Open Journal of Genetics
Vol.2 No.1(2012), Article ID:17761,7 pages DOI:10.4236/ojgen.2012.21008
Pronuclear microinjection is not suitable for RNA polymerase III promoter driven constitutive RNAi transgenesis in mice for XY male-to-female sex reversal by Sry gene knockdown
![]()
Department of Pharmacology, School of Medicine, Faculty of Medicine, Toho University, Otaku, Tokyo, Japan
Email: itomasanori@med.toho-u.ac.jp
Received 9 November 2011; revised 11 December 2011; accepted 22 January 2012
Keywords: Transgenic Mice; Pronuclear Microinjection; RNA Interference
ABSTRACT
Silencing of gene expression by RNA interference (RNAi) has become a widely used tool. For the study of mammalian gene function expression vectors for short hairpin RNA (shRNA) were developed. However the standard methods of shRNA transgenic (Tg) mice production have not been established. Sry (sexdetermining region on the Y chromosome) is a mammalian sex-determining gene on the Y chromosome. In mice, the transient expression of Sry in supporting cell precursor cells between 10.5 and 12.5 days postcoitus (dpc) triggers the differentiation of Sertoli cells from granulosa cells. Then high efficiency of Sry gene silencing in Tg mice should induce XY male-to-female sex reversal. An shRNA Tg mouse targeting Sry gene was attempted to be generated by pronuclear microinjection. A low rate (Tg pups/all pups born after microinjection = 2/154 to 7/178) of Tg pups was observed. These Tg mice showed no XY male-to-female sex reversal. The results suggest that exogenous expression of small RNA might exert a negative effect on embryonic development and another approach should be needed for RNAi transgenesis in mice.
1. INTRODUCTION
RNA interference (RNAi) has become a routine genetic tool to study gene function in mammalian cells including cancer cells [1]. Short hairpin RNA (shRNA) expression vectors have been developed which can be designed for knockdown of specific gene expression [2]. However the standard methods of shRNA transgenic (Tg) mice production have not been established. As RNA polymerase III (polIII) promoter such as U6, H1 and tRNA is ubiquitous promoter that is commonly used for shRNA expression, the phenotype of shRNA Tg mice should be similar to the phenotype of the knockout mice of the target gene. Sry (Sex determining region on the Y chromosome) is a transcription factor with a DNA-binding domain referred to as the high mobility group (HMG), which triggers a gene expression cascade required for initiating male sex differentiation in the bipotential indifferent gonads of mammals [3]. Mouse Sry is expressed for a brief period between 10.5 and 12.5 days post-coitus (dpc) in the supporting cells of undifferentiated gonads that differentiate into Sertoli cells instead of granulosa cells [4-6]. For specific purposes, shRNA targeting Sry was constructed [7] and the corresponding Tg mice were then attempted to be generated. As the pronuclear microinjection method is standard method for production of Tg mice, shRNA Tg mice were produced by pronuclear microinjection method in this study. If Sry knockdown in mice would be success, XY female-to-male sex reversal should be occurred. Then shRNA targeting Sry gene will be useful tool to establish the methods of RNAi transgenesis in mice.
2. MATERIALS AND METHODS
2.1. Animals
The following strains of mice were purchased from a commercial animal breeder (Sankyo Labo-Service Corporation, Inc., Tokyo, Japan): B6C3F1 (C57BL/6NxC3H/ He), C57BL/6J, and ICR. The mice were kept in an environment with regulated temperature (22˚C - 25˚C), humidity (40% - 50%), and illumination cycles (14-h light, 10-h dark), and were provided with food and water ad libitum. The present study was conducted according to the committee guidelines of the National Institute of Advanced Industrial Science and Technology for animal experiments and the Law for Prevention of Cruelty to Animals and the Guide for Care and Use of Laboratory Animals in Japan. The experiments complied with current Japanese laws.
