Advances in Biological Chemistry
Vol.3 No.1(2013), Article ID:28097,11 pages DOI:10.4236/abc.2013.31008
Ultradian oscillators of the circadian clock in Saccharomyces cerevisiae
![]()
1Department of Foods and Nutrition, Purdue University, West Lafayette, USA
2Department of Medicinal Chemistry, Purdue University, West Lafayette, USA
Email: dmg.secre@gmail.com
Received 5 December 2012; revised 6 January 2013; accepted 14 January 2012
Keywords: Biological Clock; ECTO-NOX (ENOX) Proteins; Ultradian Oscillator; Yeast (Saccharomyces cerevisiae)
ABSTRACT
The yeast, Saccharomyces cerevisiae, has an ENOX1 activity with a period length of 24 min similar to that of other eukaryotes. In contrast to other eukaryotes, however, Saccharomyces cerevisiae has a second ENOX1-like activity with a period length of 25 min. The latter is distinguishable from the traditional ENOX1 on the basis of the longer period length along with resistance to an ENOX1 inhibitor, simalikalactone D, and failure to be phased by melatonin. In addition, two periods are apparent in measurements of oxygen consumption indicating that the consumption of oxygen to water occurs independently by homodimers of both of the two forms of ENOX. Based on the measurements of glyceraldehyde-3- phosphate dehydrogenase, S. cerevisiae exhibits circadian activity maxima at 24 and 25 h together with a 40 h period possibly representing the 40 min metabolic rhythm of yeast not observed in our measurement of oxygen consumption and normally observed only with continuous cultures. The findings are indicative of at least three independent time-keeping systems being operative in a single cell.
1. INTRODUCTION
ECTO-NOX designates a family of cell surface proteins that exhibit a time-keeping function, oxidize reduced coenzyme Q or NAD(P)H and carry out protein disulfide-thiol interchange [1]. They are referred to as ECTONOX proteins because of their localization on the outer surface of the plasma membrane [2] and to distinguish them from the phox-Nox proteins of host defense [3]. This external location of ECTO-NOX proteins has been demonstrated both in plants and animals.
At least three members of the ECTO-NOX (ENOX) family have been described: 1) the human constitutive NADH oxidase (ENOX1 or cNOX) (GenBank Accession No. EF432052), expressed in normal cells and located on chromosome 13q14.11; 2) the human tumor-associated NADH oxidase (ENOX2 or tNOX) (GenBank Accession No. AF207881), present on the surface of invasive cancer cells and located on chromosome Xq25-q26.2 and 3) the age-related NADH oxidase (arNOX or ENOX3), which is present only in individuals after the age of 30, in late passage cultured cells and in senescent plant parts [4]. Human ENOX1 shares 64% identity and 80% similarly with human ENOX2. Members of all three ENOX families contain an adenine nucleotide-binding site, a disulfide-thiol interchange site and two copper-binding motifs [5-7].
ECTO-NOX proteins differ from all other NAD(P)H oxidases in that they are able to reduce pyridine nucleotides in the absence of bound flavin [5]. Moreover, their activity is not inhibited by cyanide [8]. Also, ECTONOX proteins have properties of prions, including stability, protease resistance and ability to form amyloid [9].
Although enzymatic activity has been most often measured by monitoring NAD(P)H oxidation at 340 nm, the physiological substrates appear to be quinols (i.e., ubiquinols or phytoquinols) [10,11]. The NADH binding site is on the outside of the cell where there is usually little or no NAD(P)H. Quinones, such as Coenzyme Q, act as trans-plasma membrane shuttles to transfer reducing equivalents from cytosolic NAD(P)H to external acceptors, including molecular oxygen and protein disulfide. ENOX1 and ENOX2 lack iron or iron sulfur clusters. However, the copper binding motifs revealed from sequence analysis and site-directed mutagenesis are necessary for electron transfer to molecular oxygen [6,7]. Based on size exclusion chromatography, the soluble forms of ENOX2 are predominantly dimers. Thereforethe ENOX proteins may be viewed as homodimeric proteins containing four coppers per dimer, thus carrying out four electron transfers to molecular oxygen as required to form water [6].
ECTO-NOX proteins exhibit a second catalytic activity that of protein disulfide-thiol interchange despite the absence of the CXXC motif common to most members of the protein disulfide isomerase family [5,6,12]. The two activities, hydroquinone oxidation and protein disulfide-thiol interchange, alternate with a period length that is characteristic for each family member. ENOX1 exhibits oscillations with a period length of 24 min, that of ENOX2 with a period length of about 22 min and that of arNOX with a period length of about 26 min [1].
Several lines of evidence demonstrate that ENOX1 and ENOX2 proteins are involved in at least two important cell functions. They drive the enlargement phase of cell growth and they are components of the biological clock. The regulation of the circadian oscillatory system is mainly ascribed to ENOX1. The two enzymatic activities catalyzed by ENOX1 alternate within a 24 min period: the first activity rests after 12 min and the second one begins in a cycle that confers a time-keeping function to the protein. This oscillation pattern is entrainable and temperature independent. Cells transfected with mutated ENOX cDNAs, which have oscillation period lengths of 22, 36 or 42 min, exhibited circadian period lengths of 22, 36 or 42 h for the cellular circadian biochemical marker, glyceraldehyde-3-phosphate dehydrogenase [13].
ENOX proteins are ubiquitous at the surface of all cells thus far studied [1]. They are protease and heat resistant [14] and respond to hormones [15]. Evidence suggests that ENOX1 serves as an ultradian oscillator of the biological clock in mammals [1] and a human protein with activity characteristics of ENOX1 has been cloned [6]. Whereas circadian rhythms in yeasts have been studied less widely than in mammals, there is no reason to suspect that Saccharomyces cerevisiae should be exempt from circadian regulation. Especially well studied in S. cerevisiae is the 40 min cycle of respiratory activity in continuous culture [Discussion].
In this report, we have identified two different constitutive oscillatory ENOX proteins of S. cerevisiae one with a period length of 24 min and a second with a period length of about 25 min. Both proteins catalyze the oxidation of both externally supplied NAD(P)H to NAD(P)+ and hydroquinone and carry out protein disulfide interchange. These two forms of ENOX activity together with an already known ultradian rhythm with a period length of 40 min in S. cerevisiae generate three independent sets of potential ultradian time keeping oscillations.
