 American Journal of Anal yt ical Chemistry, 2011, 2, 539-572 doi:10.4236/ajac.2011.25064 Published Online September 2011 (http://www.SciRP.org/journal/ajac) Copyright © 2011 SciRes. AJAC Comparative Analysis of Protein Expression Concomitant with DNA Methyltransferase 3A Depletion in a Melanoma Cell Line Shengnan Tang1,#, Xiaoyan Liu1,#, Tonghua Li1*, Haoyue Wang2, Jiangming Sun1, Qian Qiao1, Jun Yao3, Jian Fei2 1Department of Chemistry, Tongji University, Shanghai, China 2School of Life Science & Technology, Tongji University, Shanghai, China 3School of Medicine, Fudan University, Shanghai, China E-mail: *lith@tongji.edu.cn Received March 17, 2011; revised May 3, 2011; accepted June 1, 2011 Abstract DNA methyltransferase 3A (Dnmt3a), a de novo methyltransferase, has attracted a great deal of attention for its important role played in tumorigenesis. We have previously demonstrated that melanoma is unable to grow in-vivo in conditions of Dnmt3a depletion in a mouse model. In this study, we cultured the Dnmt3a depletion B16 melanoma (Dnmt3a-D) cell line to conduct a comparative analysis of protein expression con-comitant with Dnmt3a depletion in a melanoma cell line. After two-dimensional separation, by gel elec- tro-phoresis and liquid chromatography, combined with mass spectrometry analysis (1DE-LC-MS/MS), the re-sults demonstrated that 467 proteins were up-regulated and 535 proteins were down-regulated in the Dnmt3a-D cell line compared to the negative control (NC) cell line. The Genome Ontology (GO) and KEGG pathway were used to further analyze the altered proteins. KEGG pathway analysis indicated that the MAPK signaling pathway exhibited a greater alteration in proteins, an interesting finding due to the close rela- tion-ship with tumorigenesis. The results strongly suggested that Dnmt3a potentially controls the process of tu-morigenesis through the regulation of the proteins (JNK1, p38α, ERK1, ERK2, and BRAF) involved in tu-mor-related pathways, such as the MAPK signaling pathway and melanoma pathway. Keywords: Dnmt3a, Melanoma Cell Line, 1DE-LC-MS/MS; MAPK Signaling Pathway, Melanoma Pathway 1. Introduction Malignant melanoma, one of the most aggressive of all skin cancers, exhibits a high skin cancer mortality rate [1]. Upon metastasis, the disease is incurable in most affected people as melanoma does not respond to most systemic treatments and chemotherapy drugs [2]. It is well established that malignant melanoma con- tains at least 50 genes that exhibit differential expression as abnormal methylation changes emerge in the promoter region [3-5], and that DNA methylation patterns are es- tablished and maintained by the coordinated action of three DNA methyltransferases (Dnmts). It has been de- monstrated that Dnmt1 [6,7], Dnmt3a, and Dnmt3b [8] are over-expressed in many malignant tumors [9]. Dnmt- 3a plays an important role in epigenetic modification that has attracted a great deal of attention in recent years [10,11]. It has been reported that epigenetic modification is induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase-Dnmt3a [10]. Ad- ditionally, Dnmt3a has been found to maintain DNA methylation and regulate synaptic function in adult fore- brain neurons [12]. Furthermore, it has been demon- strated that Dnmt3a-dependent non-promoter DNA me- thylation facilitates the transcription of neurogenic genes [13]. Recently, studies of Dnmt3a have focused on its effect on proliferation and apoptosis of hepatocellular carcinoma, colorectal cancer and malignant melanoma [14-16] In a previous study we demonstrated that tumor growth inhibition was mediated by Dnmt3a depletion [17]. In this study we describe the proteomic experiment and comparative analysis of protein expression in the #These authors contributed equally to this work.  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 540 Dnmt3a depletion B16 melanoma (Dnmt3a-D) cell line. The mouse B16 melanoma cell line used in our study exhibited a specific down-regulation of Dnmt3a via sta- ble transfection using a Dnmt3a-RNAi construct. We obtained the negative control (NC) cell line through use of an unrelated, non-target, shRNA expression vector. Proteins of the NC cell line and Dnmt3a-D cell line were initially separated by one-dimensional gel electrophore- sis (1DE), and the two gel tracks were subsequently split into 20 proteome fractions, respectively, and digested by trypsin. The peptide fractions were analyzed by capillary liquid chromatography and high accuracy mass spectro- metric acquisition on a LTQ-Orbitrap (Thermo Scientific Germany) in the MS/MS mode using various and com- plementary fragmentation modes. The results of the comparative proteomics demonstrated that Dnmt3a de- pletion affects a large number of proteins. The Genome Ontology and KEGG pathway were used to group these altered proteins according to respective cellular compo- nents, molecular functions, and pathway. The suppres- sors of skin tumour development JNK1 and p38α [18] were found to be up-regulated in the MAPK signaling pathway of the Dnmt3a-D cell line. The oncogene BRAF was down-regulated in the melanoma pathway of the Dnmt3a-D cell line. These results strongly suggested that Dnmt3a depletion potentially inhibits melanoma tumori- genesis by regulating the proteins involved in tumor- related pathways. 