 Vol.3, No.9, 577-583 (2011) doi:10.4236/health.2011.39099 C opyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ Health In vitro examining the existing prognoses how TBP binds to TATA with SNP associated with human diseases Irina A. Drachkova1, Petr M. Ponomarenko1, Tatyana V. Arshinova1, Мikhail P. Ponomarenko1*, Valentin V. Suslov1, Ludmila K. Savinkova1, Nikolay А. Kolchanov1,2 1Institute of Cytology and Genetics, Siberian Division, Russian Academy of Sciences, Novosibirsk, Russia; *Corresponding Author: pon@bionet.nsc.ru 2Novosibirsk State University, Novosibirsk, Russia. Received June 18th, 2011; revised July 12th, 2011; accepted August 1st, 2011. ABSTRACT We in vitro examined the existing prognoses of the dissociation constant, KD, between ТАТА- Binding Protein (TBP) and ТАТА box with single nucleotide polymorphism (SNP) associated with human diseases. Five SNPs of the genes for cytochrome P450 2A6 (associated with lung cancer), β-globin (associated with β-thalassemia), mannose binding lectin (associated with vari- able immunodeficiency), superoxide dismutase 1 (associated with amyotrophic lateral sclerosis) and triosephosphate isomerase (associated with anemia) fell within the range of –ln(KD;M/KD;W T) between –1.5 and –1 (here KD;WT and KD;M denote the normal ТАТА box and with SNP). The meas- urements using EMSA demonstrated that: 1) all the predictions stating that the affinity between ТВР and ТАТА boxes with SNPs would be re- duced were correct; 2) the departures of three predictions from the measurements fell within the confidence interval; 3) all the predictions consistently underestimated actual mutational damage caused to ТАТА boxes with SNPs ( < 0.05; binomial law) and two of these predictions did so significantly ( < 0.05, Student’s t-test). This consistent underestimation seems to be associated with the damage to the context that modulates ТВP/ТА ТА affinity, for example, the contact between the nucleosomal histone H3-Н4 dimer and the core promoter immediately near ТАТА boxes. Keywords: Disease; Polymorphism; ТАТА Box; TATA-Binding Protein; Affinity; In Vitro; In Silico 1. INTRODUCTION Variome is largely composed of single nucleotide polymorphisms (SNPs). Consequently, no study of their role in ontogenesis or evolution could be efficient with- out computer-aided support, which would facilitate searches for SNPs, their documentation and systematiza- tion, and prediction of their effects on the phenotype. Over the past 10 years of the coordinated development of SNP databases and tools [1], anything outside the phenotype prediction problem has been successfully addressed [2]. In particular, SNPs responsible for the propensity for diseases, susceptibility to therapy, sensi- tivity to regulatory signals, etc. have been identified. Phenotype prediction has only been successful for the SNPs that are located in coding gene regions. The com- mon molecular mechanism that can be proposed for them is mutational damage made to the gene product [3]. It is still difficult to propose the same for SNPs in regu- latory gene regions because of the diversity of such re- gions and a multi-step sequence of assembly, rear- rangement and degradation of DNA-protein complexes in them [4]. Examination of the computer-aided methods, developed on such a diverse material, requires an ex- perimental examination in standardized conditions (the standardized examination throughout). Because different authors set different experimental conditions, the sole analysis of experimental results in databases will be of little help. For avoidance of doubt, there is a need for a coordination of bioinformatic and experimental studies of each type of site. For that purpose we conduct an in- tegrated study of regulatory SNPs, especially those in ТАТА boxes. The binding of the ТАТА-binding protein (ТВР) to the ТАТА box initiates assembly of the pre-initiation complex on the ТАТА-containing promoters of eu- karyotic genes, which is a critical step in transcription  I. A. Drachkova et al. / Health 3 (2011) 577-583 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 