2. METHODS
2.1. Saccharomyces Growth and Preparation
The yeast strains were maintained on YEPD agar plates and stored at 4˚C. Cells were grown at room temperature with shaking in rich media (YEPD) for 1 - 2 days to attain ca 2 × 108 cells/ml. ENOX proteins of yeast are exposed at the external surface of the yeast cells such that activities may be measured with unbroken cells. Additionally, the yeast cells were heated for 1 h at 70˚C to inactivate heat-sensitive enzymes prior to analysis. This was to prevent yeast contamination in the open laboratory. ENOX proteins resist heating at 70˚C for one h. Results were similar with unheated yeast cells.
2.2. NAD(P)H Oxidase Assay
Heat-inactivated yeast cells (ca. 107 cells in 50 µl) were combined with 2.5 ml 50 mM Tris-Mes buffer (pH 7.0), 2 mM KCN and 150 µM NADH. To refold and renature the ENOX proteins of the heat-inactivated yeast, reduced glutathione (GSH) was added to a final concentration of 100 µM, and incubated for 10 min to reduce disulfides to allow for refolding. Next H2O2 was added to a final concentration of 0.03% followed by incubation for 10 min to reform disulfides to stabilize the protein in a correctly folded conformation.
NADH oxidation was measured in a Hitachi Model U3210 spectrophotometer at 340 nm, continuously for one min at 1.5 min intervals for two h using an extinction coefficient of 6.21 mM−1∙cm−1. Inhibitors were added at 60 min and data were collected for a further 60 min. A second machine of the same model with the vehicle alone added at 60 min served as the control.
2.3. Hydroquinone Oxidation
Reduced coenzyme Q10 was prepared fresh by addition of an equal volume of 0.25% NaBH4 under nitrogen to a 7.5 mM stock solution of coenzyme Q10 in ethanol. After several min of incubation, 0.l volume of 0.1 N HC1 was added to degrade the excess NaBH4. The reaction mixture for measuring the oxidation of the reduced coenzyme Q10 (Q10H2) contained heat-inactivated yeast, ca. 107 cells in 2.5 ml of 50 mM Tris-Mes buffer, pH 7.0 and 0.08% Triton X-100 to maintain the Q10H2 in suspension. The reaction was started with the addition of 40 µl of 5 mM Q10H2. The disappearance of reduced CoQ was measured at both 290 and 410 nm using an extinction coefficient of 0.805 mM−1∙cm−1.
2.4. Dithiodipyridine (DTDP) Cleavage
Heat-inactivated yeast (ca. 107 cells in 50 µl) were added to 2.5 ml of 50 mM Tris-Mes, pH 7.0. The reaction was pre-incubated with 0.5 µmol of 2.2’-dithiodipyridine (DTDP) in 5 µl DMSO to oxidize residual thiols. After 10 min of incubation, a further 3.5 µmol of DTDP were added in 35 µl of DMSO to start the reaction. The increase in absorbance due to the cleavage of DTDP was monitored at 340 nm using a millimolar extinction coefficient of 6.21.
2.5. Glyceraldehyde-3-Phosphate Dehydrogenase Assay
Yeast were grown in 200 ml minimal SD minus uracil or SR minus uracil media (Open Biosystems, Huntsville, AL) on a shaker at 200 rpm at room temperature for 2 - 3 days at 25˚C. Samples of 50 µl were collected every 2 h and snap frozen at −80˚C. Time windows were used to collect the samples, with the flow between collections returned to the flask. Although this was a batch culture, the small sample sizes did not reduce the total volume of the culture significantly, and each sample had approximately the same total protein content.
To prepare the samples for the assay, the yeast were thawed and 50 µl of Y-PER (Yeast Protein Extraction Reagent, Pierce) were added and the solution was vortexed for 20 min to lyse the yeast and release the cell contents. Glyceraldehyde-3-phosphate dehydrogenase activity was assayed in a reaction solution containing 100 µl lysed yeast, 0.1 M Tris-HC1, 0.5 mM EGTA, pH 8.0, 1 mM Na2HAsO4, 2 mM NAD+ and 3 mM glyceroldehyde-3-phosphate. NAD+ reduction was determined from the increases in absorbance at 340 nm at 37˚C measured over 5 min using a millimolar extinction coefficient of 6.21.
2.6. Statistical Analyses
Results were analyzed using fast Fourier transform and decomposition fits [16]. To determine the period length of activity oscillations, the fast Fourier transforms were with a user-defined transform in Sigma Plot 8.0. Decomposition fits on both the enzyme activity and the growth determinations used MINITAB and were based on the period length established by Fourier analysis.
3. RESULTS
3.1. NAD(P)H Oxidase Assays
Standard NAD(P)H oxidase activity assays in which the disappearance of NADH was measured over one min at 1.5 min intervals in heat-inactivated yeast revealed two sets of oscillations based on the recurrence of two maxima separated by 6 min of similar but not identical period lengths. One set denoted by ① and ② in Figure 1(a) exhibited a period length of 24 min. The period length of the second set denoted by A and B in Figure 1(a) was longer and nearer to 25 min. This double period in yeast

Figure 1. Oscillatory pattern of NADH oxidation (ENOX activity) of heat killed yeast (a) and statistical analysis indicating two activities of non-identical period lengths (b)-(f). ① and ② indicate the first and second maxima of the ENOX activity producing a 24 min period and A and B indicate a second activity producing a 25 min period. ① and ② are separated from each other by 6 min as are (a) and (b); (b) Decomposition (time series) fit using a 24 min period length from Fourier analysis; (c) Decomposition fit using a 25 min period from Fourier analysis. In (b) and (c) the dashed lines are the decomposition fits and the solid lines are the values of (a); (d) Fourier analysis comparing the maxima labeled ① and ② (solid symbols and lines) yielding a period length of 24 min and those labeled A and B in Part A (open symbols and dashed lines) yielding a period length of 25 min; (e) Decomposition fit for B; (f) Decomposition fit for C. Three measures of the accuracy of the statistically fitted values are provided in Table 1.
increases the complexity of the data over that of mammalian cells and plants which normally exhibit only a single set of oscillations with a period length of 24 min.