2. Materials and Methods 2.1. Cell Culture The B16 cells were purchased from the American Type Culture Collection (Manassas, VA), cultured in DMEM supplemented with 10% FBS, and maintained in a hu- midified incubator at 37˚C and 5% CO2. The sequence of Dnmt3a shRNA was 5′ gtgcagaaacatcgaggacTTCAAG- AGAgtcctcgatgtttctg- cac 3′. A non-target shRNA with the sequence 5′gcaagtctaaccaacgcgt TTCAAGAGAacg- cgttggtt - agacttgc 3′ was used as negative control (NC). The process of small hairpin RNA (shRNA) RNAi was performed as previously described [17]. At this point in the procedure we chose the B16 cells with Dnmt3a de- pletion (Dnmt3a-D) for further experiments. 2.2. Protein Extraction and Measurement After harvesting the cells, the following experimental pro- cess was conducted (Figure 1). The cells were washed 2-3 times in ice-cold phosphate-buffered saline (PBS), lysed in 900 μL RIPA lysis buffer (Bi Yun Tian Bio- technology Research Institute, China) plus 10 μL 100 mM phenylmethanesulfonyl fluoride (PMSF) for ap- proximately 10 - 20 min. The samples were incubated for 2 s in an ice bath and exposed to an ultrasonic power of less than 70 W, and then centrifuged for 45 min at 15,000 rpm at 4˚C. The supernatants were then collected into eppendorf tubes for the next measurement. The total protein of the cells was measured using the BCA Protein Assay (Bi Yun Tian Biotechnology Re- search Institute, China) with bovine serum albumin as a standard recommended by the manufacturer. The protein was stored at –80˚C until performance of isoelectro- fo- cusing (IEF). 2.3. 1-DE and In-Gel Tryptic Digestion In order to conduct proteomic analysis on proteins expres- sed at a low level, shotgun proteomics based on 1-DE separation of total protein was evaluated in addition to the analysis of peptide mixtures produced by tryptic di- gestion of proteins in gel fragments by LC-MS/MS. Equal amounts of NC cell protein and Dnmt3a-D cell protein (40 μg) were prepared in an identical manner. Respective samples were separated using small analyti- cal immobilized pH gradient (IPG) strips (7 cm, 3 - 10 pH gradient; Bio-Rad). The proteins were electro-fo- cused by initially using a voltage of 8 V/cm for the stacking gel, and subsequently increasing the voltage to 15 V/cm for the separating gel. Figure 1. The experimental process of proteomic methods used in this study.  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 541 Gel lanes were excised from the SDS-PAGE gels us- ing a razor blade and were divided into 20 slices accord- ing to the distribution of protein (Figure 2). The proteins were separated by 12% resolving gel. Each slice was then further divided into approximately 1 mm3 pieces and placed into an eppendorf tube. The proteins were deoxidized, alkylated, dehydrated and digested. The pep- tide mixtures were extracted with frequent vortexing at 37˚C for 30 min. Samples were evaporated to dryness and stored at –20˚C until MS analysis. 2.4. LC-MS/MS Analysis In recent years, the high performance and sensitivity of the linear quadrupole ion trap-orbitrap (LTQ-Orbitrap) mass spectrometer has interested researchers due to the capacity for top-down analysis of complete protein from tissue, body fluid, and cells [19,20]. The sample was separated using online reverse-phase nanoscale capillary liquid chromatography, and then analyzed by electrospray tandem mass spectrometry. The peptide mixtures loaded and desalted on a C18 trap col- umn (0.5 mm diameter, 2 mm, MICHROM,USA) using a Tempo 1D nanoLC system, then separated on a reverse- phase MagicTM C18 column (100 µm diameter, 15 cm, MICHROM,USA), using a 120 min linear gradient of mobile phase A (0.875% ACN/0.125% FA/99% H2O) at a flow rate of 500 nL/min. The eluent was analyzed on a Figure 2. SDS-PAGE patterns of NC cell proteins and Dnmt3a - D cell proteins. LTQ-orbitrap mass spectrometer (Thermo Electron, Bre- men, Germany) equipped with a heated Desolvation Ch- amber Interface set to 200˚C and operated using XCali- bur software. The LTQ-orbitrap operated in positive ion mode to survey full scan mass spectra (from m/z 385 to 2000). The most intense ten ions were selected for tandem mass spectrometry and the lock-mass option was used as de- scribed in previously published reports [21]. 