578 initiation [5]. We had previously [6] proposed a method for in silico prediction of TBP/TATA affinity on the basis of the equation of equilibrium of ТВР/ТАТА binding over four subsequent steps: 1) non-specific ТВР/ DNA binding [7]; 2) TBP sliding along DNA [8]; 3) molecular identification of ТВР/ТАТА [9] and 4) stabi- lization of the ТВР/ТАТА complex [10] by endothermic DNA rearrangement [11] with the helix axis bent at 90˚ [10]. This equation allows the relative affinity, Δ = –ln(KD;M/KD;WT), that is, the ratio of the dissociation constant of TBP and the normal TATA box (KD;WT) to that of TBP and the mutant ТАТА box (KD;M), to be estimated in logarithms. This equation puts together the commonly accepted criterion associated with TATA boxes for arbitrary DNA [9] (step 3), which only ac- counts for 33% of the variance of the measured ТВР/ ТАТА affinity [12], original criteria associated with TBP affinity estimates for single-strand DNA [13] and double-strand DNA [14] (step 4 and step 2) and an in- dependent measurement of the non-specific affinity of ТВР and DNA [7] (step 1). Stepwise binding of ТВР to ТАТА, predicted by this equation [6], has now been confirmed experimentally [15]. Although the ТАТА box is a “semi-conservative” site, no-neutral SNPs in it are quite a common occurrence in various species. Thus, in silico analysis of the current content of the GenBank database revealed 146 SNPs of the HIV-1 TATA box, of which 63 could significantly modify the replicative potential of the virus and were associated with the regional patterns of the AIDS pan- demic in 70 countries [16]. Literature data suggest that 53 SNPs in the ТАТА boxes of various human genes are associated with the propensity for diseases [17], 38 SNPs are associated with various animal and plant traits valuable with respect to breeding purposes [18]. In both cases we predicted in silico significant departures of ТВР/ТАТА affinity. The aim of the present work was to perform a standardized experimental examination of the strongest reductions in ТВР/ТАТА affinity as predicated among 53 disease-associated SNPs in human TATA boxes [19]. The measurements of KD;M and KD;WT using EMSA demonstrated that: 1) all the predictions stating that the affinity between ТВР and ТАТА boxes with SNPs would be reduced were correct; 2) the departures of three predictions from the measurements fell within the confidence interval; 3) all the predictions consis- tently underestimated actual mutational damage caused to ТАТА boxes with SNPs ( < 0.05; binomial law) and two of these predictions did so significantly ( < 0.05, Student’s t-test). 2. EXPERIMENTAL PROCEDURES For the standardized experimental examination of the predicted ТВР/ТАТА affinity, we used recombinant human ТВР expressed in E. coli BL21 (DE3) cells from plasmid pAR3038-hTBP (courtesy of Professor B. Puhg, Center for Gene Regulation, Department of Biochemis- try and Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania, USA). E. coli BL21 (DE3) transformation was performed as per Pe- terson and the co-workers [20]. The expression and puri- fication of TBP were done as [21]. A 26-bp strand of oligodeoxyribonucleotides (Biosset, Novosibirsk) was labeled with γ32P-ATP (Biosan, Novosibirsk, Russia) using Т4 polynucleotide kinases (SibEnzime, Novosi- birsk, Russia), annealed at 95˚C with the non-labeled strand and cooled slowly to room temperature. The equi- librium dissociation constants, KD, of the ТВР/DNA complexes were measured using EMSA, titration of a fixed amount of ТВР with the oligonucleotide in in- creasing concentrations and isotopic dilution [22] as shown in Figure 1. In doing so, we used two standard tools, Gel-Pro Analyzer 3.1 for the densitometry of autoradiographs and OriginPro 8 for obtaining KD from densitometry data (Figure 1). The confidence intervals of the 5% boundary ( < 0.05) for each prediction were in silico estimated using Student’s t-test as [19]. For all the in vitro measurements, the confidence interval, KD, commonly acceptable for the above two standard tools, was set as ±0.37 in relative natural logarithms, which corresponds to a confidence interval of ±30% of the KD value in nM, commonly ac- cepted for EMSA-measurements of the parameters of the protein/DNA complex. 