Paper Menu >>
Journal Menu >>
 J. Software Engineering & Applications, 2009, 2: 150-159 doi:10.4236/jsea.2009.23022 Published Online October 2009 (http://www.SciRP.org/journal/jsea) Copyright © 2009 SciRes JSEA Data Mining in Biomedicine: Current Applications and Further Directions for Research S. L. TING1, C. C. SHUM2, S. K. KWOK1, A. H. C. TSANG1, W. B. LEE1 1Department of Industrial and Systems Engineering, The Hong Kong Polytechnic University, Hong Kong, China; 2Department of Computing, The Hong Kong Polytechnic University, Hong Kong, China. Email: jacky.ting@polyu.edu.hk Received January 16th, 2009; revised June 18th, 2009; accepted June 24th, 2009. ABSTRACT Data mining is the process of finding the patterns, associations or relationships among data using different analytical techniques involving the creation of a model and the concluded result will become useful information or knowledge. The advancement of the new medical deceives and the database management systems create a huge number of data- bases in the biomedicine world. Establishing a methodology for knowledge discovery and management of the large amounts of heterogeneous data has become a major priority of research. This paper introduces some basic data mining techniques, unsupervised learning and supervising learning, and reviews the application of data mining in biomedicine. Applications of the multimedia mining, including text, image, video and web mining are discussed. The key issues faced by the computing professional, medical doctors and clinicians are highlighted. We also state some foreseeable future developments in the field. Although extracting useful information from raw biomedical data is a challenging task, data mining is still a good area of scientific study and remains a promising and rich field for research. Keywords: Data Mining, Biomedicine 1. Introduction With the tremendous improvement in the speed of com- puter and the decreasing cost of data storage, huge vol- umes of data are created. However, data itself has no value. Only if data can be changed to information, it be- comes useful. In order to generate meaningful informa- tion, or knowledge from database, the field of data min- ing was born. The data mining field is about two decade old. Early pioneers such as U. Fayyad, H. Mannila, G. Piatetsky-Shapiro, G. Djorgovski, W. Frawley, P. Smith, and others found that the traditional statistical techn iques were not adequate to handle the mass amount of data. They recognized the need of better, faster and cheaper ways to deal with the dramatic increase in the amount of data. Nowadays, besides the numerous number of databases created and accumulated in a dramatic speed, data is no longer restricted to numeric or character only especially in the biomedicine aspect. The advanced medical de- ceives and database management systems enable the in- tegration of the different types of high dimensional mul- timedia data (e.g. text, image, audio, and video) under the same umbrella. Establishing a methodology for kno- wledge discovery and management of large amounts of heterogeneous data has therefore become a main priority. Various techniques are used in different areas of bio- medicine, including genomics, proteomics, medical di- agnosis, effective drug design and pharmaceutical indus- try. In this paper, we would first give a brief outline on what is data mining, its position or role in the kn owledge discovery process and the basic principles of some com- monly used data mining techniques. Next, we present our investigation results of the applications of the data min- ing in the biomedicine aspect, which includes the area of biology, medicine, pharmacy and health care. Lastly, we discuss some difficulties of data mining in biomedicine and the possible direction for the future development. 2. What is Data Mining? Data mining (DM) is the process of finding the patterns, associations or relationships among data using different analytical techniques involving the creation of a model and the concluded result will become useful information or knowledge. DM can also be expressed as Nontrivial extraction of implicit, previously un- known, and potentially useful information from data [1]; and Making sense of large amounts of mostly unsuper- vised data in some domain [2] 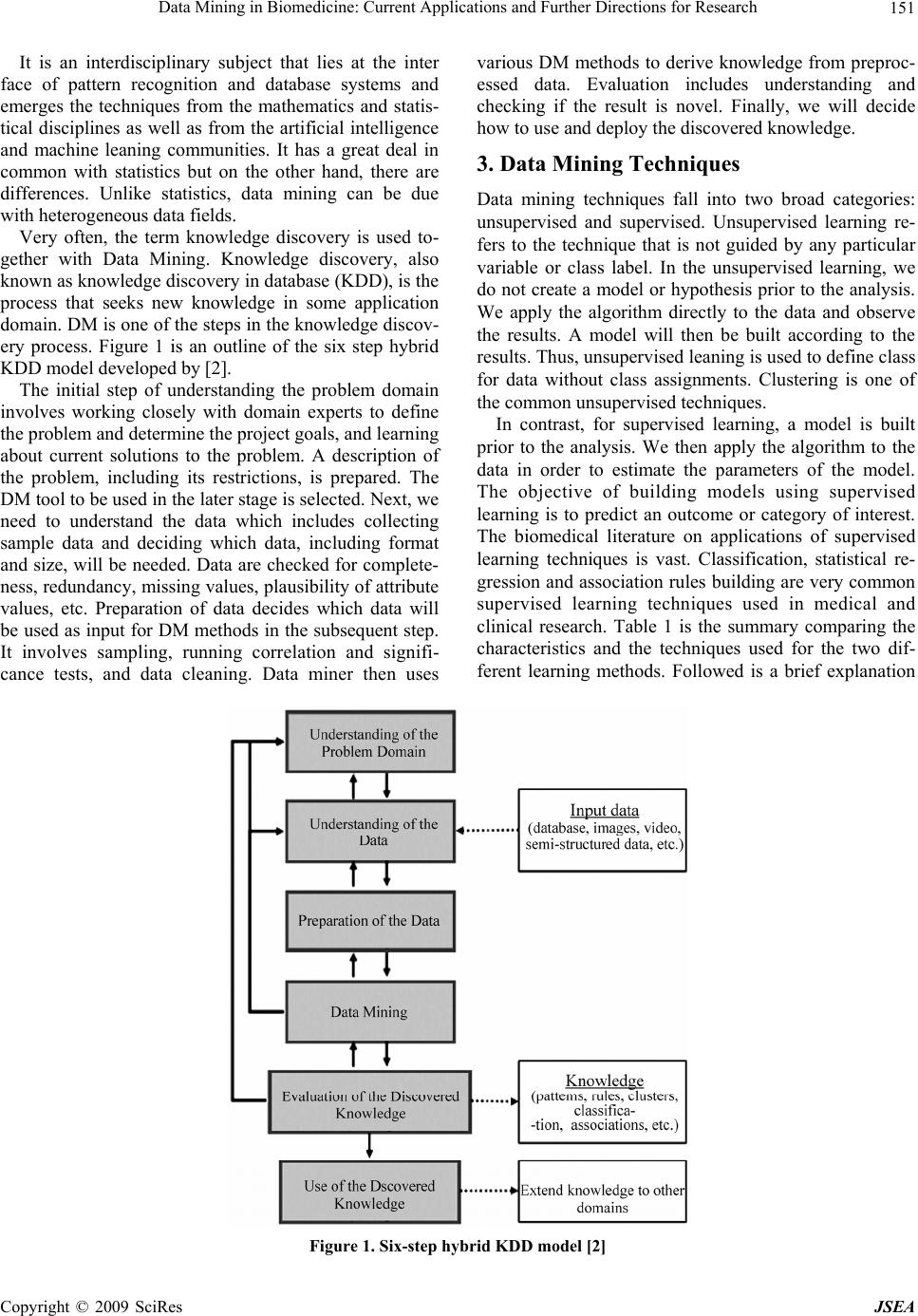 Data Mining in Biomedicine: Current Applications and Further Directions for Research151 It is an interdisciplinary subject that lies at the inter face of pattern recognition and database systems and emerges the techniques from the mathematics and statis- tical disciplines as well as from the artificial intelligence and machine leaning communities. It has a great deal in common with statistics but on the other hand, there are differences. Unlike statistics, data mining can be due with heteroge n eou s dat a field s . Very often, the term knowledge discovery is used to- gether with Data Mining. Knowledge discovery, also known as knowledge d iscover y in databa se (KDD), is the process that seeks new knowledge in some application domain. DM is one of the steps in the knowledge discov- ery process. Figure 