2.2. Tg Mouse Generation
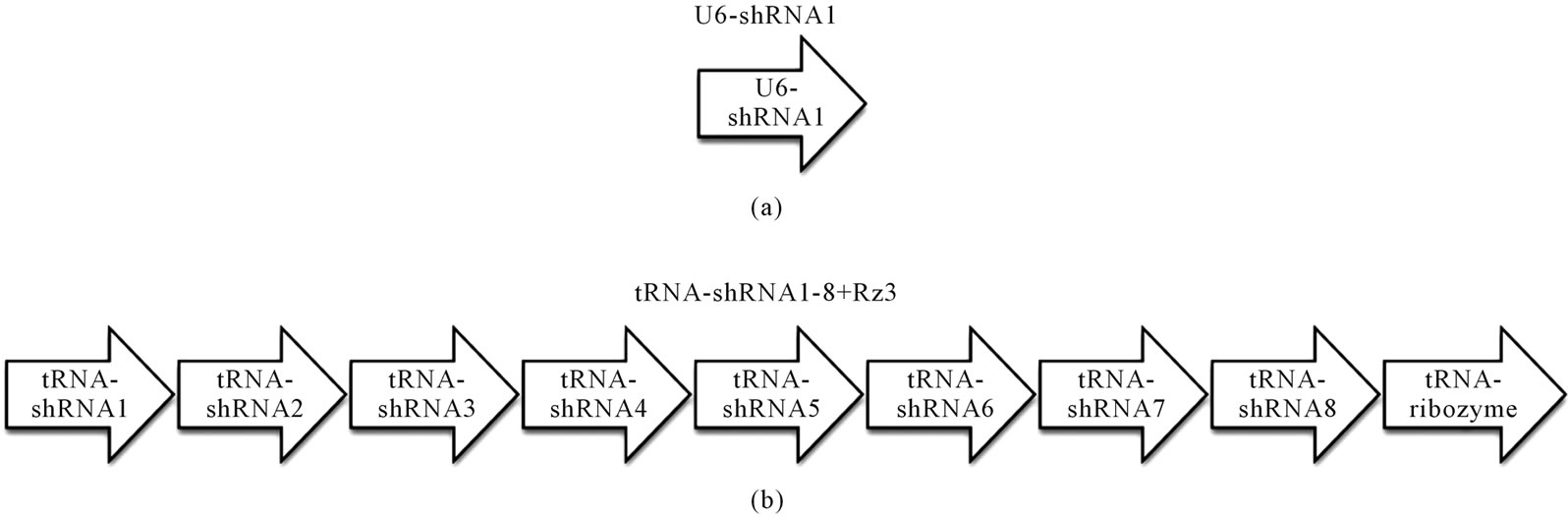
Tg mice were generated by microinjecting DNA into the pronuclei of zygotes collected from the oviducts of superovulated B6C3F1 females that were mated with B6C3F1 males. All methods for generating the Tg mice used here have been described in the protocol reported by Hogan et al. [8]. The construction of U6-shRNA1 (containing one shRNA expressing construct) and tRNAshRNA1-8 + Rz3 (eight shRNA and one ribozyme expressing constructs were connected) targeting Sry gene were described previously [7]. The EcoRI/HindIII DNA fragment for U6-shRNA1 and the EcoRI/MfeI DNA fragment for tRNA-shRNA1-8 + Rz3 were excised and separated by electrophoresis through 1% agarose gel; the fragment was then purified by CsCl ultra-centrifugation. The purified DNA fragment was dissolved in a solution containing 10 mM Tris-HCl (pH 7.4) and 0.25 mM EDTA (pH 7.4) and was used for pronuclear microinjection. To identify Tg animals and sex chromosome karyotypes (XX or XY), genomic DNA was isolated from the tip of the tail, and the genomic DNA was screened by polymerase chain reaction (PCR) amplification as previously described [9]. Transcripts of the Sry in the XY gonads at 11.5 dpc were determined by reverse transcription polymerase chain reaction (RT-PCR) as previously described [9].
2.3. Histology
For the histological analysis, tissues were fixed in 4% paraformaldehyde. After fixation, the tissues were processed for paraffin embedding as previously described [6]. The tissues were then sectioned at 6 μm, and the sections were used for hematoxylin and eosin staining.
3. RESULTS
Production of Tg Mice
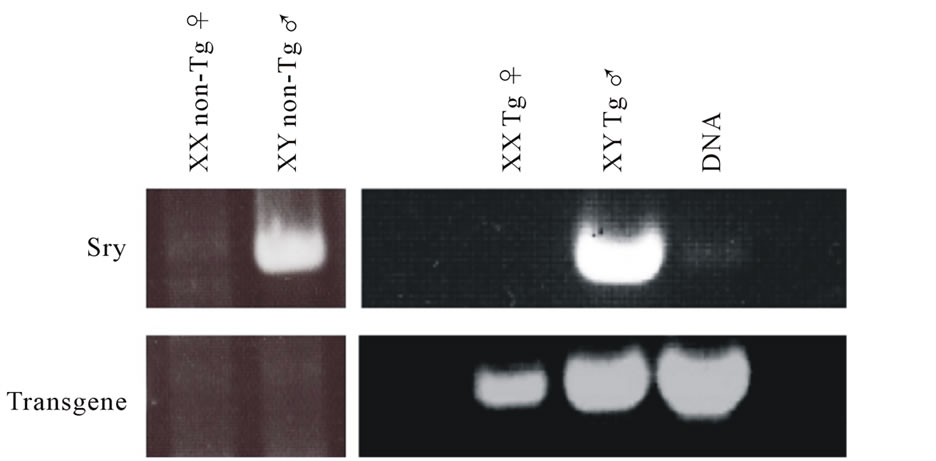
The constructs of shRNA targeting Sry gene (U6-shRNA1 or tRNA-shRNA1-8 + Rz3) were used for DNA microinjection to produce Tg mice (Figures 1(a) and (b)). The constructs were selected since these constructs were exhibited the highest silencing effect toward Sry [7]. In the analyses of DNA pups born, the percentages of Tg mice were 3.9% (Tg pups/all pups born after microinjection = 7/178) for U6-shRNA1 and 1.3% (Tg pups/all pups born after microinjection = 2/154) for tRNA-shRNA1-8 + Rz3. Seven U6-shRNA1 Tg lines (5 female and 2 male) were analyzed. All Tg mice were fertile. The female karyotype was XX, and that of the male was XY. F1 XY Tg mice from breeding with the all five F0 XX female Tg mice were male (Figure 1(c)). Two tRNA-shRNA1-8 + Rz3 Tg lines (female and male) were analyzed. The female karyotype was XX, and that of the male was XY. All Tg mice were fertile. F1 XY Tg mice from breeding with the F0 XX female Tg mouse were male. There was no histological abnormality in the testis of the Tg mice (Figures 1(e) and (f)). Sry expression levels of Tg XY gonads at 11.5 dpc were not lower than that of wild-type littermates (Figure 1(d)). Thus, shRNA was silenced at the inserted locus and not expressed in Tg tissues or Sry expression was not suppressed by the shRNA. Then, male-to-female sex reversal could not occur with the polIII promoter driven constitutive shRNA expressing transgene by pronuclear microinjection method.