3. RESULTS AND DISCUSSIONS The predicted in silico [19] and experimentally meas- ured in vitro relative affinity of ТВР for ТАТА boxes containing SNPs for the genes encoding cytochrome P450 2A6 (associated with lung cancer), β-globin (asso- ciated with β-thalassemia), superoxide dismutase 1 (as- sociated with amyotrophic lateral sclerosis), mannose binding lectin (associated with variable immunodefi- ciency) and triosephosphate isomerase (associated with anemia) are presented in Table 1. In all cases, the ex- periment confirmed the in silico predicted reduction in the affinity of ТВР to the ТАТА box containing the SNPs associated with the respective diseases. In three of the five SNPs, namely cytochrome P450 2A6 (associated with lung cancer), β-globin (associated with β-thalas- semia) and superoxide dismutase 1 (associated with amyotrophic lateral sclerosis), the departures of the pre- dicted values from those measured experimentally fell within the confidence interval. In the rightmost column of Table 1, we compared the affinity range from –3.72 ± 037 to –1.03 ± 0.14, which . 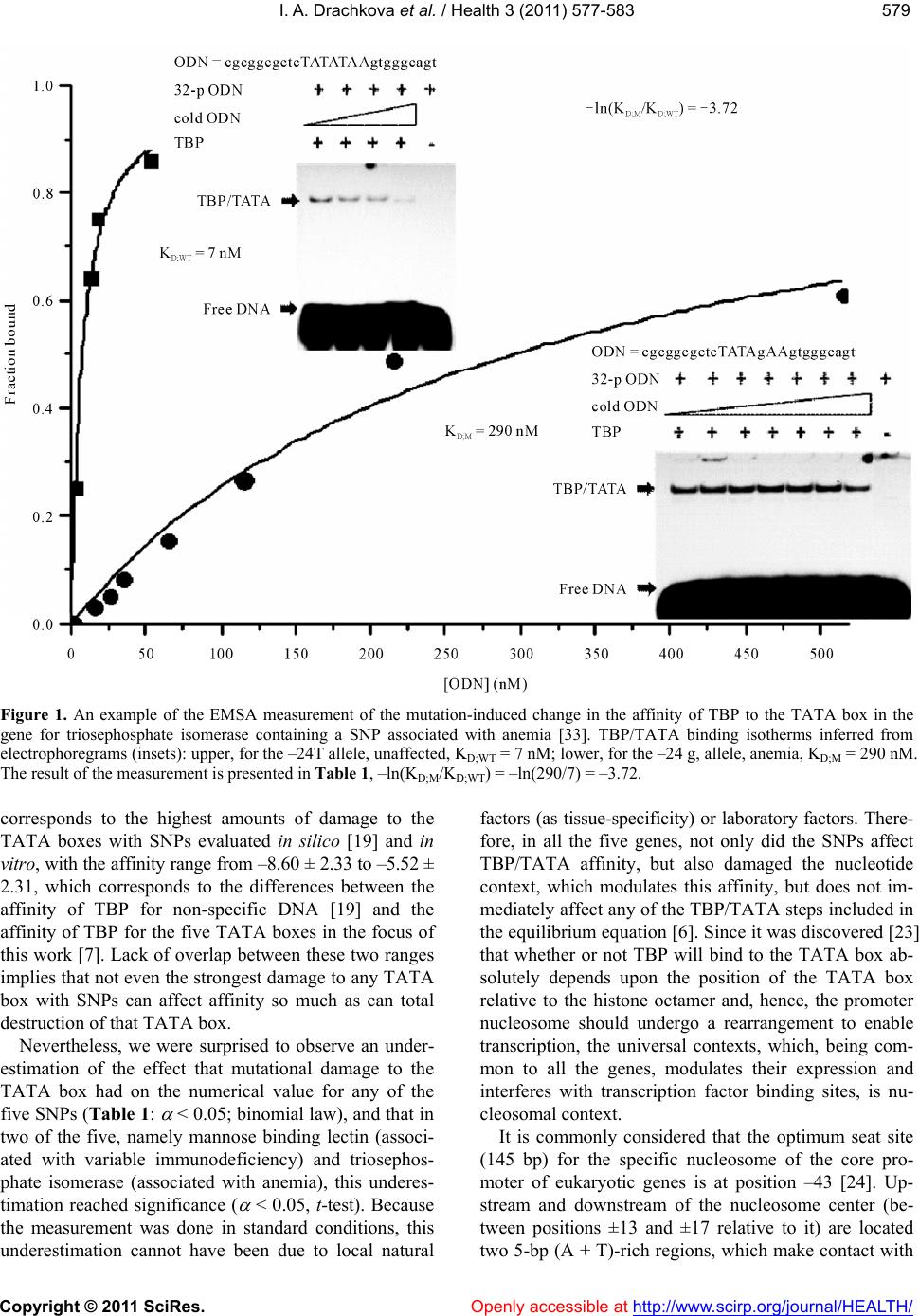 I. A. Drachkova et al. / Health 3 (2011) 577-583 Copyright © 2011 SciRes. http://www.scirp.org/journal/HEALTH/Openly accessible at 579579 Figure 1. An example of the EMSA measurement of the mutation-induced change in the affinity of ТВР to the ТАТА box in the gene for triosephosphate isomerase containing a SNP associated with anemia [33]. ТВР/ТАТА binding isotherms inferred from electrophoregrams (insets): upper, for the –24T allele, unaffected, KD;WT = 7 nM; lower, for the –24 g, allele, anemia, KD;M = 290 nM. The result of the measurement is presented in Table 1, –ln(KD;M/KD;WT) = –ln(290/7) = –3.72. corresponds to the highest amounts of damage to the TATA boxes with SNPs evaluated in silico [19] and in vitro, with the affinity range from –8.60 ± 2.33 to –5.52 ± 2.31, which corresponds to the differences between the affinity of TBP for non-specific DNA [19] and the affinity of TBP for the five TATA boxes in the focus of this work [7]. Lack of overlap between these two ranges implies that not even the strongest damage to any TATA box with SNPs can affect affinity so much as can total destruction of that TATA box. Nevertheless, we were surprised to observe an under- estimation of the effect that mutational damage to the TATA box had on the numerical value for any of the five SNPs (Table 1: < 0.05; binomial law), and that in two of the five, namely mannose binding lectin (associ- ated with variable immunodeficiency) and triosephos- phate isomerase (associated with anemia), this underes- timation reached significance ( < 0.05, t-test). Because the measurement was done in standard conditions, this underestimation cannot have been due to local natural factors (as tissue-specificity) or laboratory factors. There- fore, in all the five genes, not only did the SNPs affect ТВР/ТАТА affinity, but also damaged the nucleotide context, which modulates this affinity, but does not im- mediately affect any of the ТВP/ТАТА steps included in the equilibrium equation [6]. Since it was discovered [23] that whether or not TBP will bind to the TATA box ab- solutely depends upon the position of the TATA box relative to the histone octamer and, hence, the promoter nucleosome should undergo a rearrangement to enable transcription, the universal contexts, which, being com- mon to all the genes, modulates their expression and interferes with transcription factor binding sites, is nu- cleosomal context. It is commonly considered that the optimum seat site (145 bp) for the specific nucleosome of the core pro- moter of eukaryotic genes is at position –43 [24]. Up- stream and downstream of the nucleosome center (be- tween positions ±13 and ±17 relative to it) are located two 5-bp (А + Т)-rich regions, which make contact with 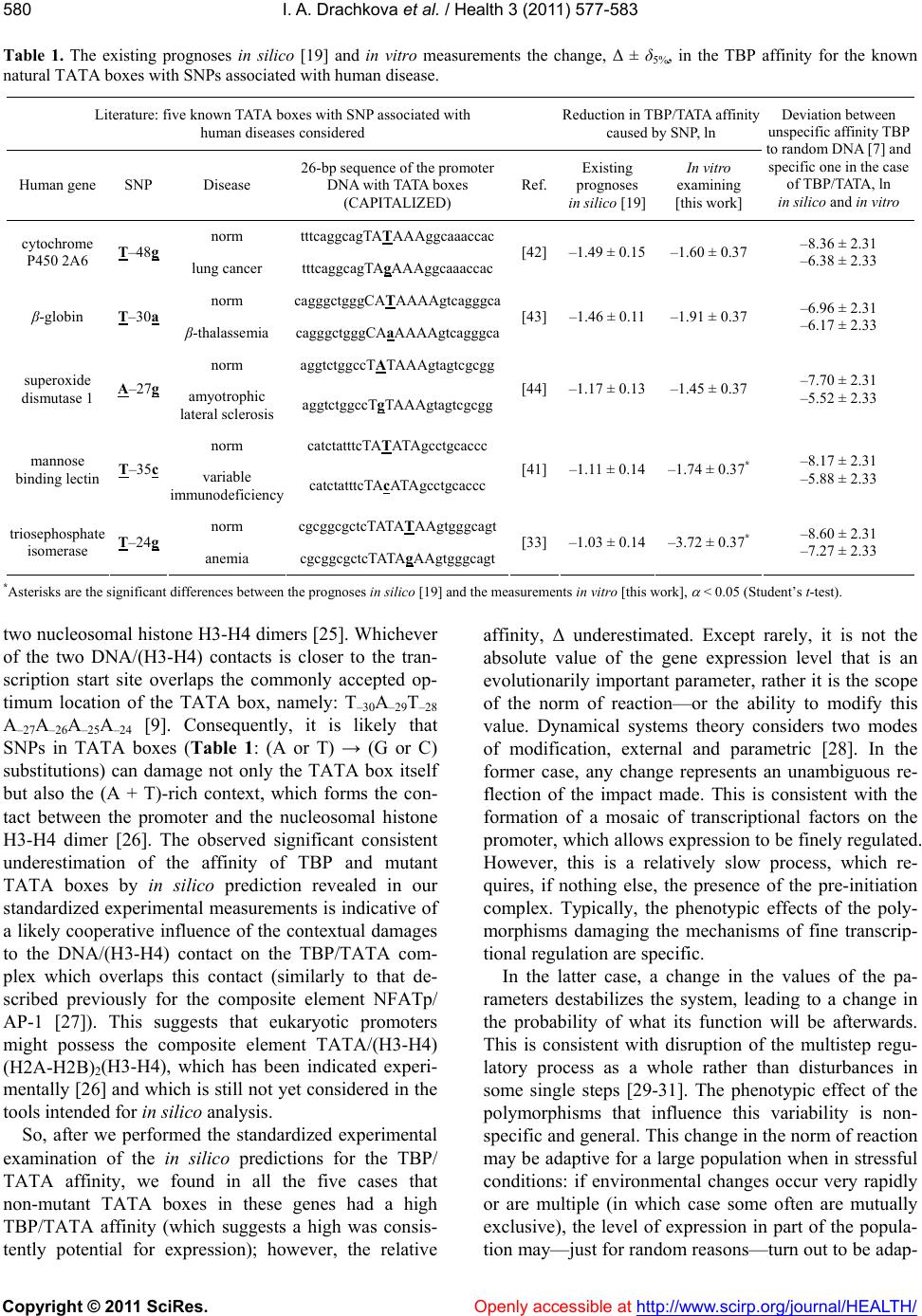 I. A. Drachkova