To resolve the two oscillatory patterns, fast Fourier (Figure 1(d)) and time series analyses (decomposition fits) (Figures 1(e) and (f)) were employed as used conventionally to resolve patterns of oscillations that are neither sinusoidal nor monotonic [16]. Fourier analyses (Figure 1(d)) partially resolved the two oscillatory patterns with the pattern labeled A and B yielding a period with a slightly lower frequency (longer period) than the pattern labeled ① and ②. Decomposition fits for a 24 min period length based on the Fourier analysis clearly superimposed the maxima labeled ① and ② but not those labeled A and B (Figure 1(b)) whereas decomposition fits for a 25 min period length based on the Fourier analysis coincided with the maxima labeled A and B but not those labeled ① and ② (Figure 1(c)). The recurring patterns predicted by the time series analysis are reproduced in Figure 1(e) for the 24 min oscillations (maxima ① and ②) and in Figure 1(f) for the 25 min oscillations (maxima A and B). Statistical measures of accuracy are summarized and explained in Table 1. With the exception of Figure 2, Figures 3-8 were analyzed in a manner identical to that for Figure 1 with comparable measurements of accuracy.
3.2. Circadian Day
To link the two ENOX activities and the extra maxima corresponding to a 40 min ultradian period to the circadian clock, glyderaldehyde-3-phosphate dehydrogenase (GAPDH) activity was used. GAPDH activity peaks once within each 24 h period making it an ideal marker of circadian day length [17]. Morré et al. [13] reported that the ENOX1 period in min equals the circadian day length in h for a series of site directed C ® A replacements in ENOX2 where ultradian period lengths of 22, 36 and 45 min resulted in circadian period lengths of 22, 36 and 45 h. Yeast cultures grown in glucose sampled every 2 h for 88 h and assayed for GAPDH activity revealed maxima of GAPDH activity at 24, 25 and 40 h (Figure 2). The two hour sampling interval was sufficient to resolve the different maxima observed as each maximum was represented by several points defining both the ascending and the descending limbs of each maximum. The observed two maxima of 24 and 25 corresponded to the 24 and 25 min ultradian rhythms based on NADH oxidation. The 24 min period is denoted by ① and ②, the 25 min period is denoted by A and B. The 40 min period, denoted by 40 in the figure, may correspond to the 40 min metabolic rhythm of yeast thus far observed only with continuous cultures.
3.3. DTDP Cleavage
As measured by cleavage of the substrate 2,2’-dithiodipyridine (DTDP), three maxia separated by 4.5 min were generated by mammalian ENOX1 [6]. Yeast also exhibited this activity (Figure 3), with two sets of 3 maxima labeled ③, ④ and ⑤ and, C, D and E, each separated by 4.5 min, an activity that alternates with the two peaks of the oxidative activity for the two sets of oscillations [1]. DTDP cleavage was analyzed statisticcally as for Figure 1 with a similar outcome and comparable accuracy values (Table 1).
3.4. Hydroquinone Oxidation
Although externally-supplied NADH was used to assay ENOX activity, hydroquinones resident in the plasma membrane are most likely the natural substrate [10]. When reduced coenzyme Q10 was assayed, two sets of oxidative maxima were seen (Figure 4) similar to those observed with NADH (Figure 1). Within each set, two maxima each separated by 6 min were observed. Statistical analyses of both Figures 4(a) and (b) resolved the two sets of oscillations into series having distinct period lengths and accuracy values similar to those of Figure 1 (Table 1).
Table 1. Accuracy measures as determined by decomposition (time series) analyses.*

*Decomposition analyses decompose a time series into trend, cyclic, and error components. The measured values of Table 1 evaluate the accuracy of the fitted values and include MAPE (mean of the average percent error), MAD (mean average deviation), and MSD (mean standard deviation). The fitted values by the additive model used agreed closely with the experimental data. Overall the accuracy measures provided show a strong positive relationship between the actual and predicted values. By the statistical criteria, the model performs exceptionally well with the r-values approaching zero for a reasonable level of precision [16].
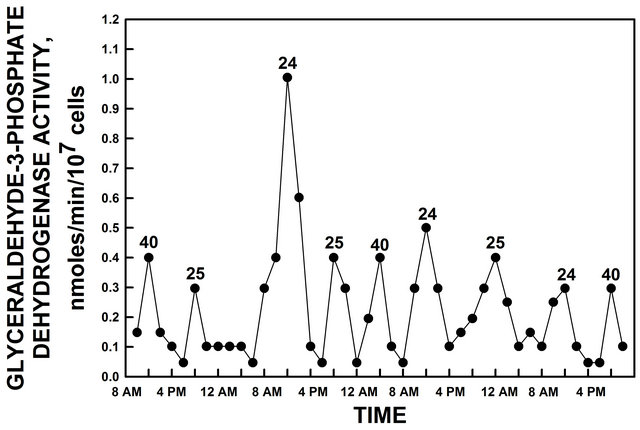
Figure 2. GAPDH activity of yeast grown in glucose. GAPDH activity peaked at intervals of 24 h, 25 h and 40 h. The 40 h maxima appear to correspond to the 40 min period normally observed only with continuous cultures [20,21].
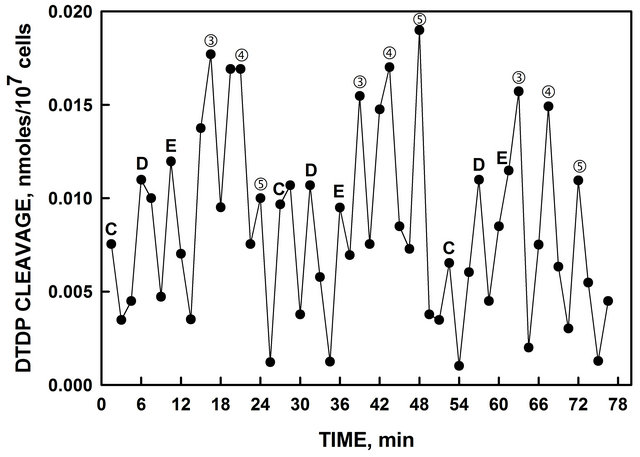
Figure 3. Heat-inactivated yeast exhibited disulfide-thiol interchange activity determined from the cleavage of a synthetic dithodipyridine DTDP substrate which alternate with the oxidative activity (Morré et al., 1999). ③, ④ and ⑤ are from the 24 min period and D, E and F are from the 25 min period. Data were analyzed as for Figure 1 and the measures of the accuracy of the statistically fitted values are provided in Table 1.