1 is an outline of the six step hybrid KDD model developed by [2]. The initial step of understanding the problem domain involves working closely with domain experts to define the problem and determine the project goals, and learning about current solutions to the problem. A description of the problem, including its restrictions, is prepared. The DM tool to be used in the later stage is selected. Next, we need to understand the data which includes collecting sample data and deciding which data, including format and size, will be needed. Data are checked for complete- ness, redundancy, missing valu es, plausibility of attribute values, etc. Preparation of data decides which data will be used as input for DM methods in the subsequent step. It involves sampling, running correlation and signifi- cance tests, and data cleaning. Data miner then uses various DM methods to derive knowledge from preproc- essed data. Evaluation includes understanding and checking if the result is novel. Finally, we will decide how to use and deploy the discovered knowledge. 3. Data Mining Techniques Data mining techniques fall into two broad categories: unsupervised and supervised. Unsupervised learning re- fers to the technique that is not guided by any particular variable or class label. In the unsupervised learning, we do not create a model or hypothesis prior to the analysis. We apply the algorithm directly to the data and observe the results. A model will then be built according to the results. Thus, unsupervised leaning is used to define class for data without class assignments. Clustering is one of the common unsupervised techniques. In contrast, for supervised learning, a model is built prior to the analysis. We then apply the algorithm to the data in order to estimate the parameters of the model. The objective of building models using supervised learning is to predict an outcome or category of interest. The biomedical literature on applications of supervised learning techniques is vast. Classification, statistical re- gression and association rules building are very common supervised learning techniques used in medical and clinical research. Table 1 is the summary comparing the characteristics and the techniques used for the two dif- ferent learning methods. Followed is a brief explanation Figure 1. Six-step hybrid KDD model [2] Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research 152 Table 1. Comparing the characteristics and the techniques of the unsupervised and supervise d le ar ning Characteristics Techniques Unsupervised Learning No guidance Use to Define the class Seldom utilized (until recently) Clustering Association Rule Supervised Learning With guidelines Class defined Common with vast literature and application Classification Statistical Regression Artificial neural networks of the four learning techniques. 3.1 Clustering Clustering is an unsupervised learning technique reveal- ing natural groupings in the data. Cluster analysis refers to the grouping of a set of data objects into clusters. A cluster is a collection of data objects wh ich are similar to one another within the same cluster but not si milar to the objects in another cluster. Clustering is also called unsu- pervised classification where no predefined classes are assigned. 3.2 Association Rule Association rule discovery is to find the relationships between the different items in a data base. It is normally express in the form X => Y, where X and Y are sets of attributes of the dataset which implies that transactions that contain X also contain Y. 3.3 Classification Classification is a supervised learning method. It is a method of categorizing or assigning class labels to a pat- tern set under the supervision. The object of classifica- tion is to develop a model for each class. Classification methods can usually be categorized as follows: a) Decision tree Decision tree classifiers divide a decision space into piecewise constant regions. It splits a dataset on the basis of discrete decisions, using certain thresholds on the at- tribute values. It is one of the most widely used classifi- cation method as it is easy to interpret and can be repre- sented under the If-then-else rule condition. b) Nearest-neighbor Nearest-neighbor classifiers [3] typically define the proximity between instances, find the neighbors if a new instance, and then assign to it the label for the majority class of its neighbors. c) Probabilistic models Probabilistic models are models which calculate prob- abilities for hypotheses base on Bayes’ theorem [3]. 3.4 Statistical Regression Regression models are very popular in the biomedical literature and have been applied in virtually every sub- specialty of medical research. Before computers were widely used, linear regression was the most popular model to find solutions of the problem of estimating the intercept and coefficients of the regression question. It has solid foundation from the statistical theory. Linear regression is similar to the task of finding the line that minimizes the total distance to a set of data. That is find the equation for line Y = a + bX. With the help of com- puters and software package, we can calculate the high complex models. 