et al. / Health 3 (2011) 577-583 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 580 Table 1. The existing prognoses in silico [19] and in vitro measurements the change, Δ ± δ5%, in the TBP affinity for the known natural ТАТА boxes with SNPs associated with human disease. Literature: five known ТАТА boxes with SNP associated with human diseases considered Reduction in ТВР/TATA affinity caused by SNP, ln Human gene SNP Disease 26-bp sequence of the promoter DNA with TATA boxes (CAPITALIZED) Ref. Existing prognoses in silico [19] In vitro examining [this work] Deviation between unspecific affinity TBP to random DNA [7] and specific one in the case of TBP/TATA, ln in silico and in vitro norm tttcaggcagТАТАААggcaaaccac cytochrome P450 2A6 T–48g lung cancer tttcaggcagТАgАААggcaaaccac [42]–1.49 ± 0.15–1.60 ± 0.37 –8.36 ± 2.31 –6.38 ± 2.33 norm cagggctgggCATAAAAgtcagggca β-globin T–30a β-thalassemia cagggctgggCAaAAAAgtcagggca [43]–1.46 ± 0.11–1.91 ± 0.37 –6.96 ± 2.31 –6.17 ± 2.33 norm aggtctggccТАТАААgtagtcgcgg superoxide dismutase 1 A–27g amyotrophic lateral sclerosis aggtctggccТgТАААgtagtcgcgg [44]–1.17 ± 0.13–1.45 ± 0.37 –7.70 ± 2.31 –5.52 ± 2.33 norm catctatttcТАТАТАgcctgcaccc mannose binding lectin T–35c variable immunodeficiency catctatttcТАcАТАgcctgcaccc [41]–1.11 ± 0.14–1.74 ± 0.37* –8.17 ± 2.31 –5.88 ± 2.33 norm cgcggcgctcТАТАTААgtgggcagt triosephosphate isomerase T–24g anemia cgcggcgctcТАТАgААgtgggcagt [33]–1.03 ± 0.14–3.72 ± 0.37* –8.60 ± 2.31 –7.27 ± 2.33 *Asterisks are the significant differences between the prognoses in silico [19] and the measurements in vitro [this work], < 0.05 (Student’s t-test). two nucleosomal histone H3-Н4 dimers [25]. Whichever of the two DNA/(Н3-Н4) contacts is closer to the tran- scription start site overlaps the commonly accepted op- timum location of the TATA box, namely: T–30A–29T–28 A–27A–26A–25A–24 [9]. Consequently, it is likely that SNPs in TATA boxes (Table 1: (A or T) → (G or C) substitutions) can damage not only the TATA box itself but also the (A + T)-rich context, which forms the con- tact between the promoter and the nucleosomal histone H3-Н4 dimer [26]. The observed significant consistent underestimation of the affinity of TBP and mutant TATA boxes by in silico prediction revealed in our standardized experimental measurements is indicative of a likely cooperative influence of the contextual damages to the DNA/(Н3-Н4) contact on the ТВР/ТАТА com- plex which overlaps this contact (similarly to that de- scribed previously for the composite element NFATp/ AP-1 [27]). This suggests that eukaryotic promoters might possess the composite element ТАТА/(Н3-Н4) (Н2А-Н2В)2(Н3- Н4), which has been indicated experi- mentally [26] and which is still not yet considered in the tools intended for in silico analysis. So, after we performed the standardized experimental examination of the in silico predictions for the ТВР/ ТАТА affinity, we found in all the five cases that non-mutant ТАТА boxes in these genes had a high ТВР/ТАТА affinity (which suggests a high was consis- tently potential for expression); however, the relative affinity, Δ underestimated. Except rarely, it is not the absolute value of the gene expression level that is an evolutionarily important parameter, rather it is the scope of the norm of reaction—or the ability to modify this value. Dynamical systems theory considers two modes of modification, external and parametric [28]. In the former case, any change represents an unambiguous re- flection of the impact made. This is consistent with the formation of a mosaic of transcriptional factors on the promoter, which allows expression to be finely regulated. However, this is a relatively slow process, which re- quires, if nothing else, the presence of the pre-initiation complex. Typically, the phenotypic effects of the poly- morphisms damaging the mechanisms of fine transcrip- tional regulation are specific. In the latter case, a change in the values of the pa- rameters destabilizes the system, leading to a change in the probability of what its function will be afterwards. This is consistent