3.5. Response to Melatonin
Melatonin shifts the period of ENOX1 in mammalian cells [18]. When melatonin (final concentration 10 µM) was added to the NADH oxidase assay of mammalian ENOX1, a new maximum of NADH oxidase activity appeared exactly 24 min after melatonin addition. In yeast, the 24 min ENOX1 period responded to melatonin in this manner, denoted by ① and ② before melatonin addition and M after melatonin addition (Figure 5(a)). The initial 24 min period denoted ① and ② was suppressed immediately following melatonin addition (Figure 5(b)). The third maximum labeled ① just prior to melatonin addition was present but no third maximum ② was observed. The 24 min periodic oscillations then re-
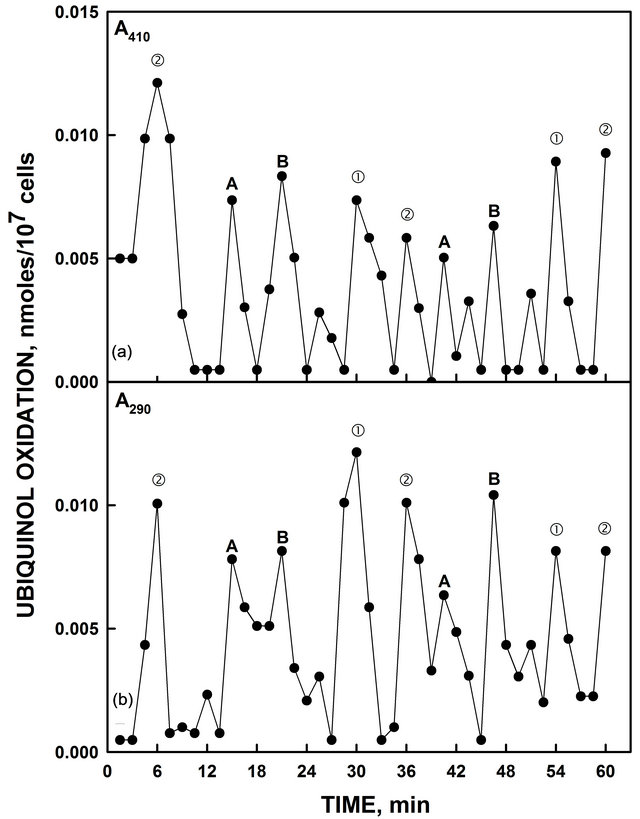
Figure 4. Heat-inactivated yeast oxidized reduced coenzyme Q measured either by an increase in A410 (a) or by a decrease in A290 (b). As with NADH oxidation of Figure 1, the activity oscillated with two sets of prominent maxima separated by 6 min. There were two maxima within each set of oscillations separated by 6 min. Those labeled ① and ② generated a period of 24 min in length. Those labeled A and B generated a period of 25 min in length. Accuracy measures of the decomposition fits are provided in Table 1.
sumed exactly 24 min following addition of melatonin 1.5 min out of phase with the original 24 min oscillations. In contrast, melatonin addition did not affect the oscillations with the 25 min period length (A and B) (Figure 5(c)).
3.6. Simalikalactone D
An extract from the leaves and bark of Quassia amara L. containing the ENOX 1 inhibitor, simalikalactone D, that inhibits the activity of ENOX1 in mammalian and plant preparations [19], when added to the NADH oxidase assay inhibited the activity represented by the 24 min ENOX period (① and ②), Figure 6(b)) but was without effect on the ENOX activity represented by the 25 min period length (A and B, Figure 6(c)).
3.7. Copper Dependence
The mammalian form of ENOX1 binds 2 moles of copper/mole of protein and copper is essential for its activity

Figure 5. Heat-inactivated yeast ENOX1 activity was phased by addition of 10 µM melatonin. The activity exhibiting a 24 min period labeled ① and ② was shifted as shown by the 24 min decomposition fits (open circles, dashed lines) (b). In contrast the activity exhibiting a 25 min period length (A and B) was not phased by melatonin addition as shown by the 25 min decomposition fits (open circles, dashed lines). (c). The two maxima observed post melatonin addition (labeled M) corresponded to neither. Accuracy measures are given in Table 1.
[6]. Alteration of the two copper sites by site-directed mutagenesis or copper removal by bathocuproine inactivated both human ENOX1 [6] and ENOX2 [7] such that little or no oscillatory activity remained. To determine if the yeast ENOX forms also required copper, to 500 µl heat-inactivated yeast were added 1 µl of trifluoroacetic acid to unfold the protein in the presence or absence of 15 µl of 10 mM bathocuproine, a copper chelator, to remove the copper. Yeast ENOX activity was diminished when bathocuproine was present (Figure 7). TFA alone did not reduce activity and activity could be restored to
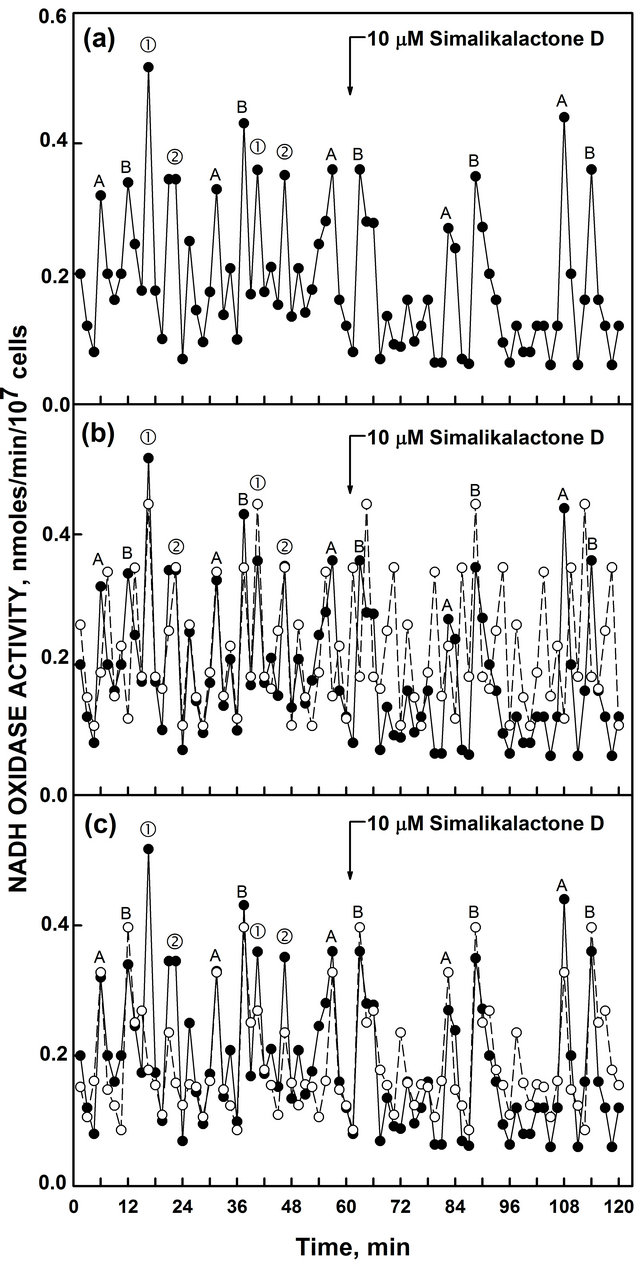
Figure 6. Inhibition of ENOX activity of heat-inactivated yeast inhibited by simalikalactone D. (a) The ENOX activity (① and ② with a period length of 24 min was inhibited by simalikalactone D as shown by the 24 min decomposition fits (open circles, dashed lines); (b) The second activity (A and B with a period length of 25 min period) was not; (c) Accuracy measures are given in Table 1.