2.5. Identification and Analysis of Proteins SEQUEST (V.28 (rev.12) 1998-2007) was used to iden- tify proteins based on MS/MS spectra with Bioworks so- ftware (Rev.3.3.1 SP1, Thermo Scientific). We searched both forward and reverse sequences against the Swiss- port Mouse 090303 database in order to estimate the number of identifications that were false-positive in the sample. The parameters for the SEQUEST search were com- prised of the following criteria: Enzyme, trypsin; Missed cleavage sites, 2; fragment tolerance, 1.0 Da; peptide to- lerance, 50.00 ppm; Numerical results, 250; Ion and Ion Series Calculated, b ion and y ion; Peptide matches, 10; Report duplicate peptide matches, 10. Oxidation of me- thionine, methylation of lysine, as well as phosphoryla- tion of serine, threonine, and tyrosine were specified as variable modifications. We inputted the DAT files from SEQUEST to the Trans-Proteomic Pipeline (TPP) v4.2 JETSTREAM rev 1 (ISB/SPC Proteomics Tools) and searched by allocat- ing all DAT files in the biological sample. The parame- ters for peptide identification probability and protein identification probability were both set at 0.95. The results were exported and the proteins found to be altered in the Dnmt3a-D cell line as compared to the NC cell line were extracted by a program developed in-house. GO terms of altered proteins for cellular component and molecular function were searched and checked by Swiss-Prot (http://expasy.org/sprot/), NCBI (http://www. ncbi.nlm.nih.gov) and PIR (http://pir.georgetown.edu/ pirwww/index.shtml). The pathways of altered proteins were established using the KEGG mapper (http://www. genome.jp/kegg/.html). Demonstration of the role of al- tered proteins in metabolic channels and signal transduc- tion pathways was considered significant. 3. Results and Discussion 3.1. Cell Harvest Growth conditions of the two cell lines were optimized in our previously reported experiment [17]. Cultured cells were harvested at a confluence level of 90% (Fig-  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 542 ure 3(a)). Western blot analysis was employed to test the effi- ciency and specificity of the Dnmt3a-RNAi. The results demonstrated that stably transfected B16 Dnmt3a-D cells exhibited a remarkably low level of Dnmt3a expression. No change in Dnmt3a expression was observed in the control NC shRNA transfected cells. The expression of Dnmt3b was not affected in the NC or Dnmt3a-D cells, which is in concordance with our findings in our previ- ous experiment (see Figure 3 (b) in [17]). 3.2. Difference of Protein Expression between NC and Dnmt3a-D Cell Lines The NC and Dnmt3a-D cell line proteins identified were compared to determine their overlap (Figure 3(b)). There were 2413 proteins in common between the NC and Dnmt3a-D cell lines, and 467 were unique to the Dnmt3a-D cell line and were considered to be up-regu- lated proteins in the Dnmt3a-D cell line (Supplementary Materials, Table S1). There were 535 proteins unique to the NC cell line considered to be down-regulated pro- teins in the Dnmt3a-D cell line (Supplementary Materials, Table S2). The evidence suggests that Dnmt3a poten- tially either directly or indirectly regulates the expression of altered proteins. 3.3. Genome Ontology of Altered Proteins In order to evaluate the effect of Dnmt3a depletion on protein expression and to explore the mechanism of Dnmt3a in tumorigenesis, the altered proteins were the principal targets for analysis. The genome ontology cel- lular localization of altered proteins was as follows: 35% of up-regulated proteins were localized to the cytoplasm, 24% to the nucleus, and 13% to the mitochondrion. The remainder of the up-regulated proteins were classified as golgi, plasma membrane, endoplasmic reticulum, cy- toskeleton, extracellular, ribosome, others, and unclassi- fied (28%). The cellular localization of down-regulated proteins was found to be similar to the cellular localiza- tion of the up-regulated proteins (Figure 4(a), (b)). This result demonstrated that the altered proteins are primarily concentrated in the cytoplasm, nucleus, and mitochon- dria. Among 467 up-regulated proteins, 161 exhibited an activity function (Figure 4(c)). Among the 535 down- regulated proteins, 178 exhibited an activity function (Figure 4(d)). There were a large number of altered proteins that ex- hibited catalytic activity and transferase activity, and they played important roles in the methylation, acetyla- tion, glycosylation, and other epigenetic modifications of (a) (b) Figure 3. Cell Harvest and overlap of proteins between two cell lines. (a)The confluence of NC and Dnmt3a-D cell was more than 90% after two days under microscope. (b)The proteins of NC and Dnmt3a-D cell line identified, and overlap of proteins between the two cell lines.  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 543 (a) (b) (c) (d) Figure. 4 Genome Ontology of altered Proteins. (A) Genome ontology cellular localization of up-regulated proteins. (B) Ge- nome ontology cellular localization of down-regulated proteins. (C) Genome ontology molecular function of up-regulated proteins. (D) Genome ontology molecular function of down-regulated proteins. the proteins. Another function that attracted our attention was phosphotransferase activity, which is essential for protein phosphorylation. It is well understood that pro- tein phosphorylation is closely related to a variety of biological processes, such as DNA damage and repair [22], transcriptional regulation [23], signal transduction, and the regulation of apoptosis [24,25]. 