4. DISCUSSION
In this study, an shRNA Tg mouse production approach was used to repress the expression of Sry. Foreign mouse Sry has been shown to induce XX female-to-male sex reversal [3]. Then it was expected that transgenesis of shRNA targeting Sry would induce XY male-to-female sex reversal. Tg mice expressing shRNA targeting Sry
 (c)
(c)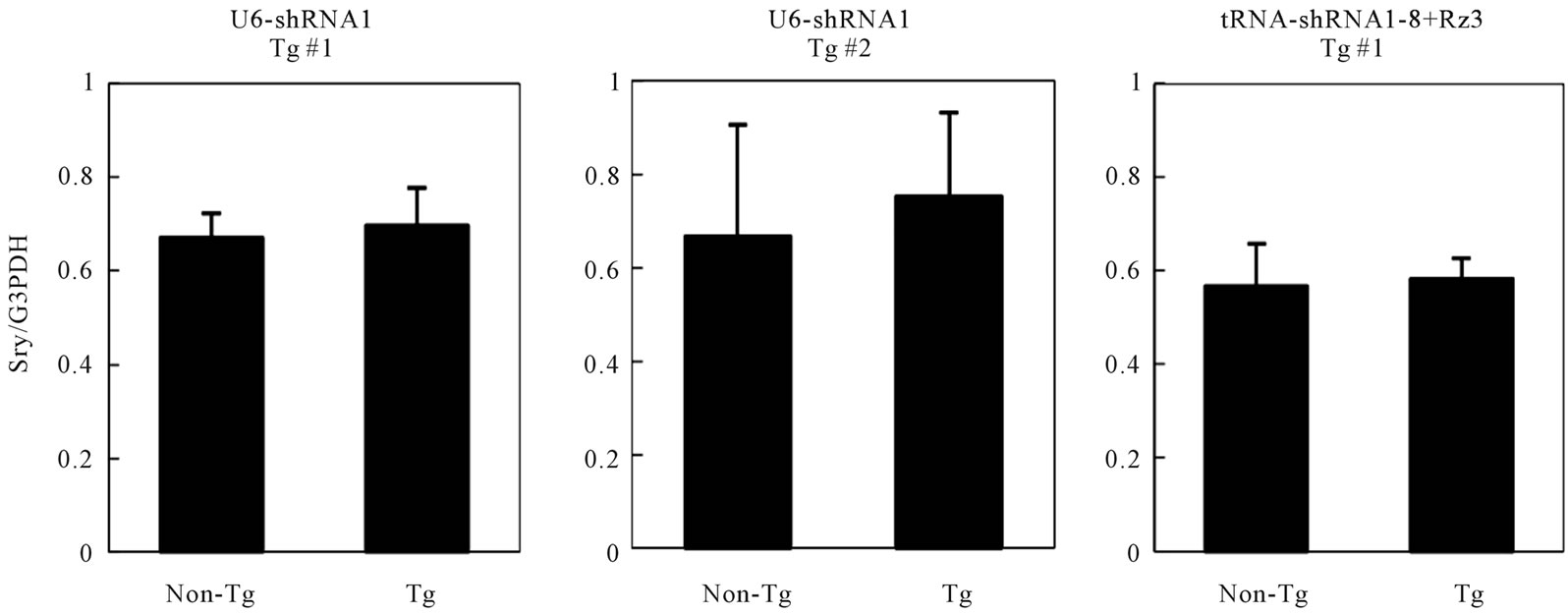 (d)
(d)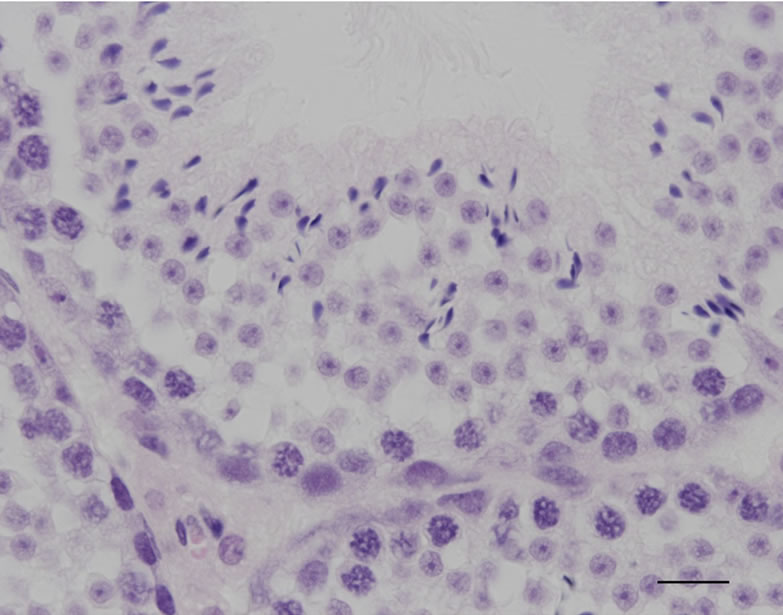 (e)
(e)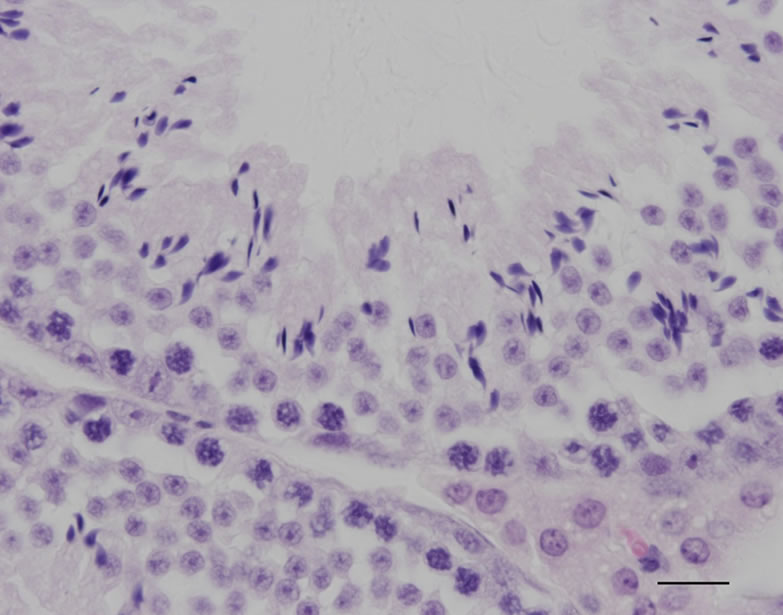 (f)
(f)
Figure 1. Production and analysis of transgenic (Tg) mice. (a), (b) A schematic representation of the transgene construct used in the microinjection; U6-shRNA1 and tRNA-shRNA1-8 + Rz3; (c) PCR analysis with the genomic DNA to identify the U6-shRNA1 Tg mice and the presence of the Y chromosome. Endogenous Sry was used to identify the presence of the Y chromosome. Tg, transgenic; non-Tg, non-transgenic; DNA, Injection DNA control; XX, XX karyotype; XY, XY karyotype; ♀, female phenotype; ♂, male phenotype; (d) Sry expression levels relative to those of G3PDH in the XY gonads of each Tg line at 11.5 dpc. Expression levels of Sry in the Tg mice were not reduced. Values are expressed as the mean ± S.E.M. (n = 3); (e), (f) Hematoxylin and eosin staining of testis from tRNA-shRNA1-8 + Rz3 Tg mice (e) and wild-type littermates (f) bar, 20 μm.
were attempted to be generated. The efficiency of transgenesis was remarkably low (1.3% to 3.9%) compared with the efficiency of usual Tg experiments [10-13]. Furthermore, no XY male-to-female sex reversal occurred with the transgene. All the female Tg mice were characterized as XX and the male Tg mice were characterized as XY. This result might suggest that ubiquitous and strong expression of shRNA yields embryonic lethality; the integration of the transgene at the locus where the transgene is silenced could explain why the XY sex reversal was not occurred in the Tg mice. It is also possible that the design of shRNA sequences was not suitable for suppression of Sry expression. However the shRNA constructs exhibited the strong silencing effect toward Sry gene in the cultured cells [7]. The knockdown efficiency of the shRNA constructs in vivo may be confirmed by organ culture on agar blocks and magnetically induced transfection using embryonic genital ridges [14] or by injection into tail vein of pregnant mice [15].