3.5 Artificial Neural Networks Artificial neural networks [4] are signal processing sys- tems that try to emulate the behavior of human brain by providing a mathematical model of combination of nu- merous neurons conn ected in a network. It learns through examples and discriminate the characteristics among various pattern classes by reducing the error and auto- matically discovering inherent relationships in a data-rich environment. No rules or programmed information is need beforehand. It composes of many elements, called nodes which are connected in between. The connection between two nodes is weighted and by the adjustment of these weights, the training of the network is performed. The weights are network parameters and their values are obtained after the training procedure. There are usually several layers of nodes. During the training procedure, the inputs are directed in the input layer with the desir- able output values as targets. A comparison mechanism will operates between the out and th e target valu e and the weights are adjusted in order to reduce error. The proce- dure is repeated until the network output matches the targets. There are many advantages of neural networks like adaptive learning ability, self-o rganization, real-time operation and insensitivity to noise. However, it also has a huge disadvantage that it is highly dependence on the training data and it does not provide an explanation for the decisions they make, just like working in the ‘black box’. 3.6 Advanced Data Mining Techniques During the past few years, researchers have tried to com- bine both unsupervised and supervised methods for the Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research153 analysis [5]. Some examples of advanced unsupervised learning models are hierarchical clustering, c-means clustering self-organizing maps (SOM) and multidimen- sional scaling techniques. Advanced examples of the supervised learning models classification and regression trees (CART) and support vector machines [6]. 4. Applications of Data Mining in Biomedicine 4.1 Data Mining Models Data mining applies in descriptive modeling for under- standing. In [7], Tseng and Yang use Gene Ontology (GO) to group genes in advance in order to show the po- tential relations among gene groups and discover the hidden relations between genes set in association with GO terms. It can also be used to predict the outco me of a future observation or to assess the potential risk in a dis- ease situation. Regarding the predictive power, data mining algorithms can learn from past examples in clini- cal data and model the oftentimes non-linear relation- ships between the independent and dependent variables, thereby the resulting model representing the formalized knowledge that can often provide a good diagnostic op- tion [8]. Data mining techniques have been widely used to find new patterns and knowledge from biomedical data. 4.2 Recent Development The typical data mining process involves transferring data originally collected in production systems (such as electronic medical records) into data warehouse, cleaning or scrubbing the data to remove errors and check for format consistency, and then searching the data using statistical model, artificial intelligence (such as neural networks), and other machine learning methods [9]. In [10], Prather et al. employs the KDD for identifying the factors that will improve the quality and cost effective- ness of perinatal care in an ex tensive clinical database of obstetrical patients. Given the data warehouse of diabetic patients, Breault et al. employ the CART to investigate the factors affecting the occurrence of diabetics [11]. They are surprisingly discovered that younger age pre- dicts bad diabetic control, in which explore a new area to manage the diabetic control in younger age. Similar ap- plicati ons of data m ining can al s o be f ound in Ta ble 2. Apart from the diagnostic prediction, the knowledge discovery ability in data mining also demonstrated a good detector in adverse drug events (ADE). In [12], Wilson et al. utilize the KDD techniques in pharma- covigilance for detecting signals earlier than using exist- ing methods. In [13], Lian et al. has pointed out that the prescription is specified by a preference function based on the user's preference in prior clinical experience. Thus, they propose a dose optimization framework based on probability theory. In [14], Susan and Warren have demonstrated that the cond itional probability (CP) model is superior in optimizing the drug lists over the multiple linear regression and discriminant analysis models. Con- cerning the strong relationship between the diagnosis and medication, it formulates a posterior probability (what medication is needed) b ased on a priori probability (wh at diagnosis has been made). This approach aligns with the Mediface as purposed by [15]. Table 2. Recent applications of data mining Author Description Megalooikonomou et al. [20] They introduce statistical methods that aid the discovery of interesting associations and patterns between brain images and other clinical data Brossette et al. [21] They design a Data Mining Surveillance System (DMSS) that uses novel data mining techniques to discover unsuspected, useful patterns of nosocomial infections and antimicrobial resistance from the analysis of hospital laboratory data Antonie et al. [22] They investigate the use of different data mining techniques for anomaly detection and classification of medi- cal images Coulter et al. [23] They examine the relation between antipsychotic drugs and myocarditis and cardiomyopathy Li et al.