TFA and bathocuproine treated preparations by addition of copper. In the presence of TFA, both ENOX1 activities were present (Figure 7(a)). In the presence of bathocuproine (Figure 7(b)), both ENOX1 activities were reduced or eliminated. All that remained were two maxima separated by 40 min which may correspond to the fundamental respiratory oscillation of 40 min normally seen only with continuous cultures [20,21].
3.8. Temperature Independence of Period Length
The period lengths of the two ENOX forms in yeast were
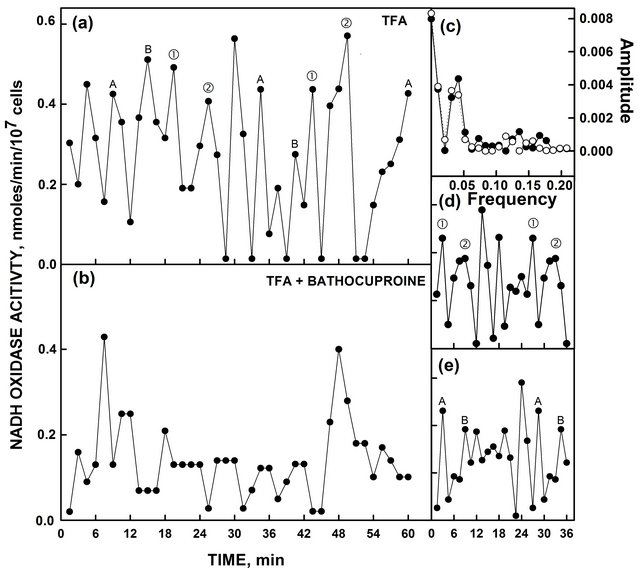
Figure 7. Heat-inactivated yeast ENOX activity with TFA ± bathocuproine. (a) In the presence of TFA, the activity with both the period length of 24 min (① and ②) as determined from the corresponding Fourier analysis and decomposition fits and the activity with the 25 min (A and B) periodiodic activities were unaffected; (b) When assayed with TFA and bathocuproine, the 24 min and 25 min periods were greatly reduced or absent. Two maxima separated by 40 min remained, however. (c) Fourier analysis comparing the maxima labeled (① and ②) in part A (solid symbols and lines) and those labeled A and B in part A (open symbols and dashed lines) yielded a period length of 24 min. (d) Decomposition fit for the maxima labeled (① and ②); (e) Decomposition fit for the data labeled A and B. Accuracy measures are given in Table 1.
temperature independent (Table 2), a hallmark of the biological clock and a characteristic of mammalian and plant ENOX1. The period length of the oscillations did not change over the range of 17˚C to 37˚C. In contrast, as is typical of both biochemical and chemical reactions, the amplitude doubled with a 10˚C increase in temperature.
3.9. Protease Resistance
A hallmark of the ENOX family of proteins is that of protease resistance. When the yeast ENOX forms were assayed after digestion with proteinase K or trypsin, the activity was unaffected even when unfolded in the presence of trifluoroacetic acid prior to addition of protease (Table 3).
3.10. Period Length in Deuterium Oxide
Organisms grown in heavy water (deuterium oxide or D2O) exhibit a circadian day lengthened in period length by about 25% [23,24]. When mammalian ENOX was assayed in heavy water, it exhibited a period length of about 30 min [25]. Heat-inactivated yeast assayed in heavy water yielded period lengths of approximately 30
Table 2. ENOX1 period length of Saccharomyces cerevisiae is independent of temperature (n = 4).
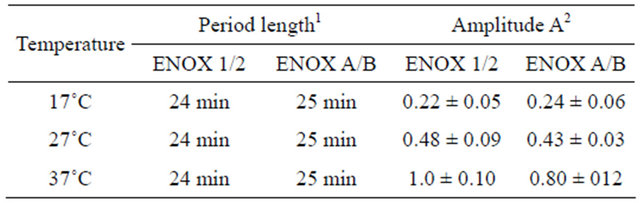
1Period length monitored over time intervals as long as 6 h varied with a cumulative error of less than one min; 2Nanomoles/min/106 cells.
Table 3. Protease resistance of yeast ENOX forms of heatinactivated Saccharomyces cerevisiae (n = 4).
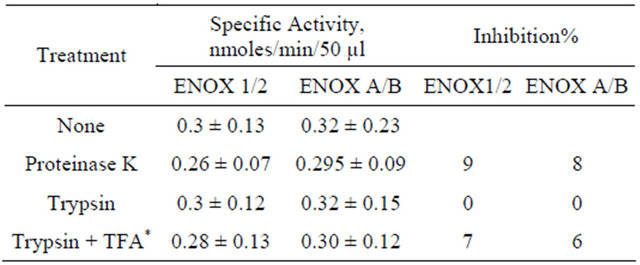
*Trifluoroacetic acid.
min for the maxima labeled ① and ② and of approximately 32 min (31.25 min) for the maxima labeled A and B (Figure 8).