For transgenic RNAi in mice, one-cell embryos [16-29] or embryonic stem (ES) cells [30-37] have been used as the target (Table 1). The methods of ES cells need several months for many steps such as vector construction, clone isolation, chimera mouse production and breeding. However ES cells have made possible a ubiquitously active locus (e.g., ColA1 and Rosa26) targeting transgene integration [34-37]. One-cell embryos have been used for pronuclear microinjection [16-25] or lentivirus infection [17,26-29]. Long double strand RNA (dsRNA) (around 500bp) expressing vectors were constructed for microinjection [19-23]. However these constructs were effective only oocytes or early embryos as expression of dsRNA induces apoptosis by dsRNA-dependent protein kinase (PKR) in most mammalian cell types [38]. In the case of shRNA expressing constructs, Carmell et al. reported that Tg mice produced by pronuclear microinjection did not show a phenotype that was expected for the targeted genes such as tyrosinase (albino), myosin VIIa (shaker), Bmp-5 (crinkled ears), Hox a10 (limb defects), homogentisate 1,2-dioxygenase (urine turns black upon exposure to air), Hairless (hair loss) and melanocortin 1 receptor (yellow) [17]. The report agrees with the present study in that pronuclear microinjection is not suitable for RNAi transgenesis. On the other hand Peng et al. reported that pronuclear microinjection of shRNA-expressing constructs was used for genetic screening in mice. In the study one of the transgene targeting p57 was effective and the knockdown effect was transmitted for many generations [18]. Then it might depend on the targeting gene whether the shRNA-expressing constructs have knockdown effect in the Tg mice produced by pronuclear microinjection. The method of lentivirus infection has some advantages over the pronuclear microinjection method [26]. First, the injection of the lentivirus into the perivitelline space is less invasive than the pronuclear microinjection. Second, the efficiency of transgenesis is much higher than that of pronuclear microinjection. Third, single copy insertions are occurred by lentivirus infection. On the other hand multiple copies are inserted into one locus in the most cases of transgenesis by the pronuclear microinjection method. Single copy insertion is attractive for RNAi transgenesis since it is expected that too high expression of shRNA should induce embryonic lethality in this study. Moreover high efficiency of transgenesis would make it easy to analyze F0 Tg embryos and it should be useful tool when the

Table 1. Application of RNAi for genetics of mice.
gene knockdown causes embryonic lethality. F0 Tg embryos also can be analyzed using tetraploid aggregation technique and ES cells containing shRNA-expressing construct [30-33]. Kirilov et al. reported that lentivirus mediated F0 Tg mice were genetically mosaic and the mosaicism led to a reduced rate of germ line transmission [29]. It is possible that the overexpression of shRNA might have interfered with the processing of endogenous micro RNA (miRNA) and should be cause of the high mosaicism and the low degree of germ line transmission.
To overcome the problem, inducible shRNA expression system should be useful such as Cre/loxP system [24,32, 33] and tetracycline system [25,34-37]. In the Cre/loxP system, a loxP-flanked stuffer sequence should be inserted into the shRNA expression construct and can be removed by Cre. After the recombination, shRNA can be expressed and work for target gene knockdown [24,32, 33]. Tissue and stage specific promoter can be used for the control of Cre expression and the Cre Tg mouse will be used for the spatiotemporally controlled shRNA expression. Chemical inducible Cre recombination system also has been developed using doxycycline (a member of the tetracycline antibiotics) dependent transcription or tamoxifen (an agonist/antagonist of the estrogen receptor) dependent nuclear translocation. The tetracycline system should need two Tg lines—the tetracycline transactivator (tTA; for Tet-Off) or reverse tTA (rtTA; for Tet-On) expressing line and the line containing the construct of shRNA under the control of tetracycline responsive promoter. Dickins et al. established Tg lines expressing miRNA based shRNA controlled by tetracycline responsive–cytomegalovirus promoter using pronuclear microinjection method, and the doxycycline-dependent reversible induction was worked effectively [25]. More-