[24] They explore a novel analytic cancer detection method with different feature selection methods and to compare the results obtained on different datasets and that reported by Petricoin et al. in terms of detection performance and selected proteomic patterns Delen et al.[25] They use two popular data mining algorithms (artificial neural networks and decision trees) along with a most commonly used statistical method (logistic regression) to develop the prediction models on breast cancer using a large dataset. Su et al. [26] They use four different data mining approaches to select the re levant features from the data to predict diabetes Phillips-Wren et al. [27] They assess the utilization of healthcare resources by lung cancer patients related to their demographic charac- teristics, socioeconomic markers, ethnic backgrounds, medical histories, and access to healthcare resources in order to guide medical decision making and pu bl i c p ol icy Copyright © 2009 SciRes JSEA 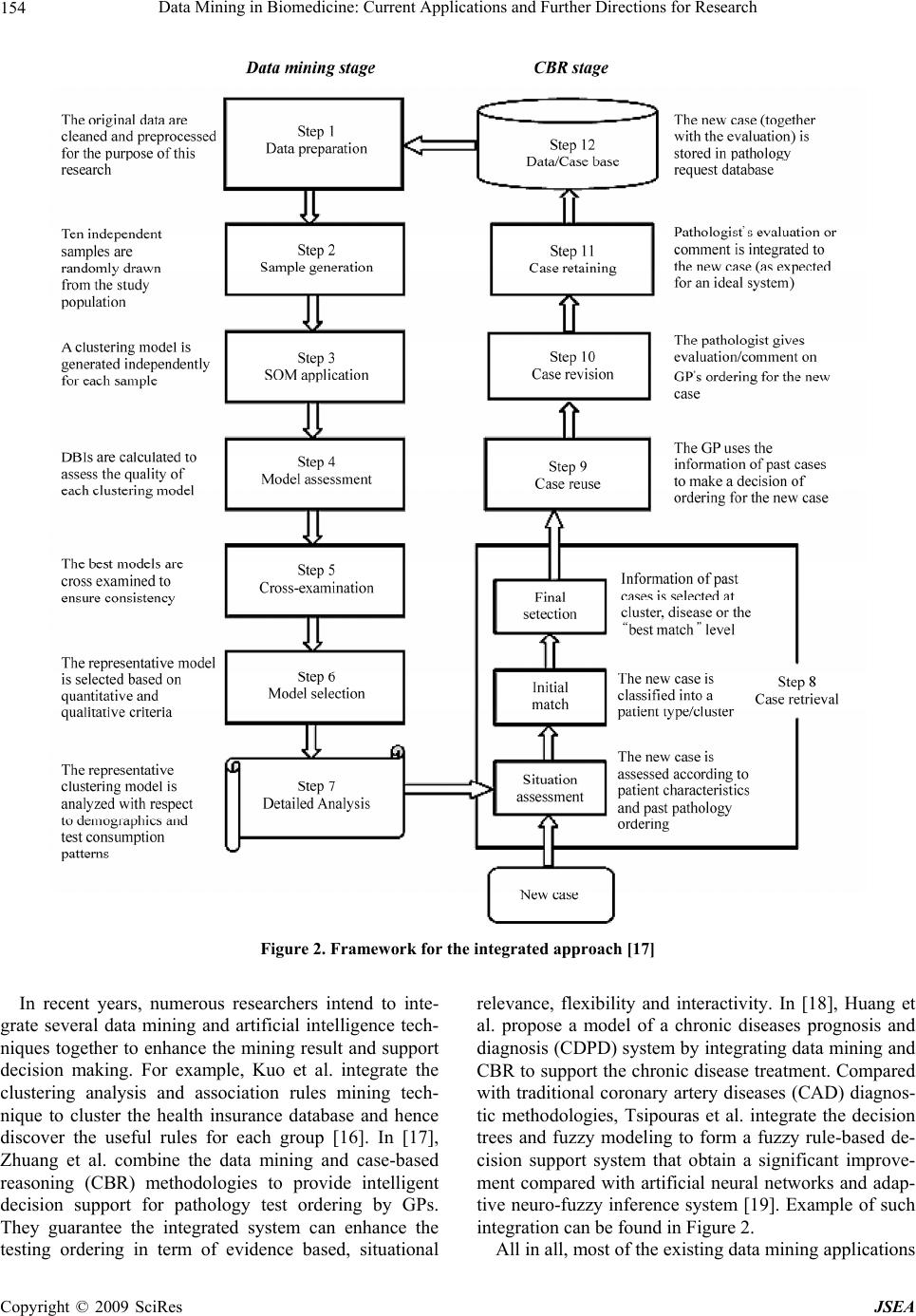 Data Mining in Biomedicine: Current Applications and Further Directions for Research 154 Figure 2. Framework for the integrated approach [17] In recent years, numerous researchers intend to inte- grate several data mining and artificial intelligence tech- niques together to enhance the mining result and support decision making. For example, Kuo et al. integrate the clustering analysis and association rules mining tech- nique to cluster the health insurance database and hence discover the useful rules for each group [16]. In [17], Zhuang et al. combine the data mining and case-based reasoning (CBR) methodologies to provide intelligent decision support for pathology test ordering by GPs. They guarantee the integrated system can enhance the testing ordering in term of evidence based, situational relevance, flexibility and interactivity. In [18], Huang et al. propose a model of a chronic diseases prognosis and diagnosis (CDPD) system by integrating data mining and CBR to support the chronic d isease treatment. Compared with traditional coronary artery diseases (CAD) diagnos- tic methodologies, Tsipouras et al. integrate the decision trees and fuzzy modeling to form a fuzzy rule-based de- cision support system that obtain a significant improve- ment compared with artificial neural networks and adap- tive neuro-fuzzy inference system [19]. Example of such integration can be found in Figure 2. All in all, most of the existing data mining app lications Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research155 are focused on exploring the pattern in sound biomedical databases. With proper structure of the data collected via different medical devices, data mining techniques can serve as a promising tool to convert the information into useful and valuable knowledge to physicians and re- searchers. 4.3 Current Trend 4.3.1 Mul timedi a Mining Classically, databases were formed by tuples of numeric and alphanumeric contents, but with the widespread use of medical information systems, information absorption are now expands to different data types including text, document, image, graphics, speech, audio, hypertext, etc. At the same time, the growth in Internet information can also be considered as a new dimension as a distributed multimedia database of the largest useful information. Concerning the tremendous amount of visual information, it is obvious that the development of data mining tech- niques in these multimedia data is the next generation in biomedicine. With the widely advanced in digital multi- media technology, numerous researchers introduce sev- eral novel data mining techniques, namely image mining, text mining, video mining, and web mining. Below we will discuss these four technology revolution and how does it impact the biomedicine area. 4.3.2 Text Mining Apart from the medical images and signals, another clinical data that physicians would like to interpret is the unstructured free-text. Regarding there is a lot of infor- mation presented in text or document databases, in form of electronic books, research articles, digital libraries, medical dictionaries, etc., several researchers developed a novel data mining approach in extracting useful knowledge from textual data or documents, so called the text mining [28,29]. For example, we can employs text mining techniques to extract the information of pro- tein-protein interaction within three different documents. In addition to the traditional data mining techniques, text mining uses techniques from many multidisciplinary scientific fields (e.g. text