3.4. Pathway of Altered Proteins The altered proteins were grouped according to their respective KEGG pathway (Figure 5). The pathways were arranged according to the number of altered pro- teins. The pathways that included less than 10 altered proteins were not shown. The four pathways with the largest number of altered proteins were metabolic path- ways (71 proteins), endocytosis (21 proteins), mitogen activated protein kinase signaling pathway (MAPK sig- naling pathway, 19 proteins) and pathways in cancer (17 proteins). Among these four pathways, the MAPK sig- naling pathway, closely related to the cancer and mela- noma pathway, and which plays a key role in melanoma tumorigenesis, attracted our attention. The locations of altered proteins in the two pathway maps were estab- lished using the KEGG mapper. 3.4.1. MAPK Signaling Pathway The altered proteins involved in the MAPK signaling pathway are listed in Table 1. The locations of the al- tered proteins involved in the MAPK signaling pathway are marked by pink boxes (up-regulated proteins) and yellow boxes (down-regulated proteins) as shown in Figure 6. The MAPKs belong to a family of highly conserved kinases that convert extracellular signals to intracellular responses. MAPKs are unique to eukaryotes and are im- portant signal transducing enzymes regulated by a phos- phorylation cascade. Two upstream protein kinases are activated in the series that leads to the activation of a MAP kinase; additional kinases may also be required  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 544 upstream of this three-kinase module. After activation, MAPKs phosphorylate specific serine and threonine residues of target substrates, including other protein kinases and many transcription factors. MAPKs are switched off by both generic phosphatases and dual- specificity phosphatases and are further regulated by scaffold proteins, which are usually specific for each of the three major mammalian MAPK pathways, including extracellular signal-regulated kinase (ERK), c-Jun N- terminal kinase (JNK), and p38 MAPK [18,26,27]. Figure 5. The altered proteins were grouped by KEGG pathway. Table 1. The altered proteins involved in MAPK signaling pathway. Location in Pathway Gen Bank ID Gene name Synonyms Name of Altered Protein; ↑: Up-regulated protein; ↓: Down-regulated protein 18747 Prkaca Pkaca cAMP-dependent protein kinase catalytic subunit alpha ↑ PKA 18749 Prkacb Pkacb cAMP-dependent protein kinase catalytic subunit beta ↑ 26413 Mapk1 Erk2, Mapk, Prkm1Mitogen-activated protein kinase 1/ERK2 ↑ ERK 26417 Mapk3 Erk1, Prkm3 Mitogen-activated protein kinase 3/ERK1 ↑ Ppp3c 19058 Ppp3r1 Cnb Calcineurin subunit B type 1 ↑ CrkII 12929 Crkl Crkol Crk-like protein ↑ HGK 26921 Map4k4 Nik Mitogen-activated protein kinase kinase kinase kinase 4 ↑ JNK 26419 Mapk8 Jnk1, Prkm8 Mitogen-activated protein kinase 8/JNK1 ↑ Cdc42Rac 12540 Cdc42 / Cell division control protein 42 homolog precursor ↑ MKK3 26397 Map2k3 Mkk3, Prkmk3 Dual specificity mitogen-activated protein kinase kinase 3 ↑ P38 26416 Mapk14 Crk1,Csbp1,Csbp2Mitogen-activated protein kinase 14/ p38α↑ PP2CB 19043 Ppm1b Pp2c2, Pppm1b Protein phosphatase 1B ↑ TAB1 66513 Tab1 Map3k7ip1 TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 ↑ Rap1 109905 Rap1a Krev-1 Ras-related protein Rap-1A precursor ↓ RafB 109880 Braf B-raf Serine/threonine-protein kinase B-raf ↓ cPLA2 18783 Pla2g4a Cpla2, Pla2g4 Cytosolic phospholipase A2 ↓ CASP 12367 Casp3 Cpp32 Caspase-3 precursor ↓ ECSIT 26940 Ecsit Sitpec mitochondrial precursor ↓ MKK6 26399 Map2k6 Prkmk6, Sapkk3 Dual specificity mitogen-activated protein kinase kinase 6 ↓  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 545 Figure 6. The location of altered proteins involved in MAPK signaling pathway from KEGG database. In recent years, it has been demonstrated that the MAPK signaling pathway plays a key role in the cell cycle and gene expression regulation of various cells. MAP kinases are major components of the pathways that control embryogenesis, cell differentiation, cell prolif- eration, and cell death. Current research on the three pathways is described in detail, and offers insight into the mechanism of the pathways. Much of the review highlights research into the JNK and p38 MAPK path- ways, stress activated protein kinase pathways that are also often deregulated in cancer. JNKs and p38 MAPKs are activated by environmental and genotoxic stress and are associated with tumorigenesis in humans and mice. The function of JNKs and p38 MAPKs in cancer devel- opment are complex and correlate with the wide range of cellular responses that they modulate [28,29]. 