4. DISCUSSION
Mammalian ENOX1 exhibits several defining characteristics to indicate that yeast contain not one but two ENOX forms similar to the mammalian ENOX1. The ENOX activities of yeast are at the external surface of the plasma membrane so that activity may be measured with intact cells using external NADH, a membrane impermeant substrate. The activities are protease and heat (80˚C) resistant in keeping with data from mammalian sources [26-28]. In mammalian and plant preparations of ENOX1, 5 maxima are seen, in which the first two dominant maxima are separated by 6 min from each other, and the remaining 3 subordinate maxima are separated from each other and from the first two maxima by 4.5 min [1]. Because of the presence of two ENOX1-like activities, one with a 24 min period length and one with a 25 min period length, only the first two dominant maxima of each of the two ENOX cycles are clearly resolved in the experiments with wild type yeast. However, the existence of the subordinate intervening maxima is indicated in the decomposition fits averaged from the primary data (Figures 1(e) and (f); Figures 7(d) and (e)).
Analysis of the statistical validity of asymmetric, nonsinusoidal oscillatory patterns such as those exhibited by ENOX proteins is limited primarily to Fast Fourier and decomposition (seasonal forecasting) types of analyses [16]. When applied to the data presented herein, the data

Figure 8. ENOX activity of heat-inactivated yeast assayed in D2O. (a) Heat-inactivated yeast exhibited two increased period lengths as is characteristic of ENOX1 proteins when assayed in the presence of heavy water. ① and ② exhibited a period of ca. 30 min as compared to 24 min in H2O and A and B exhibited a period of ca. 32 min as compared to 25 min in H2O; (b) Decomposition fits (open circles, dashed lines) using a 30 min period length from Fourier analysis; (c) Decomposition fits (open circles, dashed lines) using a 32 min period length; (d) Fourier analysis comparing the maxima labeled (① and ②) (solid symbols and lines) and the maxima labeled A and B (open symbols, dashed lines); (e) Decomposition fit for B; (f) Decomposition fit for C. Accuracy measures are given in Table 1.
were resolved into two recurrent patterns, one with a period length of 24 min and one with a period length of 25 min. Three statistical measures of reproducibility MAPE (mean absolute percentage error), MAD (mean average deviation) and MSD (mean standard deviation) comparing successive periods from the same data sets revealed a mean standard deviation of 5% or less for each of the several data sets amenable to this type of analysis.
Both of the two patterns of ENOX activity, one with a period length of approximately 24 min and one with a period length of approximately 25 min share features in common with mammalian ENOX1. In addition to NADH oxidation, both activities cleave the dithiodipyridine (DTDP) substrate indicative of protein disulfidethiol exchange and oxidize hydroquinone indicating that both proteins share the dual functionality of other ECTONOX family members. These alternating activities are unprecedented in the biochemical literature and, thus far, are unique to the ENOX family of proteins [1,15].
More compelling evidence came from measurements of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity as an indicator of circadian day length [17]. GAPDH activity cycles with an activity maximum every 24 h in mammalian cells and is controlled by components of the circadian clock. Previous work has shown that the ENOX1 period in minutes equals the circadian day length in h [13]. When GAD(P)H activity of a yeast culture was measured over time, maxima indicating circadian day lengths of 24 h, 25 h and 40 h were revealed corresponding to the 24 min, 25 min and 40 min period lengths of yeast ultradian oscillators. This indicates that yeast contain at least three separate clock rhythms, each regulating GAPDH activity independently of the other. In fact, Saccharomyces cerevisiae has three genes encoding GAPDH proteins, TDH1, TDH2 and TDH3 [29]. TDH2 and TDH3 appear to have redundant activities but TDH1 is active in stationary phase and in response to stress [30]. Which of these several GADPH proteins are clock regulated has not been determined.
Despite important similarities, the two yeast ENOX activities are not identical. Upon addition of melatonin, mammalian ENOX1 exhibits a new maximum exactly 24 min after melatonin addition as in characteristic of ENOX1 of other organisms [18]. In yeast, the activity with the 24 min period length but not that with the 25 min period was phased by melatonin. Following melatonin addition neither the maxima with a 24 min period (Figure 5(b)) nor the maxima with a 25 min period (Figure 5(c)) aligned with the new maxima labeled M that occurred exactly 24 min post melatonin addition. The maxima labeled M based on previous experience with melatonin phasing of mammalian ENOX1, represent the previous maxima of the 24 min period now missing from the post melatonin data as evidenced from the decomposition fits. Yeast are not known to synthesize melatonin. However, the dose response to melatonin was similar to that of other plant and vertebrate ENOX1 proteins such that a response of the ENOX1 of yeast to melatonin was to the melatonin of the preparation and not to a contaminant.
The 24 h rhythm in GAPDH activity corresponded to the 24 min ENOX1 activity as observed in other organisms, and accounted for one of the two ENOX activities demonstrated here for heat-inactivated yeast. It was blocked by simalikalactone D and phased by melatonin whereas the activity with the 25 min period length was not. Thus we suggest that the 25 h rhythm corresponds to a second ENOX activity, which in addition to a slightly longer (25 min) period length, responded differently from ENOX1 to ENOX modulators.
In contrast to melatonin and simalikalactone D which affected only the ENOX1 with the 24 min period, the periods of both ENOX forms of S. cerevisiae were lengthened by assay in D2O. The latter underscores the vital role of water associated with the ENOX1 proteinbound copper hexahydrate in the oscillatory process [31].
Among the first defined clock-related oscillation in yeast, known as the respiratory oscillation, was a 40 min cycle of dissolved oxygen levels in continuous culture [20-22,32-35]. After establishing that the oscillations were driven by some aspect of ethanol metabolism [33], acetaldehyde and sulfide were found to be responsible for synchronizing the cell population in continuous culture [22]. Also, the period length of this oscillation was temperature independent, indicating a clock function [22]. In addition, a mutant in which the GTS1 gene, which has homology to period genes in other organisms [36] was deleted, produced a shortened period, about 20 min, with a destabilization in the amplitude of the oscillation every 11 h [34].
Maxima separated by 40 min and not assignable to either the ENOX form with the 24 min period or the ENOX form with the 25 min period were normally not observed. Only in Figure 7 following bathocuproine inhibition of both ENOX activities was the presence of maxima corresponding to the 40 min metabolic rhythm [20-22] observed.