analysis techniques) to gain insight and automatically rev eal useful info rmation to the human users. In [30], Cohen and Hunter describe text mining is “the use of automated methods for exploiting the enormous amount of knowledge available in the biomedical literature”. One of the examples of text min- ing is to manage the health information in Internet and response the needs for those who have health information inquiry in HIV/AIDS [31]. Another common application of text mining is used to extract the information of pro- tein-protein interaction. When given the unstructured text, Zhou et al. employ the semantic parsing and hidden vec- tor state model to mine the knowledge within the text [32]. By setting the annotation PROTEIN_NAME (AC- TIVATE(PROTEIN_NAME), the system will automati- cally generate the result as shown in Figure 3. Figure 3. Semantic parsing employe d in protein documents [32] Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research 156 4.3.3 Image Mining More and more medical procedures employ imaging as a preferred diagnostic tool. Thus, there is a need to develop methods for efficient mining in images databases, which is completely different and more difficult than mining in structured datatypes. Therefore, mining of image data is a challenge problem. Meanwhile, with numerous imag- ing techniques (such as SPECT, MRI, PET, and collec- tion of ECG or EEF signals) can generate gigabytes of data per day, and heterogeneous nature of image data (like a single cardiac SPECT procedure of one patient may contain dozens of 2D images), image mining has become one of the emerging field in biomedical study. Typically, most of the activities in mining image data are based on the searching, retrieving and comparing of query image with the stored image by its degree of simi- larity or feature(s). In [22], Antonie et al. present the use of different data mining techniques for tumor classifica- tion in digital mammograph y and they find that associate rule obtains a better result than neural networks. Fur- thermore, in order to tackle the issue of complicated na- ture of surrounding of breast tissue, the variation of MCs in shape, orientation, brightness and size, Peng et al. propose knowledge-discovery incorporated genetic algo- rithm (KD-GA) to search for the bright spots in mam- mogram and hence evaluate the possibility of a bright spot being a true MC, and adaptively adjust the associ- ated fitness values [34]. Another example, which intro- duces a notion of image sequence similarity patterns (ISSP) for discovering the possible Space-Occupying Lesion (PSO) in brain images, is presented by [35]. 4.3.4 Vi deo Mining With the advancement in streaming audio and digital TV, more and more video data are stored in which this brings the interest of researchers to discover and explore inter- esting patterns in the audio-visual content. In order to meet such demand, video mining is developed. In the biomedicine area, it is observed that specialists intend to use cameras to take the video in each operation, which imply there are ample opportunities of applying data mining principles in conjunction with the video retrieval techniques. For example, Zhu et al. introduce a video database management framework and strategies for video content structure and events mining [36]. They first seg- mented the video shot into groups and hence organized the video shots into a hierarchical structure using clus- tered scenes, scenes, groups, and shots, in increasing granularity from top to bottom. With a sound structure, audio and video processing techniques are integrated to mine event information, such as dialog, presentation and clinical operation, from the detected scenes. 4.3.5 Web Mining Internet is growing at a tremendous speed. World Wide Web (WWW) becomes the largest database that ever existed. In particular, many medical literatures are writ- ten in electronic format which are widely available and accessible in the Internet nowadays. Therefore, the capa- bility of knowledge discovery and retrieving information from WWW is important to physicians. But, the com- plexity of web pages and the dynamic nature of data stored in the Internet make adoption of data mining tech- niques difficult. In [37], web mining is the use of data mining techniques to automatically retrieve, extract and evaluate information for knowledge discovery from the Internet. With its exploratory of hidden information abil- ity, Yu and Jonnalagadda present an approach regarding Semantic Web and mining that can improve the quality of Web mining results and enhance the functions and services and the interoperability of medical information systems and standards in the healthcare field [38]. 