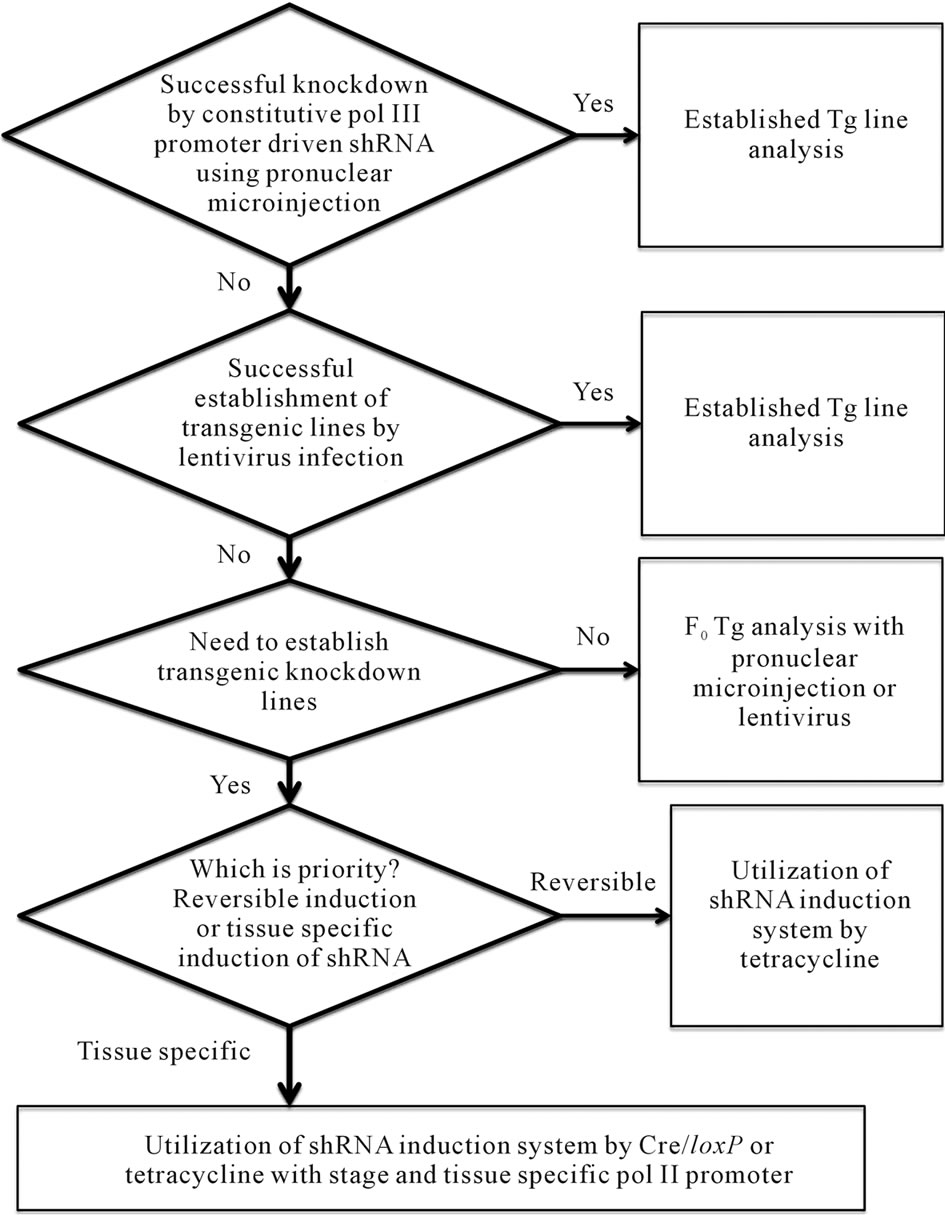
Figure 2. Flowchart to assist selecting the most appropriate method for RNAi transgenesis. As summarized in the text, strong ubiquitous expression of small RNA might induce embryonic lethality. It makes it impossible to establish constitutive shRNA expressing Tg lines. When establishment of Tg lines were failed for such a reason inducible shRNA system should be tried. The tetracycline system and Cre/loxP system should be suitable.
over, tissue specific polII promoter would be applied for the tetracycline-dependent shRNA expression system [25]. However, Cre/loxP system might be more useful than tetracycline system since strong transgene expression can be switched on by Cre/loxP system when weak polII promoter such as Sry promoter would be used [6, 39,40]. As blood-brain barrier and blood-testis barrier have low permeability to doxycycline, the tetracycline inducible system may not be applicable for the expression in brain and testis [41]. As for the knockdown of Sry gene at the embryonic stage the tetracycline inducible system should be applicable since shRNA expression was induced in Tg embryos when the water or food pellets containing doxycycline were supplied to the pregnant mothers [35,36]. In conclusion the pronuclear microinjection method is not suitable for the generation of constitutive RNAi Tg mice for Sry gene knockdown. Recently, methods of conditional RNAi transgenesis in mice have been developed [42,43]. Figure 2 shows flowchart for selection of an appropriate RNAi transgenesis method. As male-to-female sex reversal is one of the most obvious phenotype, Sry is useful as a target gene to test the RNAi transgenesis method.
5. ACKNOWLEDGEMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- Ito, M., Zhao, N., Zeng, Z, Chang, C.C. and Zu, Y. (2010) Synergistic growth inhibition of anaplastic large cell lymphoma cells by combining cellular ALK gene silencing and a low dose of the kinase inhibitor U0126. Cancer Gene Therapy, 17, 633-644. doi:10.1038/cgt.2010.20
- Ito, M., Kawano, K., Miyagishi, M. and Taira, K. (2005) Genome-wide application of RNAi to the discovery of potential drug targets. FEBS Letters, 579, 5988-5995. doi:10.1016/j.febslet.2005.08.015
- Koopman, P., Gubbay, J., Vivian, N., Goodfellow, P. and Lovell-Badge, R. (1991) Male development of chromosomally female mice transgenic for Sry. Nature, 351, 117- 121. doi:10.1038/351117a0
- Bullejos, M. and Koopman, P. (2001) Spatially dynamic expression of Sry in mouse genital ridges. Developmental Dynamics, 221, 201-205. doi:10.1002/dvdy.1134
- Albrecht, K.H. and Eicher, E.M. (2001) Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Developmental Biology, 240, 92-107. doi:10.1006/dbio.2001.0438
- Ito, M., Yokouchi, K., Yoshida, K., Kano, K., Naito, K., Miyazaki, J. and Tojo, H. (2006) Investigation of the fate of Sry-expressing cells using an in vivo Cre/loxP system. Development Growth and Differentiation, 48, 41-47. doi:10.1046/j.1440-169X.2002.00668.x
- Ito, M., Miyagishi, M., Murata, C., Kawasaki, H., Baba, T., Tachi, C. and Taira, K. (2006) Down-regulation of endogenous Wt1 expression by Sry transgene in the murine embryonic mesonephros-derived M15 cell line. Journal of Reproduction and Development, 52, 415-427. doi:10.1262/jrd.17094
- Hogan, B., Constantini, F. and Lacy, E. (1994) Manipulating the mouse embryo. 2nd Edition, Cold Spring Harbor Laboratory Press, New York.