3.4.2. JNK Pathway Three genes encode the JNK proteins: MAPK8 encodes JNK1/ MAPK8, MAPK9 encodes JNK2/ MAPK9, and MAPK10 encodes JNK3/ MAPK10 [30]. Various JNKs differ substantially in their ability to interact with JUN, a well-established regulator of cell cycle progression [31]. A JNK2 deficiency results in elevated c-Jun phosphory- lation and stability, whereas the absence of JNK1 re- duces c-Jun phosphorylation and stability. JNK2 prefer- entially binds to c-Jun in unstimulated cells, thereby con- tributing to c-Jun degradation. In contrast, JNK1 be- comes the major c-Jun interacting kinase after cell stimulation [32]. It has been recently demonstrated that the JNK pathway is linked to p53-dependent senescence via a conditional JNK1 allele [33]. The contribution of JNK1 to tumour development has also been investigated in mouse skin carcinogenesis. JNK1-knockout mice were shown to be more susceptible to tumours of the skin [34]. These results indicate that JNK1 may act as a suppressor of skin tumour development. In this study, JNK1 was found to be up-regulated in the Dnmt3a-D cell line which is unable to grow in-v i v o, which potentially illustrates the fact that Dnmt3a depletion affects melanoma tumori- genesis by regulation of JNK1 expression. 3.4.3. p38 MAPK Pathway The p38 MAPKs are activated by the upstream MKK3 and MKK6 kinases as shown in Figure 6. There are four genes that encode p38 MAPKs: MAPK14 encodes p38α/MAPK14; MAPK11 encodes p38β/MAPK11; MA- PK12 encodes p38γ/MAPK12; and MAPK13 encodes p38δ/MAPK13 [35]. Among these, p38α was found to be up-regulated in the Dnmt3a-D cell line. Most of the pub- lished studies that have investigated p38 MAPKs refer to p38α, as p38α was highly abundant in most cell types [36]. A stress-activated protein kinase, p38α suppresses tumor formation by negatively regulating cell cycle pro-  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 546 gression and proliferation, or by inducing apoptosis [37-40]. There is evidence that indicates that p38α also may be an important regulator of differentiation programs in many cell types, including epithelial lung cells and em- bryonic stem cells [41,42]. Moreover, p38α directly ph- osphorylates and modulates the activity of several tran- scription factors involved in tissue-specific differentia- tion. The differentiation-inducing activity of p38α may be related to tumour suppression, as p38α activation triggers a high level of specific differentiation and lowers transformed phenotypes in renal carcinoma and colon cancer cell lines compared to cancer cell lines in which p38α has not been activated [43-45]. The function of p38α as a tumour suppressor in-vivo has been observed in a mouse model [18]. These results provide a reason- able explanation as to why the Dnmt3a-D cell line with p38α up-regulated is unable to grow in-vivo. Dnmt3a depletion potentially directly or indirectly regulates the expression of p38α. 3.4.4. ERK Pathway ERK1 and ERK2 are ubiquitously expressed, although their relative abundance in tissues is variable. They are stimulated to a certain extent by a vast number of ligands and cellular perturbations, and there is evidence of some cell type specificity [46]. They are highly expressed in postmitotic neurons and other highly differentiated cells [47], and in these cells they are often involved in adap- tive responses, such as long-term potentiation [48,49]. The receptor tyrosine kinase uses the ERK1 and ERK2 cell membrane signaling pathway [50-52]. Stimulation of these receptors by the appropriate ligand results is an increase in the catalytic activity of the receptor and sub- sequent autophosphorylation on tyrosine residues. Phos- phorylation of these receptors results in the formation of multiprotein complexes whose organization dictates fur- ther downstream signaling events. The ERK pathway is a major pathway involved in the control of growth signals, cell survival, and invasion. ERK acts as a central point where multiple signaling pathways coalesce to drive transcription, and it plays a critical role in the pathway downstream of Ras, Raf, and MEK. Melanomas are known to harbour activation mu- tations for both Ras and BRAF, suggesting that the downstream effector ERK may be playing a key role in the oncogenic behaviour of these tumours. In-vitro stud- ies have demonstrated that melanoma cell lines and tu- mour tissues exhibit high constitutive ERK activity. The high constitutive ERK activity in melanoma is most likely a consequence of mutations in the upstream com- ponents of the MAPK pathway [53,54]. In this study, we found that tumour cells cannot grow normally with ERK1 and ERK2 up-regulated while upstream compo- nents B-raf are concurrently down-regulated. Accord- ingly, we suspected that the activity of ERK1 and ERK2 was limited by the down-regulation of upstream compo- nents B-raf. This may account for the ability of Dnmt3a to affect the growth of melanoma by regulating the activ- ity of ERK. 3.4.5. Melanoma Pathway The altered proteins involved in the melanoma pathway are listed in Table 2. The locations of the altered proteins involved in the melanoma pathway are indicated with pink boxes (up-regulated proteins) and yellow boxes (down-regulated proteins) in Figure 7. The figure was manually edited. BRAF plays an important role in repli- cative and oncogene-induced senescence, as indicated by the red oval. Frequent somatic mutation of BRAF is described in melanoma cell lines, such as the B16 mouse melanoma cell line, and other tumors [55,56]. BRAF is a serine/ threonine kinase that