5. Discussions Biomedicine has been evolved as a new application area for data mining in recent year. As reflected by the brief literature survey in this study, the current data mining research concentrates on applying the data mining tech- niques to manage the complex and unstructured data, and in particular in form of visual and textual nature. Al- though numerous studies resulting satisfactory result of data mining adoption, it is found that data quality is one of the major challenges on impacting the performance in the biomedicine industry. In theory, data mining is a data driven approach as the outcome of data mining heavily depends on the quality and quantity of available data. However, the data in the biomedicine area is rather com- plex in nature. Thus, in order to enhance the performance of data mining adoption in the domain area, concerns are raised as follow: a) Huge volume of data Because of the sheer size of databases, it is unlikely that any of the data mining methods will succeed with raw data. In the field of biomedicine, it is particular true that particular medical experts are required to pre-process the data before adopting data mining. As different medi- cal experts are professional in different medical aspects, therefore it is time consuming and labor intensive to handle the data beforehand. b) Dynamic nature of data Databases are constantly updated and adding new in- formation at an alarming rate. For example, new SPECT images (for the same or a new patient), or by replacement of the existing ones (a SPECT had to be repeated because of technical problems). This requires methods that are able to incrementally update the knowledge learned so far. c) Incomplete or imprecise data The information collected in a database can be either incomplete or imprecise. To address this problem, fuzzy sets and rough sets were developed explicitly. Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research157 d) Noisy data It is very difficult for any data collection technique to entirely eliminate noise. This implies that data mining methods should be made less sensitive to noise, or care should be taken that the amount of noise in data to be collected in the future will be approximately the same as that in the current da ta. e) Missing attribute values Missing values create a problem for most data mining methods, since nearly all the methods require a fixed dimension for each data object. In fact, this problem is widely encountered in the medical databases because most medical data are collected as a byproduct of patient care activities, rather than for organized research proto- cols; even in some large medical databases such as breast cancer data set from University of Wisconsin Hospitals, this problem are still existed. Typically, one approach to remedy this problem is to ignore the missing values, or omit any records containing missing values; whereas another approach is to substitute missing values with mostly likely values from obtaining values in the mode or mean, or directly infer missing values from existing values via artificial intelligence method (e.g. case-based reasoning). f) Redundant, insignificant data, or inconsistent d ata The data set may contain redundant, insignificant, or inconsistent data objects and attributes. Generally, medi- cal data can be stored in numeric and textual format; in which a large amount of preprocessing is required in or- der to make the data useful. For example, misspelled of medical terms is frequently occurred and one medication or condition may be commonly referred to by a variety of names (i.e. stomach and abdominal pain). In addition to the data quality perspectives, several considerations are also been made: a) Quality of learning mechanism Over- and under-learning will affect the performance of data mining in which the learning mechanism will misunderstand the human’s preferences and require hu- man to adjust for achieving the goal state. b) Quality of knowledge representation Knowledge representation is an important element to represent knowledge in an understandable manner to facilitate the conclusions drawn from knowledge. If the machine is insufficient to store the k now ledge d iscov ered, it is also incapable to represent them; thus, such insuffi- cient knowledge will make the machine less intelligent. c) Nature o f p roblem When the problem is too complex, chaos, or has not encountered before, the intelligent machine do not have enough knowledge or time to deduce an appropriate re- sult. Using the case of diagnostic decision support as an example, if most of the learning cases and rules are re- lated to some general diagnosis, wh en there is a n ew case related to specific diagnosis encountered, the system cannot provide a good solution since there are no rules triggered inside in the system. As a result, with this study at hand, we can conclude that opportunities to use data mining truly in bio medicine will happen only when the data quality is committed to the level of standard and there are new methods or algo- rithms to handle the complex data types. Furthermore, adoption of data mining in biomedicine is quite a young field with many issues that still need to be researched and explored in depth. Some further research directions and questions are summarized as follow: a) An absurd and false model may fit perfectly if the model has enough complexity by comparison to the amount of data available. When the degrees of freedom in parameter selection exceed the information content of the data, this leads to arbitrariness in