- Yoshida, K., Ito, M., Yokouchi, K., Kano, K., Naito, K. and Tojo H. (2010) Ubiquitous expression of Sry induces embryonic lethality related to suppression of Tie2/Tek expression. Advances in Bioscience and Biotechnology, 1, 444-452. doi:10.4236/abb.2010.15058
- Kato, M., Yamanouchi, K., Ikawa, M., Okabe, M., Naito, K. and Tojo, H. (1999) Efficient selection of transgenic mouse embryos using EGFP as a marker gene. Molecular Reproduction and Development, 54, 43-48. doi:10.1002/(SICI)1098-2795(199909)54:1<43::AID-MRD6>3.0.CO;2-N
- Seo, B.B., Kim, C.H., Yamanouchi, K., Takahashi, M., Sawasaki, T., Tachi, C. and Tojo, H. (2000) Co-injection of restriction enzyme with foreign DNA into the pronucleus for elevating production efficiencies of transgenic animals. Animal Reproduction Science, 63, 113-122. doi:10.1016/S0378-4320(00)00148-2
- Kubo, J., Yamanouchi, K., Naito, K. and Tojo, H. (2002) Expression of the gene of interest fused to the EGFP-expressing gene in transgenic mice derived from selected transgenic embryos. Journal of Experimental Zoology, 293, 712-718. doi:10.1002/jez.10163
- Ito, M., Yamanouchi, K., Naito, K. Calos, M.P. and Tojo, H. (2011) Site-specific integration of transgene targeting an endogenous lox-like site in early mouse embryos. Journal of Applied Genetics, 52, 89-94. doi:10.1007/s13353-010-0011-3
- Svingen, T., Wilhelm, D., Combes, A.N., Hosking, B., Harley, V.R., Sinclair, A.H. and Koopman, P. (2009) Ex vivo magnetofection: A novel strategy for the study of gene function in mouse organogenesis. Developmental Dynamics, 238, 956-964. doi:10.1002/dvdy.21919
- Wu, N., Lin, X.K., Liao, B., Du, W.H., Han, F.T. and Zhao, J.H. (2008) Effect of Sry silencing by siRNA on the expression of sex determining genes in mouse embryos. Yi Chuan, 30, 195-202. doi:10.3724/SP.J.1005.2008.00195
- Hasuwa, H., Kaseda, K., Einarsdottir, T. and Okabe, M. (2002) Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Letters, 532, 227-230. doi:10.1016/S0014-5793(02)03680-3
- Carmell, M.A., Zhang, L., Conklin, D.S., Hannon, G.J. and Rosenquist, T.A. (2003) Germline transmission of RNAi in mice. Nature Structural Biology, 10, 91-92. doi:10.1038/nsb896
- Peng, S., York, J.P. and Zhang, P. (2006) A transgenic approach for RNA interference-based genetic screening in mice. Proceedings of the National Academy of Sciences of USA, 103, 2252-2256. doi:10.1073/pnas.0511034103
- Shinagawa, T. and Ishii, S. (2003) Generation of Skiknockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes & Development, 17, 1340-1345. doi:10.1101/gad.1073003
- Stein, P., Svoboda, P. and Schultz, R.M. (2003) Transgenic RNAi in mouse oocytes: A simple and fast approach to study gene function. Developmental Biology, 256, 187-193. doi:10.1016/S0012-1606(02)00122-7
- Yu, J., Deng, M., Medvedev, S., Yang, J., Hecht, N.B. and Schultz, R.M. (2004) Transgenic RNAi-mediated reduction of MSY2 in mouse oocytes results in reduced fertility. Developmental Biology, 268, 195-206. doi:10.1016/j.ydbio.2003.12.020
- Stein, P., Zeng, F., Pan, H. and Schultz, R.M. (2005) Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Developmental Biology, 286, 464-471. doi:10.1016/j.ydbio.2005.08.015
- Sarnova, L., Malik, R., Sedlacek, R. and Svoboda, P. (2010) Shortcomings of short hairpin RNA-based transgenic RNA interference in mouse oocytes. Journal of Negative Results in Biomedicine, 9, 8. doi:10.1186/1477-5751-9-8
- Coumoul, X., Shukla, V., Li, C., Wang, R.H. and Deng, C. (2004) Conditional knockdown of Fgfr2 in mice using Cre-LoxP induced RNA interference. Nucleic Acids Research, 33, e102. doi:10.1093/nar/gni100
- Dickins, R.A., McJunkin, K., Hernando, E., Premsrirut, P.K., Krizhanovsky, V., Burgess, D.J., Kim, S.Y., Cordon-Cardo, C., Zender, L., Hannon, G.J. and Lowe, S.W. (2007) Tissue-specific and reversible RNA interference in transgenic mice. Nature Genetics, 39, 914-921. doi:10.1038/ng2045
- Pfeifer, A. (2004) Lentiviral transgenesis. Transgenic Research, 13, 513-522. doi:10.1007/s11248-004-2735-5
