbiology & Biotechnology, Vol. 26, No. 2, 2010, pp. 233-240. doi:10.1007/s11274-009-0165-5
- V. Valaskova, W. de Boer, P. Gunnewiek, M. Pospisek and P. Baldrian, “Phylogenetic Composition and Properties of Bacteria Coexisting with the Fungus Hypholoma fasciculare in Decaying Wood,” ISME Journal, Vol. 3, No. 10, 2009, pp. 1218-1221. doi:10.1038/ismej.2009.64
- S. Samadi, L. Corbari, J. Lorion, S. Hourdez, T. Haga, J. Dupont, M.-C. Boisselier and B. R. de Forges, “Biodiversity of Deep-Sea Organisms Associated with SunkenWood or Other Organic Remains Sampled in the Tropical Indo-Pacific,” Cahiers de Biologie Marine, Vol. 51, No. 4, 2010, pp. 459-466.
- J. P. N. Rosazza, Z. Huang, L. Dostal, T. Volm and B. Rousseau, “Review: Biocatalytic Transformations of Ferulic Acid: An Abundant Aromatic Natural Product,” Journal of Industrial Microbiology, Vol. 15, No. 6. 1995, pp. 457-471. doi:10.1007/BF01570016
- Z. X. Huang, L. Dostal and J. P. N. Rosazza, “Purification and Characterization of a Ferulic Acid Decarboxylase from Pseudomonas fluorescens,” Journal of Bacteriology, Vol. 176, No. 19, 1994, pp. 5912-5918.
- G. Degrassi, P. P. Delaureto and C. V. Bruschi, “Purification and Characterization of Ferulate and p-Coumarate Decarboxylase from Bacillus pumilus,” Applied and Environmental Microbiology, Vol. 61, No. 1, 1995, pp. 326- 332.
- J. M. Salgado, R. Rodríguez-Solana, J. A. Curiel, B. de L. Rivas, R. Muñoz, J. M. Domínguez, “Production of Vinyl Derivatives from Alkaline Hydrolysates of Corn Cobs by Recombinant Escherichia coli Containing the Phenolic Acid Decarboxylase from Lactobacillus plantarum CECT 748T,” Bioresource Technology, Vol. 117, 2012, pp. 274- 285. doi:10.1016/j.biortech.2012.04.051
- S. Mathew and T. E. Abraham, “Bioconversions of Ferulic Acid, an Hydroxycinnamic Acid,” Critical Reviews in Microbiology, Vol. 32, No. 3, 2006, pp. 115-125. doi:10.1080/10408410600709628
- S. Fetzner, “Ring-Cleaving Dioxygenases with a Cupin Fold,” Applied and Environmental Microbiology, Vol. 78, No. 8, 2012, pp. 2505-2514. doi:10.1128/AEM.07651-11
- A. Buchan, E. L. Neidle and M. A. Moran, “Diversity of the Ring-Cleaving Dioxygenase Gene pcaH in a Salt Marsh Bacterial Community,” Applied and Environmental Microbiology, Vol. 67, No. 12, 2001, pp. 5801- 5809. doi:10.1128/AEM.67.12.5801-5809.2001
- Y. Wang, J. K. Yang, O. O. Lee, S. Dash, S. C. K. Lau, A. Al-Suwailem, T. Y. H. Wong, A. Danchin and P. Y. Qian, “Hydrothermally Generated Aromatic Compounds Are Consumed by Bacteria Colonizing in Atlantis II Deep of the Red Sea,” ISME Journal, Vol. 5, No. 10, 2011, pp. 1652-1659. doi:10.1038/ismej.2011.42
NOTES
*Corresponding author.