the final (fitted) model parameters which reduces or destroys the ability of the model to generalize beyond the fitting data. If you've got a learning algorithm in one hand and a dataset in the other hand, to what extent can you decide whether the learning algorithm is in danger of over-fitting or un- der-fitting? Almost all of the data mining research is done on an ad-hoc base. The techniques are designed for an individual problem. There is no unifying theory. b) The storage of large multimedia databases is often required to be in compressed form. Data compression if the techniques to reduce the redundancies in data repre- sentation. Reducing the storage requirement is equivalent to increasing the capacity of the storage medium. The development of the data compression technology will play a significant role in terms of the performance of data mining. However, it seems the data compression field has so far been neglected by the data mining community. c) In today’s network ed society, data care not stored in a single place. Internet has no doubt being the greatest and largest databases that we have ever had. Information inside the internet is often a mixed of text, image, audio, speech, hypertext, graphics and video components. In many cases, databases spread over multiple files in dif- ferent disks or in different geographical locations. How to handle or collaborate all kind of heterogeneous data in a distributed environment will open up a newer area of development. d) More and more multimedia data mining systems will be used by medical doctors or clinicians. Th e design of the system needs to take into consideration of the hu- man perceptual. How to develop a system work synergis- tically is a subject of ongoing research. In order to achieve the goal, biologist, medical doctors, clinicians and the computing professional all need to work closely together. Any little part missing may lead to the failure of the system design. Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research 158 6. Conclusions The well use of the data mining tools in the biomedicine should bring revolutionary impact to the field. The study of biomedical processes is heavily based on the identifi- cation of understandable patterns which are present in the data. These patterns may be used for diagnostic or prog- nostic purpose as well as the analysis of microarrays. Data mining is at the care of the pattern recognition process. Biologist, medical doctors, clinicians and com- puting professionals should collaborate so that the two fields can contribute to each other. The challenge is for each to widen its focus to attain harmonious and produc- tive collaboration to develop the best practices. 7. Acknowledgement The authors would like to express their sincere thanks to the Research Committee of The Hong Kong Polytechnic University for financial support of the research work presented in this paper. REFERENCES [1] W. Frawley, G. Piatetsky-Shapiro, and C. Matheus, “Knowledge discovery in databases: An overview,” AI Magazine, pp. 213–228, 1992. [2] K. J. Cios, W. Pedrycz, R. W. Swiniarski, and L. A. Kur- gan, “Data mining: A knowledge discovery approach,” Springer, New York, 2007. [3] J. T. Tou and R. C. Gonzalez, “Pattern recognition prin- ciples,” Addison-Wesley, London, 1974. [4] R. O. Duda, P. E. Hart, and D. G. Stork, “Pattern classi- fication,” Wiley, 2001. [5] T. Hastie, R. Tibshirani, and J. Friedman, “The elements of statistical learning: Data mining, inference, and predic- tion,” Springer, New York, 2001. [6] J. W. Lee, J. B. Lee, M. Park, and S. H. Song, “An exten- sive comparison of recent classification tools applied to microarray data,” Computational Statistics & Data Analysis, Vol. 48, No. 4, pp. 869–885, 2005. [7] V. S. Tseng and S. C. Yang, “Mining multi-level associa- tion rules from gene expression profiles and gene ontol- ogy,” in Proceedings IEEE Workshop Life Science Data Mining (held with IEEE ICDM), UK, November 2004. [8] H. Chen, S. S. Fuller, C. Friedman, and W. Hersh, “Medical informatics–knowledge management and data mining in biomedicine,” Springer, 2005. [9] C. D. Krivda, “Data-Mining Dynamine,” Byte, 1995. [10] J. C. Prather, D. F. Lobach, L. K. Goodwin, J. W. Hales, M. L. Hage, and W. E. Hammond, “Medical data mining: Knowledge discovery in a clinical data warehouse,” in Proceedings AMIA Annual Fall Symposium, pp. 101– 105, 1997. [11] J. L. Breault, C. R. Goodall, and P. J. Fos, “Data mining a diabetic data warehouse,” Artificial Intelligence in Medi- cine, Vol. 26, pp. 37–54, 2002. [12] A. M. Wilson, L. Thabane, and A. Holbrook, “Applica- tion of data mining techniques in pharmacovigilance,” British Journal of Clinical Pharmacology, Vol. 57, No. 2, pp. 127–134, 2004. [13] J. Lian, C. Cotrutz, and L. Xing, “Therapeutic treatment plan optimization with probability density-based dose prescription,” Medical Physics, Vol. 30, No. 4, pp. 655– 666, 2003. [14] E. G. Susan and J. M. Warren, “Statistical modelling of general practice medicine for computer assisted data entry in electronic medical record systems,” International Jour- nal of Medical Informatics, Vol. 57, No. 