is commonly activated by a somatic point mutation in human cancer, and may provide new therapeutic opportunities for malignant melanoma [57]. Thus, we focused on BRAF in the melanoma pathway. It has been reported that PTEN deficiency combined with BRAF activation induces a melanoma in-situ like phenotype without dermal invasion [58]. In our study, Table 2. The altered proteins involved in melanoma pathway. Location in Pathway Gen Bank ID Gene name Synonyms Name of Altered Protein; ↑: Up-regulated protein; ↓: Down-regulated protein 26413 Mapk1 Erk2, Mapk, Prkm1 Mitogen-activated protein kinase 1 ↑ ERK 26417 Mapk3 Erk1, Prkm3 Mitogen-activated protein kinase 3 ↑ BRAF 109880 Braf B-raf Serine/threonine-protein kinase B-raf ↓ Raf 109880 Braf B-raf Serine/threonine-protein kinase B-raf ↓ CDK4/6 12571 Cdk6 Crk2 Cell division protein kinase 6 ↓  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 547 Figure 7. The location of altered proteins involved in melanoma pathway from KEGG database. BRAF was found to be down-regulated in the melanoma pathway of the Dnmt3a-D cell line (Figure 7). Most likely, the down-regulated expression of BRAF resulted in the suppression of tumorigenesis. Dnmt3a depletion inhibited the growth of melanoma by directly or indi- rectly regulating the expression of BRAF. 4. Conclusions This study demonstrates the varied changes in protein level, and that in the Dnmt3a-D cell line 467 proteins were up-regulated, while 535 proteins were down-regu- lated as compared to the NC cell line. GO analysis indi- cated that the altered proteins were primarily concen- trated in the cytoplasm, nucleus, and mitochondrion, and that the altered proteins exhibiting activity function were primarily classified as exhibiting catalytic activity, tran- sferase activity, and phosphotransferase activity. KEGG pathway analysis demonstrated that the MAPK signaling pathway exhibited a greater level of altered proteins, a fact that attracted our attention due to the close relation- ship with tumorigenesis. Taken together, our results strongly suggested that Dnmt3a depletion has a great impact on the expression of melanoma cell proteins. Ad- ditionally, Dnmt3a depletion may affect tumorigenesis through regulation of the proteins involved in tumor- related pathways, such as the MAPK signaling pathway and the melanoma pathway. These results indicated that the tumor-related pathways may be potentially valuable for the treatment of malignant melanoma in the future. 5. Acknowledgements The authors would like to acknowledge the financial support by the National Natural Science Foundation of China (20675057, 20705024). 6. References [1] A. J. Miller and M. C. Jr Mihm, “Melanoma,” The New England Journal of Medicine, Vol. 355, 2006, pp. 51-65. [2] A. D. Sasse, E. C. Sasse, L. G. Clark, L. Ulloa and O. A. Clark, “Chemoimmunotherapy Versus Chemotherapy for Metastatic Malignant Melanoma,” Cochrane Database System Reviews, 2007, p. D5413. [3] M. F. Paz, M. F. Fraga, S. Avila, M. Guo, M. Pollan, J. G. Herman and M. Esteller, “A Systematic Profile of Dna Methylation in Human Cancer Cell Lines,” Cancer Re- search, Vol. 63, 2003, pp. 1114-1121. [4] W. M. Gallagher, O. E. Bergin, M. Rafferty, Z. D. Kelly, I. M. Nolan, E. J. Fox, A. C. Culhane, L. McArdle, M. F. Fraga, L. Hughes, C. A. Currid, F. O’Mahony, A. Byrne, A. A. Murphy, C. Moss, S. McDonnell, R. L. Stallings, J. A. Plumb, M. Esteller, R. Brown, P. A. Dervan and D. J. Easty, “Multiple Markers for Melanoma Progression Regulated by Dna Methylation: Insights From Transcrip- tomic Studies,” Carcinogenesis, Vol. 26, No. 11, 2005, pp. 1856-1867. [5] P. A. Van der Velden, W. Zuidervaart, M. H. Hurks, S. Pavey, B. R. Ksander, E. Krijgsman, R. R. Frants, C. P. Tensen, R. Willemze, M. J. Jager and N. A. Gruis, “Ex- pression Profiling Reveals that Methylation of Timp3 is Involved in Uveal Melanoma Development,” Interna-  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 548 tional Journal of Cancer, Vol. 106, No. 4, 2003, pp. 472-479. [6] E. Li, T. H. Bestor, and R. Jaenisch, “Targeted Mutation of the Dna Methyltransferase Gene Results in Embryonic Lethality,” Cell, Vol. 69, No. 6, 1992, pp. 915-926. [7] H. Lei, S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch and E. Li, “De Novo Dna Cytosine Methyl- transferase Activities in Mouse Embryonic Stem Cells,” Development, Vol. 122, 1996, pp. 3195-3205. [8] M. Okano, D. W. Bell, D. A. Haber and E. Li, “Dna Me- thyltransferases Dnmt3a and Dnmt3B are Essential for De Novo Methylation and Mammalian Development,” Cell, Vol. 99, No, 3, 1999, pp. 247-257. [9] K. D. Robertson, E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales and P. A. Jones, “The Human Dna Methyltransferases (Dnmts) 1, 3a and 3B: Coordi- nate Mrna Expression in Normal Tissues and Overex- pression in Tumors,” Nucleic Acids Research, Vol. 27, No. 11, 1999, pp. 2291-2298. 0doi:org/10.1093/nar/27.11.2291 [10] D. L. Zheng, L. Zhang, N. Cheng, X. Xu, Q. Deng, X. M. Teng, K. S. Wang, X. Zhang, J. Huang and Z. G. Han, “Epigenetic Modification Induced by Hepatitis B Virus X Protein Via Interaction with De Novo Dna Methyltrans- ferase Dnmt3a,” Journal of Hepatology, Vol. 50, No. 2, 2009, pp. 377-387. 