with disruption of the multistep regu- latory process as a whole rather than disturbances in some single steps [29-31]. The phenotypic effect of the polymorphisms that influence this variability is non- specific and general. This change in the norm of reaction may be adaptive for a large population when in stressful conditions: if environmental changes occur very rapidly or are multiple (in which case some often are mutually exclusive), the level of expression in part of the popula- tion may—just for random reasons—turn out to be adap-  I. A. Drachkova et al. / Health 3 (2011) 577-583 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 581581 tive. In particular, a change in nucleosome packaging is capable of non-specifically changing the probability of the gene expressing in many tissues at a time. A poly- morphism that changes nucleosome packaging affects at least two parameters: the “layout” of the transcription- ally active genomic regions [23] and the order of bind- ing/affinity of transcriptional factors for DNA due to chemical modification of histones [32]. It is commonly accepted that the housekeeping gene promoters have a special nucleosomal context, which ensures a looser nu- cleosome packaging and thus makes the promoter acces- sible by various regulatory proteins in a large variety of tissues. Destruction of one of the few nucleosomes should considerably affect regulation. True is, it was a housekeeping gene, the only one in our check, for which we observed the largest departure of Δ from the in silico prediction (Table 1: triosephosphate isomerase associ- ated with anemia [33]). The MBL2 gene, too, is characterized by a large de- parture of Δ from the in silico prediction. Mannose binding lectin (MBL) is a key protein in the develop- ment of the innate immune response. The polymor- phisms that reduce MBL expression are associated with variable immunodeficiency, which is a risk factor (espe- cially in the tender age [34,35]) for a variety of infec- tious diseases [34-36]. In lower primates, both copies of the MBL gene are under stabilizing selection [37]. In the anthropoid lineage, one of the copies has underwent pseudogenization [38], and man has additionally ac- quired a high frequency of polymorphisms that reduce the MBL level in the tissues, disrupt folding (codon 52 Arg → Cys, codon 54 Gly → Asp, codon 57 Gly → Glu) or transcription (–2550 - H/L polymorphism, –2221 - X/Y polymorphism, –2427, –2349, –2336, –270, +4 - P/Q polymorphism; –2324 - –2329 deletion) [34,36,38]. A low level of MBL eases the after-effects of the stroke [39] and pre-eclampsia [40]. Thus, it is adaptive for the humanoids, with their actively working brain and diffi- cult child-birth, to have strong variation of the within-tissue level of MBL across populations by com- bination of these polymorphisms in the heterozygotes. In addition to the common polymorphisms mentioned above, local human populations may have other, inde- pendently fixed polymorphisms [41], the effect of one of which, located in the area of the ТАТА box (T-35c, Ta- ble 1), has been reported here. This polymorphism is likely to serve the same purpose as the common SNPs; specifically, it expands the norm of reaction, but does it somewhat differently, namely, by modulation of a trig- ger-like regulator based on the composite ТАТА/(Н3-Н4) (Н2А-Н2В)2(Н3-Н4) unit. Importantly, this modulation of the norm of reaction is mild. The polymorphisms that disrupt folding or tran- scription [34,36,38] inhibit MBL gene expression. Con- sequently, the norm of reaction widens only at a popula- tion-wide level. Individuals homozygous for such poly- morphisms are vulnerable to infection at all times, that is, the individual norm of reaction is narrow and these indi- viduals will not survive any attack by viruses or mi- crobes. The lack of overlap between the affinity of ТВР for the TATA box in the MBL gene with the T-35c polymorphism and the affinity of TBP for non-specific DNA indicates that the expression of this gene is not totally suppressed even in the individuals that were ho- mozygous for this