It is interesting that in yeast, ENOX1 inhibitors do not inhibit growth, as they do in mammalian and plant cells [19]. This attribute is probably due to the difference in the mechanism of enlargement. Plant and mammalian cells enlarge by physical membrane displacement [37-39] while yeast enlarge by tip growth, or budding [40,41]. In mammalian cells, the cell enlargement function of ENOX1 is central to its activity and essential for cell viability. In yeast, the enlargement function is no longer a primary function, so the other activities of ENOX1 may be studied separated from cell growth. As ENOX1 activity was blocked without affecting growth, deletion mutants to identify the genes encoding the two ENOX forms of S. cerevisiae provide a route to ENOX identification independent of protein purification and peptide sequencing [Dick et al. in preparation]. In continuing studies, mutant yeast strains exhibiting only the ENOX activity with the 24 min period and lacking the ENOX activity with the 25 min period have been investigated along with mutant yeast strains with the 25 min period and lacking the 24 min period. The ENOX1 form with the 24 min period has been identified as YML117W and the ENOX form with the 25 min period has been identified as YDR005C. These findings will be the subject of subsequent publications.
REFERENCES
- Morré, D.J. and Morré, D.M. (2003) Cell surface NADH oxidases (ECTO-NOX proteins) with roles in cancer, cellular time-keeping, growth, aging and neurodegenerative disease. Free Radical Research, 37, 9795-808. doi:10.1080/1071576031000083107
- Morré, D.J. (1995) NADH oxidase activity of HeLa plasma membranes inhibited by the anticancer sulfonylurea N-(4-methylphenylsulfonyl)-N-(4chlorophenyl)urea (LY181984) at an external site. Biochimica et Biophysica Acta, 1240, 201-208. doi:10.1016/0005-2736(95)00199-9
- Lambeth, J.D., Cheng, G., Arnold, R.S. and Edens, W.A. (2000) Novel homologs of gp91phox. Trends in Biological Sciences, 25, 459-461. doi:10.1016/S0968-0004(00)01658-3
- Morré, D.J., Guo, F. and Morré, D.M. (2003) An aging-related cell surface NADH oxidase (arNOX) generates superoxide and is inhibited by coenzyme Q. Molecular and Cellular Biochemistry, 254, 101-109. doi:10.1023/A:1027301405614
- Chueh, P.J., Kim, C., Cho, N., Morré, D.M. and Morré, D.J. (2002) Molecular cloning and characterization of a tumor-associated, growth-related and time-keeping hydroquinone (NADH) oxidase (NOX) of the HeLa cell surface. Biochemistry, 41, 3732-3741. doi:10.1021/bi012041t
- Jiang, Z., Gorenstein, N.M., Morré, D.M. and Morré, D.J. (2008) Molecular cloning and characterization of a candidate human growth-related and time-keeping constitutive cell surface hydroquinone (NADH) oxidase. Biochemistry, 47, 14028-14038. doi:10.1021/bi801073p
- Tang, X., Chueh, P.J., Jiang, Z., Layman, S., Martin, B, Kim, C., et al. (2010) Essential role of copper in the activity and regular periodicity of a recombinant, tumorassociated, cell surface, growth-related and time-keeping hydroquinone (NADH) oxidase with protein disulfidethiol interchange activity (ENOX2). Journal of Bioenergetics and Biomembrances, 42, 355-360. doi:10.1007/s10863-010-9305-8
- Brightman, A.O., Wang, J., Miu, R.K., Sun, I.L., Barr, R., Crane, F.L., et al. (1992) A growth factorand hormonestimulated NADH oxidase from rat liver plasma membrane. Biochimica et Biophysica Acta, 1105, 109-117. doi:10.1016/0005-2736(92)90168-L
- Kelker, M., Kim, C., Chueh, P.J., Guimont, R., Morré, D.M. and Morré, D.J. (2001) Cancer isoform of a tumorassociated cell surface NADH oxidase (tNOX) has properties of a prion. Biochemistry, 40, 7351-7354. doi:10.1021/bi010596i
- Kishi, T., Morré, D.M. and Morré, D.J. (1999) The plasma membrane NADH oxidase of HeLa cells has hydroquinone oxidase activity. Biochimica et Biophysica Acta, 1412, 66-77. doi:10.1016/S0005-2728(99)00049-3
- Bridge, A., Barr, R. and Morré, D.J. (2000) The plasma membrane NADH oxidase of soybean has vitamin K1 hydroquinone oxidase activity. Biochimica et Biophysica Acta, 1463, 448-458. doi:10.1016/S0005-2736(99)00239-4
- Morré, D.J., Chueh, P.J., Lawler, J. and Morré, D.M. (1998) The sulfonylureas-inhibited NADH oxidase activity of HeLa plasma membranes has properties of a protein disulfide-thiol oxido-reductase with protein disulfidethiol interchange activity. Journal of Bioenergetics and Biomembrances, 30, 477-487. doi:10.1023/A:1020594214379
- Morré, D.J., Chueh, P.J., Pletcher, J., Tang, X., Wu, L.Y. and Morré, D.M. (2002) Biochemical basis for the biological clock. Biochemistry, 41, 11941-11945. doi:10.1021/bi020392h
- Kim, C. and Morré, D.J. (2004) Prion proteins and ECTO-NOX proteins exhibit similar oscillating redox activities. Biochemical and Biophysical Research Communications, 315, 1140-1146. doi:10.1016/j.bbrc.2004.02.007
- Morré, D.J. (1998) NADH oxidase: A multifunctional ectoprotein of the eukaryotic cell surface. In: Asard, H., Bérczi, A. and Caubergs, R., Eds., Plasma Membrane Redox Systems and Their Role in Biological Stress and Disease, Kluwer Academic Publishers, Dordrecht, 121- 156.