1doi:org/10.1016/j.jhep.2008.10.019 [11] D. V. Maltseva, A. A. Baykov, A. Jeltsch, and E. S. Gro- mova, “Impact of 7,8-Dihydro-8-Oxoguanine On Methy- lation of the Cpg Site by Dnmt3a,” Biochemistry, Vol. 48, No. 6, 2009, pp. 1361-1368. [12] J. Feng, Y. Zhou, S. L. Campbell, Le T, E. Li, J. D. Sweatt, A. J. Silva and G. Fan, “Dnmt1 and Dnmt3a Maintain Dna Methylation and Regulate Synaptic Func- tion in Adult Forebrain Neurons,” Nature Neuroscience, Vol. 13, 2010, pp. 423-430. 2doi:org/10.1038/nn.2514 [13] H. Wu, V. Coskun, J. Tao, W. Xie, W. Ge, K. Yoshikawa, E. Li, Y. Zhang and Y. E. Sun, “Dnmt3a-Dependent Nonpromoter Dna Methylation Facilitates Transcription of Neurogenic Genes,” Science, Vol. 329, No. 5990, 2010, pp. 444- 448. [14] J. Nawarak, R. Huang-Liu, S. H. Kao, H. H. Liao, S. Sinchaikul, S. T. Chen and S. L. Cheng, “Proteomics Analysis of a375 Human Malignant Melanoma Cells in Response to Arbutin Treatment,” Biochim Biophys Acta, Vol. 1794, No. 2, 2009, pp. 159-167. [15] Y. Zhang, Y. Gao, G. Zhang, S. Huang, Z. Dong, C. Kong, D. Su, Du J, S. Zhu, Q. Liang, J. Zhang, J. Lu, and B. Huang, “Dnmt3a Plays a Role in Switches Between Doxorubicin-Induced Senescence and Apoptosis of Co- lorectal Cancer Cells,” International Journal of Cancer, Vol. 128, No. 3, 2010, pp. 551-561. [16] Z. Zhao, Q. Wu, J. Cheng, X. Qiu, J. Zhang and H. Fan, “Depletion of Dnmt3a Suppressed Cell Proliferation and Restored Pten in Hepatocellular Carcinoma Cell,” Jour- nal of Biomedicine and Biotechnology, Vol. 2010, Vol. 2010, pp. 737535-737545. 3doi:org/10.1155/2010/737535 [17] T. Deng, Y. Kuang, L. Wang, J. Li, Z. Wang, and J. Fei, “An Essential Role for Dna Methyltransferase 3a in Melanoma Tumorigenesis,” Biochemical and Biophysical Reseasch Communications, Vol. 387, No. 3, 2009, pp. 611-616. 4doi:org/10.1016/j.bbrc.2009.07.093 [18] E. F. Wagner and A. R. Nebreda, “Signal Integration by Jnk and P38 Mapk Pathways in Cancer Development,” Nature Reviews Cancer, Vol. 9, 2009, pp. 537-549. 5doi:org/10.1038/nrc2694 [19] A. Zougman, B. Pilch, A. Podtelejnikov, M. Kiehntopf, C. Schnabel, C. Kumar and M. Mann, “Integrated Analysis of the Cerebrospinal Fluid Peptidome and Proteome,” Journal of Proteome Research, Vol. 7, 2008, pp. 386-399. 6doi:org/10.1021/pr070501k [20] B. Macek, L. F. Waanders, J. V. Olsen and M. Mann, “Top-Down Protein Sequencing and Ms3 On a Hybrid Linear Quadrupole Ion Trap-Orbitrap Mass Spectrome- ter,” Mol Cell Proteomics, Vol. 5, 2006, pp. 949-958. [21] J. V. Olsen, L. M. de Godoy, G. Li, B. Macek, P. Mortensen, R. Pesch, A. Makarov, O. Lange, S. Horning and M. Mann, “Parts Per Million Mass Accuracy On an Orbitrap Mass Spectrometer Via Lock Mass Injection Into a C-Trap,” Mol Cell Proteomics, Vol. 4, 2005, pp. 2010-2021. 7doi:org/10.1074/mcp.T500030-MCP200 [22] S. K. Binz, A. M. Sheehan and M. S. Wold, “Replication Protein a Phosphorylation and the Cellular Response to Dna Damage,” DNA Repair (Amst), Vol. 3, No. 8-9, 2004, pp. 1015-1024. 8doi:org/10.1016/j.dnarep.2004.03.028 [23] M. S. Kobor and J. Greenblatt, “Regulation of Transcrip- tion Elongation by Phosphorylation,” Biochim Biophys Acta, Vol. 1577, No. 2, 2002, pp. 261-275. [24] H. Ohkura, “Phosphorylation: Polo Kinase Joins an Elite Club,” Current Biology, Vol. 13, 2003, pp. R912-R914. 9doi:org/10.1016/j.cub.2003.11.012 [25] P. P. Ruvolo, X. Deng and W. S. May, “Phosphorylation of Bcl2 and Regulation of Apoptosis,” Leukemia, Vol. 15, No. 4, 2001, pp. 515-522. [26] L. Chang and M. Karin, “Mammalian Map Kinase Sig- nalling Cascades,” Nature, Vol. 410, No. 6824, 2001, pp. 37-40. [27] J. M. Kyriakis and J. Avruch, “Mammalian Mitogen- Activated Protein Kinase Signal Transduction Pathways Activated by Stress and Inflammation,” Physiological Reviews, Vol. 81, No. 2, 2001, pp. 807-869. [28] A. R. Nebreda and A. Porras, “P38 Map Kinases: Beyond the Stress Response,” Trends in Biochemical Sciences, Vol. 25, No. 6, 2000, pp. 257-260. 1doi:org/10.1016/S0968-0004(00)01595-4 [29] M. Karin and E. Gallagher, “From Jnk to Pay Dirt: Jun Kinases, their Biochemistry, Physiology and Clinical Importance,” Iubmb Life, Vol. 57, No. 4-5, 2005, pp. 283-295.1doi:org/10.1080/15216540500097111 [30] S. Gupta, T. Barrett, A. J. Whitmarsh, J. Cavanagh, H. K. Sluss, B. Derijard and R. J. Davis, “Selective Interaction of Jnk Protein Kinase Isoforms with Transcription Fac- tors,” EMBO Journal, Vol. 15, 1996, pp. 2760-2770. [31] R. Eferl and E. F. Wagner, “Ap-1: A Double-Edged  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 549 Sword in Tumorigenesis,” Nature Reviews Cancer, Vol. 3, 2003, pp. 859-868. 1doi:org/10.1038/nrc1209 [32] K. Sabapathy, K. Hochedlinger, S. Y. Nam, A. Bauer, M. Karin and E. F. Wagner, “Distinct Roles for Jnk1 and Jnk2 in Regulating Jnk Activity and C-Jun-Dependent Cell Proliferation,” Molecular Cell, Vol. 15, No. 5, 2004, pp. 713-725. 