polymorphism. Decrease in ТВР af- finity means decrease in the probability that the expres- sion of the MBL gene will be initiated; however, if ini- tiation does take place, the gene will be expressed to the extent that it will be in any wild-type individual. In other words, if some individuals in a population carry the T–35c polymorphism, the norm of reaction will be wid- ening not only at a population-wide level, but also at an individual level―specifically, in those carriers. Thus, even when under a viral or microbial attack, such indi- viduals are given a chance. Identification of the previously unidentified source of the consistent in silico underestimation of the amount of damage caused to the regulatory regions of genes with SNPs makes us expect that a standardized examination of all the 237 TATA box SNPs, associated with human diseases [17], regional patterns of the AIDS pandemic [16], animal and plant traits, which are valuable with respect to breeding purposes [18], and newly discovered mutations in TATA boxes will allow us to look into the mechanisms of ТВР/ТАТА binding and improve the research quality of the computer-aided tools used for analysis and prediction of SNPs in the regulatory regions of human genes. 4. ACKNOWLEDGEMENTS We are grateful to Prof. Tatyana Merkulova for her valuable advice. This work was supported by grants 10-04-00462, 11-04-01254, and 11-04-01888 from Russian Foundation for Basic Research, grant 119 (Siberian Branch of the Russian Academy of Sciences), grant B.27 “Biological diversity” (Russian Academy of Sciences), “Molecular and Cell Biology” and “Origin and evolution of biosphere” programs (Rus- sian Academy of Sciences), grant 2447.2008.4 from the Scientific School Program, State contract 10104-37/P-18/110-327/180608/015, contract 07.514.11.4003 (Russian Ministry of Education and Science). REFERENCES [1] Sherry, S.T., Ward, M. and Sirotkin, K. (1999) DbSNP database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Research, 9, 677-679.  I. A. Drachkova et al. / Health 3 (2011) 577-583 Copyright © 2011 SciRes. Openly accessible at http://www.scirp.org/journal/HEALTH/ 582 [2] Day, I.N. (2010) DbSNP in the detail and copy number complexities. Human Mutation, 31, 2-4. doi:10.1002/humu.21149 [3] Batley, J. and Edwards, D. (2009) Mining for SNPs and SSRs using SNPServer, dbSNP and SSR taxonomy tree. Methods in Molecular Biology, 537, 303-321. doi:10.1007/978-1-59745-251-9_15 [4] Collaco, J.M. and Cutting, G.R. (2008) Update on gene modifiers in cystic fibrosis. Current Opinion in Pulmo- nary Medicine, 14, 559-566. doi:10.1097/MCP.0b013e3283121cdc [5] Faiger, H., Ivanchenko, M., Cohen, I. and Haran, T.E. (2006) TBP flanking sequences: Asymmetry of binding, long-range effects and consensus sequences. Nucleic Ac- ids Research, 34, 104-119. doi:10.1093/nar/gkj414 [6] Ponomarenko, P.M., Savinkova, L.K., Drachkova, I.A., Lysova, M.V., Arshinova, T.V., Ponomarenko, M.P. and Kolchanov, N.A. (2008) A step-by-step model of TBP/TATA box binding allows predicting human heredi- tary diseases by single nucleotide polymorphism. Dok- lady Biochemistry and Biophysics, 419, 88-92. doi:10.1134/S1607672908020117 [7] Hahn, S., Buratowski, S., Sharp, P.A. and Guarente, L. (1989) Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and non nonconsensus DNA sequences. Proceedings of the National Academy of Sciences of USA, 86, 5718-5722. doi:10.1073/pnas.86.15.5718 [8] Coleman, R.A. and Pugh, B.F. (1995) Evidence for func- tional binding and stable sliding of the TATA binding protein on nonspecific DNA. The Journal of Biological Chemistry, 270, 13850-13859. doi:10.1074/jbc.270.23.13850 [9] Bucher, P. (1990) Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements de- rived from 502 unrelated promoter sequences. Journal of Molecular Biology, 212, 563-578. doi:10.1016/0022-2836(90)90223-9 [10] Juo, Z.S., Chiu, T.K., Leiberman, P.M., Baikalov, I., Berk, A.J. and Dickerson, R.E. (1996) How proteins recognize the TATA box. Journal of Molecular Biology, 261, 239-254. doi:10.1006/jmbi.1996.0456 [11] Powel, R., Parkhurst, K. and