2-3, pp. 77–89, 2000. [15] J. R. Warren, A. Davidovic, S. Spenceley, and P. Bolton, “Mediface: Anticipative data entry interface for general practitioners,” in Proceedings Computer Human Interac- tion Conference 1998, pp. 192–199, 1998. [16] R. J. Kuo, S. Y. Lin, and C. W. Shih, “Mining association rules through integration of clustering analysis and ant colony system for health insurance database in Taiwan,” Expert Systems with Applications, Vol. 33, pp. 794–808, 2007. [17] Z. Y. Zhuang, L. Churilov, F. Burstein, and K. Sikaris, “Combining data mining and case-based reasoning for intelligent decision support for pathology ordering by general practitioners,” European Journal of Operational Research, Vol. 195, No. 3, pp. 662–675, 2009. [18] M. J. Huang, M. Y. Chen, and S. C. Lee, “Integrating data mining with case-based reasoning for chronic dis- eases prognosis and diagnosis,” Expert Systems with Ap- plications, Vol. 32, No. 3, pp. 856–867, 2007. [19] M. G. Tsipouras, T. P. Exarchos, D. I. Fotiadis, A. P. Kotsia, K. V. Vakalis, K. K. Naka, and L. K. Michalis, “Automated diagnosis of coronary artery disease based on data mining and fuzzy modeling,” IEEE Transactions on Information Technology in Biomedicine, Vol. 12, No. 4, pp. 447–457, 2008. [20] V. Megalooikonomou, J. Ford, L. Shen, F. Makedon, and A. Saykin, “Data mining in brain imaging,” Statistical Methods in Medical Research, Vol. 9, No. 4, pp. 359–394, 2000. [21] S. E. Brossette, A. P. Sprague, W. T. Jones, and S. A. Moser, “A data mining system for infection control sur- veillance,” Methods of Information in Medicine, Vol. 39, No. 4-5, pp. 303–310, 2000. [22] M. L. Antonie, O. R. Zaiane, and A. Coman, “Application of data mining techniques for medical image classifica- tion,” in Proceedings Second International Workshop on Multimedia Data Mining, pp. 94–101, 2001. [23] D. M. Coulter, A. Bate, R. H. B. Meyboom, M. Lindquist, and R. Edwards, “Antipsychotic drugs and heart muscle disorder in international pharmacovigilance: Data mining study,” British Medical Journal, Vol. 322, pp. 1207–1209, 2001. [24] L. Li, H. Tang, Z. Wu, J. Gong, M. Gruidl, J. Zou, M. Tockman, and R. Clark, “Data mining techniques for cancer detection using serum proteomic profiling,” Arti- Copyright © 2009 SciRes JSEA  Data Mining in Biomedicine: Current Applications and Further Directions for Research Copyright © 2009 SciRes JSEA 159 ficial Intelligence in Medicine, Vol. 32, No. 2, pp. 71–83, 2004. [25] D. Delen, G. Walker, and A. Kadam, “Predicting breast cancer survivability: A comparison of three data mining methods,” Artificial Intelligence in Medicine, Vol. 34, No. 2, pp. 113–27, 2005. [26] C. T. Su, C. H. Yang, K. H. Hsu, and W. K. Chiu, “Data mining for the diagnosis of type II diabetes from three- dimensional body surface anthropometrical scanning data,” Computers & Mathematics with Applications, Vol. 51, No. 6–7, pp. 1075–1092, 2006. [27] G. Philips-Wren, P. Sharkey, and S. Morss, “Mining lung cancer patient data to assess healthcare resource utiliza- tion,” Expert Systems with Applications: An International Journal, Vol. 35, No. 4, pp. 1611–1619, 2008. [28] M. Hearst, “Untangling text data mining,” in the Pro- ceedings ACL’99: The 37th annual meeting of the asso- ciation for computational linguistics, University of Mary- land, June 1999. [29] H. Chen, “Knowledge management systems: A text min- ing perspective,” Tucson, AZ, The University of Arizona, 2001. [30] K. B. Cohen and L. Hunter, “Getting started in text min- ing,” PLoS Computational Biology, Vol. 4, No. 1, doi: 10.1371/journal.pcbi.0040020, 2008. [31] Y. Ku, C. Chiu, B. H. Liou, J. H. Liou, and J. Y. Wu, “Applying text mining to assist people who inquire HIV/AIDS information from Internet,” in Proceedings ISI 2008 Workshops, pp. 440–448, 2008. [32] D. Zhou, Y. He, and C. K. Kwoh, “Validating text mining results on protein-protein interactions using gene expres- sion profiles,” in Proceedings International Conference on Biomedical and Pharmaceutical Engineering 2006, pp. 580–585, 2006. [33] Y. Peng, B. Yao, and J. Jiang, “Knowledge-discovery incorporated evolutionary search for microcalcification detection in breast cancer diagnosis,” Artificial Intelli- gence in Medicine, Vol. 37, No. 1, pp. 43–53, 2006. [34] H. Pan, Q. Han, X. Xie, Z. Wei, and J. Li, “A Similarity retrieval method in brain image sequence database,” Ad- vanced Data Mining and Applications, Vol. 4632, pp. 352–364, 2007. [35] X. Zhu, W. G. Aref, J. Fan, A.C. Catlin, and A. K. Elma- garmid, “Medical video mining for efficient database in- dexing, management and access,” in Proceedings 19th International Conference on Data Engineering, pp. 569–580, 2003. [36] R. Kohavi, B. Masand, M. Spilipoulou, and J. Srivastava, “Web mining,” Data Mining and Knowledge Discovery, Vol. 6, pp. 5–8, 2002. [37] W. D. Yu and S. R. Jonnalagadda, “Semantic web and mining in healthcare,” in Proceedings 8th International Conference on e-Health Networking, Applications and Services, pp. 198–201, 2006. [38] S. Mitra and T. Acharya, “Data mining: Multimedia, soft computing and bioinformatics,” John Wiley & Sons, Inc., New Jersey, 2003. |