- Foster, K., Anward, N., Pogue, R., Morré, D.M., Keenan, T.W. and Morré, D.J. (2003) Decomposition analyses applied to a complex ultradian biorhythm: The oscillating NADH oxidase activity of plasma membranes having a potential time-keeping (clock) function. Nonlinearity in Biology, Toxicology and Medicine, 1, 51-70. doi:10.1080/15401420390844465
- Shinohara, M.L., Loros, J.J. and Dunlap, J.C. (1998) Glyceraldhyde-3-phosphate is regulated on a daily basis by the circadian clock. Journal of Biological Chemistry, 273, 446-452. doi:10.1074/jbc.273.1.446
- Morré, D.J. and Morré, D.M. (2003) The plasma membrane-associated NADH oxidase (ECTO-NOX) of mouse skin responds to blue light. Journal of Photochemistry and Photobiology B, 70, 7-12. doi:10.1016/S1011-1344(03)00023-X
- Morré, D.J. and Greico, P.A. (1999) Glaucarubolone and simalikalactone D, respectively, preferentially inhibit auxin-induced and constitutive components of plant cell enlargement and the plasma membrane NADH oxidase. International Journal of Plant Science, 160, 291-297. doi:10.1086/314133
- Lloyd, D., Eshantha, L., Salgado, J., Turner, M.P. and Murray, D.B. (2002) Respiratory oscillations in yeast: Clock-driven mitochondrial cycles of energization. FEBS Letters, 519, 41-44. doi:10.1016/S0014-5793(02)02704-7
- Murray, D.B., Roller, S., Kuriyama, H. and Lloyd, D. (2001) Clock control of ultradian respiratory oscillation found during yeast continuous culture. Journal of Bacteriology, 183, 7253-7259. doi:10.1128/JB.183.24.7253-7259.2001
- Murray, D.B., Klevecz, R.R. and Lloyd, D. (2003) Generation and maintenance of synchrony in Saccharomyces cerevisiae continuous culture. Experimental Cell Research, 287, 10-15. doi:10.1016/S0014-4827(03)00068-5
- Enright, J.T. (1997) Heavy water slows biological timing processes. Zeitschrift für vergleichende Physiologie, 72, 1-16. doi:10.1007/BF00299200
- Bruce, V.G. and Pittendrigh, C.S. (1960) Temperature independence in unicellular “clock”. Journal of Cellular and Comparative Physiology, 56, 25-31. doi:10.1002/jcp.1030560105
- Kim, C., Layman, S., Morré, D.M. and Morré, D.J. (2006) Structural changes revealed by Fourier transform infrared and circular dichroism spectroscopic analyses underlie tNOX periodic oscillations. Dose Response, 3, 391-413. doi:10.2203/dose-response.003.03.008
- Chueh, P.J., Morré, D.J., Wilkinson, F.E., Gibson, J. and Morré, D.M. (1997) A 33.5 kDa heat-and protease resistant NADH oxidase inhibited by capsaicin from sera of cancer patients. Archives of Biochemistry and Biophysics, 342, 38-47. doi:10.1006/abbi.1997.9992
- del Castillo-Olivares, A., Yantiri, F., Chueh, P.J., Wang, S., Sweeting, M., Sedlak, D., et al. (1998) A drug-responsive and protease-resistant peripheral NADH oxidase complex from the surface of HeLa S cells. Archives of Biochemistry and Biophysics, 358, 125-140. doi:10.1006/abbi.1998.0823
- Sedlak, D., Morré, D.M. and Morré, D.J. (2001) A drugunresponsive and protease-resistant CNOX protein from human sera. Archives of Biochemistry and Biophysics, 386, 106-116. doi:10.1006/abbi.2000.2180
- McAlister, L. and Holland, M.J. (1985) Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. Journal of Biological Chemistry, 260, 15019-15027.
- Boucherie, H., Bataille, N., Fitch, I.T., Perrot, M. and Tuite, M.F. (1995) Differential synthesis of glyceraldehyde-3-phosphate dehydrogenase polypeptides in stressed yeast cells. FEMS Microbiology Letters, 125, 127-133. doi:10.1111/j.1574-6968.1995.tb07348.x
- Morré, D.J., Orczyk, J., Hignite, H. and Kim, C. (2008) Regular oscillatory behavior of aqueous solutions of CuII salts related to effects on equilibrium dynamics of ortho/para hydrogen spin isomers of water. Journal of Inorganic Biochemistry, 102, 260-267. doi:10.1016/j.jinorgbio.2007.08.008
- Satroutdinov, A.D., Kuriyama, H. and Kobayashi, H. (1992) Oscillatory metabolism of Saccharomyces cerevisiae in continuous culture. FEMS Microbiology Letters, 98, 261-268. doi:10.1111/j.1574-6968.1992.tb05525.x
- Keulers, M., Suzuki, T., Satroutinov, A.D. and Duriyama, H. (1996) Autonomous metabolic oscillation in continuous culture of Saccharomyces cerevisiae grown on ethanol. FEMS Microbiology Letters, 142, 253-258. doi:10.1111/j.1574-6968.1996.tb08439.x
- Adams, C.A., Kuriyama, H., Lloyd, D. and Murray, D.B. (2003) The Gts1 protein stabilizes the autonomous oscillator in yeast. Yeast, 20, 463-470. doi:10.1002/yea.976
- Klevecz, R.R., Bolen, J., Forrest, G. and Murray, D.B. (2004) A genome wide oscillation in transcription gates DNA replication and cell cycle. Proceedings of the National Academy of Sciences USA, 101, 1200-1205. doi:10.1073/pnas.0306490101
- Mitsui, K., Taguchi, S. and Tsurugi, K. (1994) The GTS1gene, which contains a Gly-Thre repeat, affects the timing of budding and cell size of the yeast Saccharomyces cerevisiae. Molecular and Cellular Biology, 14, 5569-5578.
- Morré, D.J., Kim, C. and Hicks-Berger, C. (2006) ATPdependent and drug inhibited vesicle enlargement reconstituted using synthetic lipids and recombinant proteins. BioFactors, 28, 105-117. doi:10.1002/biof.5520280205
- Hicks-Berger, C. and Morré, D.J. (2006) Inside-out but not right side-out plasma membrane vesicles from soybean enlarge when treated with ATP + 2,4-D as determined by electron microscopy and light scattering: evidence for involvement of a plasma membrane AAAATPase. BioFactors, 28, 91-104. doi:10.1002/biof.5520280204
- Auderset, G. and Morré, D.J. (2006) ATPand growth substance-dependent cell-free enlargement of plasma membrane vesicles from soybean. BioFactors, 28, 83-90. doi:10.1002/biof.5520280203
- Adams, A.E. and Pringle, J.R. (1984) Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. Journal of Cellular Biology, 98, 934-945. doi:10.1083/jcb.98.3.934
- Cabib, E., Bowers, B., Sburlati, A. and Silverman, S.J. (1988) Fungal cell wall synthesis: The construction of a biological structure. Microbiology Sciences, 5, 370-375.
NOTES
*Present Address: Valparaiso University, Department of Biology, NSC- 105A, Valparaiso, IN.
#Present Address: Mor-NuCo, LLC, Purdue Research Park, 1291C Cumberland Avenue, West Lafayette, IN 47906.