Parkhurst, L. (2002) Com- parison of TATA-binding protein recognition of a variant and consensus DNA promoters. The Journal of Biologi- cal Chemistry, 277, 7776-7784. doi:10.1074/jbc.M110147200 [12] Savinkova, L.K., Drachkova, I.A., Ponomarenko, M.P., Lysova, M.V., Arshinova, T.V. and Kolchanov, N.A. (2007) Interaction between the recombinant TATA-binding protein and the TATA-boxes of the mam- malian gene promoters. Ecological Genetics, 5, 44-49. [13] Sokolenko, A.A., Sandomirski, I.I. and Savinkova, L.K. (1996) Interaction of yeast TATA-binding protein with promoters short sites. Molecular Biology, 30, 279-285. [14] Ponomarenko, M.P., Ponomarenko, J.V., Frolov, A.S., Podkolodny, N.L., Savinkova, L.K., Kolchanov, N.A. and Overton, G.C. (1999) Identification of sequence de- pendent DNA features correlating to activity of DNA sites interacting with proteins. Bioinformatics, 15, 687-703. doi:10.1093/bioinformatics/15.7.687 [15] Delgadillo, R.F., Whittington, J.E., Parkhurst, L.K. and Parkhurst, L.J. (2009) The TATA-binding protein core domain in solution variably bends TATA sequences via a three-step binding mechanism. Biochemistry, 48, 1801- 1809. doi:10.1021/bi8018724 [16] Suslov, V.V., Ponomarenko, P.M., Efimov, V.M., Sav- inkova, L.K., Ponomarenko, M.P. and Kolchanov, N.A. (2010) SNPs in the HIV-1 TATA box and the AIDS pan- demic. Journal of Bioinformatics and Computational Bi- ology, 8, 607-625. doi:10.1142/S0219720010004677 [17] Savinkova, L.K., Ponomarenko, M.P., Ponomarenko, P.M., Drachkova, I.A., Lysova, M.V., Arshinova, T.V. and Kolchanov, N.A. (2009) TATA box polymorphisms in human gene promoters and associated hereditary pa- thologies. Biochemistry, 74, 117-129. doi:10.1134/S0006297909020011 [18] Suslov, V.V., Ponomarenko, P.M., Ponomarenko, M.P., Drachkova, I.A., Arshinova, T.V., Savinkova, L.K. and Kolchanov, N.A. (2010) TATA box polymorphisms in genes of commercial and laboratory animals and plants associated with selectively valuable traits. Russian Jour- nal of Genetics, 46, 394-403. doi:10.1134/S1022795410040022 [19] Ponomarenko, P.M., Ponomarenko, M.P., Drachkova, I.A., Lysova, M.V., Arshinova, T.V., Savinkova, L.K. and Kolchanov, N.A. (2009) Prognosis of affinity change of the TATA-binding protein to TATA-boxes upon poly- morphisms of the human gene promoter TATA boxes. Molecular Biology (Mosc), 43, 512-520. [20] Peterson, M.G., Tanese, N., Pugh, B.F. and Tjian, R. (1990) Functional domains and upstream activation properties of cloned human TATA-binding protein. Sci- ence, 248, 1625-1630. doi:10.1126/science.2363050 [21] Pugh, F. (1995) Purification of the human TATA-binding protein, TBP. Methods in Molecular Biology, 37, 359-367. doi:10.1385/0-89603-288-4:359 [22] Drachkova, I.A., Lysova, M.V., Repkova, M.N., Prokuda, O.V., Sokolenko, A.A., Arshinova, T.V., Kobzev, V.F., Iamkovoi, V.I. and Savinkova, L.K. (2005) Interaction of proteins from general transcription complex RNA poly- merase II with oligoribonucleotides. Molecular Biology, 39, 139-146. [23] Imbalzano, A.N., Kwon, H., Green, M.R. and Kingston, R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481-485. doi:10.1038/370481a0 [24] Ioshikhes, I., Trifonov, E.N. and Zhang, M.Q. (1999) Periodical distribution of transcription factor sites in pro- moter regions and connection with chromatin structure. Proceedings of the National Academy of Sciences of USA, 96, 2891-2895. doi:10.1073/pnas.96.6.2891 [25] Richmond, T.J. and Davey, C.A. (2003) The structure of DNA in the nucleosome core. Nature, 423, 145-150. doi:10.1038/nature01595 [26] Godde, J.S., Nakatani, Y. and Wolffe, A.P. (1995) The amino-terminal tails of the core histones and the transla- tional position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Re- search, 23, 4557-4564. doi:10.1093/nar/23.22.4557 [27] Kel, A.E., Kel-Margoulis, O.V., Babenko, V. and Win- gender, E. (1999) Recognition of NFATp/AP-1 compos- ite elements within genes induced upon the activation of immune cells. Journal of Molecular Biology, 288,  I. A. Drachkova et al. / Health 3 (2011) 577-583 Copyright © 2011 