- Tiscornia, G., Singer, O., Ikawa, M. and Verma, I.M. (2003) A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proceedings of the National Academy of Sciences of USA, 100, 1844-1848. doi:10.1073/pnas.0437912100
- Rubinson, D.A., Dillon, C.P., Kwiatkowski, A.V., Sievers, C., Yang, L., Kopinja, J., Rooney, D.L., Zhang, M., Ihrig, M.M., McManus, M.T., Gertler, F.B., Scott, M.L. and Van Parijs, L. (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genetics, 33, 401-406. doi:10.1038/ng1117
- Kirilov, M., Chai, M., van der Hoeven, F., Kloz, U., Schmid, W. and Schütz, G. (2007) Germ line transmission and expression of an RNAi cassette in mice generated by a lentiviral vector system. Transgenic Research, 16, 783- 793. doi:10.1007/s11248-007-9119-6
- Kunath, T., Gish, G., Lickert, H., Jones, N., Pawson, T. and Rossant, J. (2003) Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nature Biotechnology, 21, 559-561. doi:10.1038/nbt813
- Lickert, H., Cox, B., Wehrle, C., Taketo, M.M., Kemler, R. and Rossant, J. (2005) Dissecting Wnt/beta-catenin signaling during gastrulation using RNA interference in mouse embryos. Development, 132, 2599-2609. doi:10.1242/dev.01842
- Ventura, A., Meissner, A., Dillon, C.P., McManus, M., Sharp, P.A., Van Parijs, L., Jaenisch, R. and Jacks, T. (2004) Cre-lox-regulated conditional RNA interference from transgenes. Proceedings of the National Academy of Sciences of USA, 101, 10380-10385. doi:10.1073/pnas.0403954101
- Stern, P., Astrof, S., Erkeland, S.J., Schustak, J., Sharp, P.A. and Hynes, R.O. (2008) A system for Cre-regulated RNA interference in vivo. Proceedings of the National Academy of Sciences of USA, 105, 13895-13900. doi:10.1073/pnas.0806907105
- Seibler, J., Kleinridders, A., Küter-Luks, B., Niehaves, S., Brüning, J.C. and Schwenk, F. (2007) Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Research, 35, e54. doi:10.1093/nar/gkm122
- Vidigal, J.A., Morkel, M., Wittler, L., Brouwer-Lehmitz, A., Grote, P., Macura, K. and Herrmann, B.G. (2010) An inducible RNA interference system for the functional dissection of mouse embryogenesis. Nucleic Acids Research, 38, e122. doi:10.1093/nar/gkq199
- Premsrirut, P.K., Dow, L.E., Kim, S.Y., Camiolo, M., Malone, C.D., Miething, C., Scuoppo, C., Zuber, J., Dickins, R.A., Kogan, S.C., Shroyer, K.R., Sordella, R., Hannon, G.J. and Lowe, S.W. (2011) A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell, 145, 145-158. doi:10.1016/j.cell.2011.03.012
- McJunkin, K., Mazurek, A., Premsrirut, P.K., Zuber, J., Dow, L.E., Simon, J., Stillman, B. and Lowe, S.W. (2011) Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proceedings of the National Academy of Sciences of USA, 108, 7113-7118. doi:10.1073/pnas.1104097108
- Svoboda, P. (2004) Long dsRNA and silent genes strike back: RNAi in mouse oocytes and early embryos. Cytogenetic and Genome Research, 105, 422-434. doi:10.1159/000078215
- Ito, M., Yokouchi, K., Naito, K., Endo, H., Hakamata, Y., Miyazaki, J. and Tojo, H. (2002) In vitro Cre/loxP system in cells from developing gonads: Investigation of the Sry promoter. Development Growth and Differentiation, 44, 549-557. doi:10.1046/j.1440-169X.2002.00668.x
- Ito, M., Yokouchi, K., Naito, K., Endo, H., Hakamata, Y., Miyazaki, J. and Tojo, H. (2005) Detection of elements responsible for stageand tissue-specific expression of mouse Sry using an in vitro Cre/loxP system. Biochemical and Biophysical Research Communications, 337, 264- 270. doi:10.1016/j.bbrc.2005.09.047
- Beard, C., Hochedlinger, K., Plath, K., Wutz, A. and Jaenisch, R. (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis, 44, 23-28. doi:10.1002/gene.20180
- Kleinhammer, A., Wurst, W. and Kühn, R. (2011) Constitutive and conditional RNAi transgenesis in mice. Methods, 53, 430-436. doi:10.1016/j.ymeth.2010.12.015
- Kleinhammer, A., Deussing, J., Wurst, W. and Kühn, R. (2011) Conditional RNAi in mice. Methods, 53, 142-150. doi:10.1016/j.ymeth.2010.08.003