SciRes. http://www.scirp.org/journal/HEALTH/Openly accessible at 583583 353-376. doi:10.1006/jmbi.1999.2684 [28] Gunbin, K.V., Suslov, V.V. and Kolchanov, N.A. (2008) Molecular-genetic systems of development: Functional dynamics and molecular evolution. Biochemistry, 73, 219-230. doi:10.1134/S0006297908020144 [29] Strunnikov, V.A. and Vyshinsky, I.M. (1991) Realization variation in silkworm. In: Problems in Genetics and the Theory of Evolution, Nauka, Novosibirsk, 99-114. [30] Trut, L.N. (1997) D. K. Beliaev’s evolutionary con- cept—Ten years later. Russian Journal of Genetics, 33, 1060-1068. [31] De Jong, H. (2002) Modeling and simulation of genetic regulatory systems: A literature review. Journal of Com- putational Biology, 9, 67-103. doi:10.1089/10665270252833208 [32] Agalioti, T., Chen, G. and Thanos, D. (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111, 381-392. doi:10.1016/S0092-8674(02)01077-2 [33] Watanabe, M., Zingg, B.C. and Mohrenweiser, H.W. (1996) Molecular analysis of a series of alleles in humans with reduced activity at the triosephosphate isomerase locus. The American Journal of Human Genetics, 58, 308-316. [34] Koch, A., Melbye, M., Sørensen, P., Homoe, P., Madsen, H.O., Molbak, K. Hansen, C.H., Andersen, L.H., Hahn, G.W. and Garred, P. (2001) Acute respiratory tract infec- tions and mannose-binding lectin insufficiency during early childhood. The Journal of the American Medical Association, 285, 1316-1321. doi:10.1001/jama.285.10.1316 [35] Israels, J., Frakking, F.N., Kremer, L.C., Offringa, M., Kuijpers, T.W. and Van de Wetering, M.D. (2009) Man- nose-binding lectin and infection risk in newborns: A systematic review. Archives in Disease in Childhood: Fetal & Neonatal Edition, 5, F452-F461. [36] Eisen, D.P. and Minchinton, R.M. (2003) Impact of man- nose-binding lectin on susceptibility to infectious dis- eases. Clinical Infectious Diseases, 37, 1496-1505. doi:10.1086/379324 [37] Verga Falzacappa, M.V., Segat, L., Puppini, B., Amoroso, A. and Crovella, S. (2004) Evolution of the man- nose-binding lectin gene in primates. Genes & Immunity, 5, 653-661. doi:10.1038/sj.gene.6364140 [38] Seyfarth, J., Garred, P. and Madsen, H.O. (2005) The “involution” of mannose-binding lectin. Human Molecu- lar Genetics, 14, 2859-2869. doi:10.1093/hmg/ddi318 [39] Cervera, A., Planas, A.M., Justicia, C., Urra, X., Jensen- ius, J.C., Torres, F., Lozano, F. and Chamorro, A. (2010) Genetically-defined deficiency of mannose-binding lectin is associated with protection after experimental stroke in mice and outcome in human stroke. PLoS ONE, 5, e8433. doi:10.1371/journal.pone.0008433 [40] Sziller, I., Babula, O., Hupuczi, P., Nagy, B., Rigó, B., Szabó, G., Papp, Z., Linhares, I.M. and Witkin, S.S. (2007) Mannose-binding lectin (MBL) codon 54 gene polymor- phism protects against development of pre-eclampsia, HELLP syndrome and pre-eclampsia-associated intrauter- ine growth restriction. Molecular Human Reproduction, 13, 281-285. doi:10.1093/molehr/gam003 [41] Boldt, A.B., Culpi, L., Tsuneto, L.T., De Souza, R., Kun, J.F. and Petzl-Erler, M.L. (2006) Diversity of the MBL2 gene in various Brazilian populations and the case of se- lection at the mannose-binding lectin locus. Human Im- munology, 67, 722-734. doi:10.1016/j.humimm.2006.05.009 [42] Pitarque, M., Von Richter, O., Oke, B., Berkkan, H., Oscarson, M. and Ingelman-Sundberg, M. (2001) Identi- fication of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: Impairment of its promoter ac- tivity. Biochemical and Biophysical Research Communi- cations, 292, 455-460. doi:10.1006/bbrc.2001.4990 [43] Fei, Y.J., Stoming, T.A., Efremov, G.D., Battacharia, R., Gonzalez-Redondo, J.M., Altay, C., Gurgey, A. and Hu- isman, T.H. (1988) Beta-thalassemia due to a T―A mu- tation within the ATA box. Biochemical and Biophysical Research Communications, 153, 741-747. doi:10.1016/S0006-291X(88)81157-4 [44] Niemann, S., Broom, W.J. and Brown, R.H. Jr. (2007) Analysis of a genetic defect in the TATA box of the SOD1 gene in a patient with familial amyotrophic lateral sclerosis. Muscle & Nerve, 36, 704-707. doi:10.1002/mus.20855
|