1doi:org/10.1016/j.molcel.2004.08.028 [33] M. Das, F. Jiang, H. K. Sluss, C. Zhang, K. M. Shokat, R. A. Flavell and R. J. Davis, “Suppression of P53-De- pendent Senescence by the Jnk Signal Transduction Pathway,” Proceedingof the Natlonal Academy Sciences of the U S A, Vol. 104, 2007, pp. 15759-15764. 1doi:org/10.1073/pnas.0707782104 [34] D. Morse, L. E. Otterbein, S. Watkins, S. Alber, Z. Zhou, R. A. Flavell, R. J. Davis and A. M. Choi, “Deficiency in the C-Jun Nh2-Terminal Kinase Signaling Pathway Con- fers Susceptibility to Hyperoxic Lung Injury in Mice,” American Journal of Physiology-Lung Cellular Molecu- lar Physiology, Vol. 285, No. 1, 2003, pp. L250-L257. [35] J. C. Lee, J. T. Laydon, P. C. McDonnell, T. F. Gallagher, S. Kumar, D. Green, D. McNulty, M. J. Blumenthal, J. R. Heys, S. W. Landvatter and Al. Et, “A Protein Kinase Involved in the Regulation of Inflammatory Cytokine Biosynthesis,” Nature, Vol. 372, 1994, pp. 739-746. [36] K. Ono and J. Han, “The P38 Signal Transduction Path- way: Activation and Function,” Cellular Signalling, Vol. 12, No. 1, 2000, pp. 1-13. 1doi:org/10.1016/S0898-6568(99)00071-6 [37] D. V. Bulavin and AJ Jr Fornace, “P38 Map Kinase’s Emerging Role as a Tumor Suppressor,” Advances In Cancer Research, Vol. 92, 2004, pp. 95-118. 1doi:org/10.1016/S0065-230X(04)92005-2 [38] I. Dolado, A. Swat, N. Ajenjo, G. De Vita, A. Cuadrado and A. R. Nebreda, “P38Alpha Map Kinase as a Sensor of Reactive Oxygen Species in Tumorigenesis,” Cancer Cell, Vol. 11, No. 2, 2007, pp. 191-205. [39] F. B. Engel, M. Schebesta, M. T. Duong, G. Lu, S. Ren, J. B. Madwed, H. Jiang, Y. Wang and M. T. Keating, “P38 Map Kinase Inhibition Enables Proliferation of Adult Mammalian Cardiomyocytes,” Genes Development, Vol. 19, 2005, pp. 1175-1187. 1doi:org/10.1101/gad.1306705 [40] L. J. Hui, L. Bakiri, A. Mairhorfer, N. Schweifer, C. Haslinger, L. Kenner, V. Komnenovic, H. Scheuch, H. Beug and E. F. Wagner, “P38X3B1; Suppresses Normal and Cancer Cell Proliferation by Antagonizing the JnkX2013;C-Jun Pathway,” Nature Genetics, Vol. 39, 2007. [41] A. Jones and A. Charlton, “Determination of Metalde- hyde in Suspected Cases of Animal Poisoning Using Gas Chromatography-Ion Trap Mass Spectrometry,” Journal Agric Food Chemistry, Vol. 47, No. 11, 1999, pp. 4675-4677. 1doi:org/10.1021/jf990026d [42] X. Wang, C. H. Goh and B. Li, “P38 Mitogen-Activated Protein Kinase Regulates Osteoblast Differentiation through Osterix,” Endocrinology, Vol. 148, 2007, pp. 1629-1637. [43] C. Hikita, S. Vijayakumar, J. Takito, H. Erdjument- Bromage, P. Tempst and Q. Al-Awqati, “Induction of Terminal Differentiation in Epithelial Cells Requires Po- lymerization of Hensin by Galectin 3,” Journal of Cell Biology, Vol. 151, No. 6, 2000, pp. 1235-1246. 1doi:org/10.1083/jcb.151.6.1235 [44] P. Ordonez-Moran, M. J. Larriba, H. G. Palmer, R. A. Valero, A. Barbachano, M. Dunach, A. G. de Herreros, C. Villalobos, M. T. Berciano, M. Lafarga and A. Munoz, “Rhoa-Rock and P38Mapk-Msk1 Mediate Vitamin D Effects On Gene Expression, Phenotype and Wnt Path- way in Colon Cancer Cells,” Journal of Cell Biology, Vol. 183, No. 4, 2008, pp. 697-710. 2doi:org/10.1083/jcb.200803020 [45] G. J. Finn, B. S. Creaven and D. A. Egan, “Daphnetin Induced Differentiation of Human Renal Carcinoma Cells and its Mediation by P38 Mitogen-Activated Protein Kinase,” Biochemical Pharmacology, Vol. 67, 2004, pp. 1779-1788. 2doi:org/10.1016/j.bcp.2004.01.014 [46] T. S. Lewis, P. S. Shapiro and N. G. Ahn, “Signal Trans- duction through Map Kinase Cascades,” Advances In Cancer Research, Vol. 74, 1998, pp. 49-139. 2doi:org/10.1016/S0065-230X(08)60765-4 [47] T. G. Boulton, S. H. Nye, D. J. Robbins, N. Y. Ip, E. Radziejewska, S. D. Morgenbesser, R. A. DePinho, N. Panayotatos, M. H. Cobb and G. D. Yancopoulos, “Erks: A Family of Protein-Serine/Threonine Kinases that are Activated and Tyrosine Phosphorylated in Response to Insulin and Ngf,” Cell, Vol. 65, 1991, pp. 663-675. [48] C. M. Atkins, J. C. Selcher, J. J. Petraitis, J. M. Trzas- kosand J. D. Sweatt, “The Mapk Cascade is Required for Mammalian Associative Learning,” Nature Neuroscience, Vol. 1, 1998, pp. 602-609. 2doi:org/10.1038/2836 [49] R. Brambilla, N. Gnesutta, L. Minichiello, G. White, A. J. Roylance, C. E. Herron, M. Ramsey, D. P. Wolfer, V. Cestari, C. Rossi-Arnaud, S. G. Grant, P. F. Chapman, H. P. Lipp, E. Sturani and R. Klein, “A Role for the Ras Signalling Pathway in Synaptic Transmission and Long- Term Memory,” Nature, Vol. 390, 1997, PP. 281-286. [50] G. Pearson, F. Robinson, Gibson T. Beers, B. E. Xu, M. Karandikar, K. Berman and M. H. Cobb, “Mitogen-Ac- tivated Protein (Map) Kinase Pathways: Regulation and Physiological Functions,” Endocrine Reviews, Vol. 22, No. 2, 2001, pp. 153-183. 2doi:org/10.1210/er.22.2.153 [51] T. Pawson and J. D. Scott, “Signaling through Scaffold, Anchoring, and Adaptor Proteins,” Science, Vol. 278, No. 5346, 1997, pp. 2075-2080. [52] T. Hunter, “Protein Kinases and Phosphatases: The Yin and Yang of Protein Phosphorylation and Signaling,” Cell, Vol. 80, No. 2, 1995, pp. 225-236. [53] A. M. Aronov, C. Baker, G. W. Bemis, J. Cao, G. Chen, P. J. Ford, U. A. Germann, J. Green, M. R. Hale, M. Ja- cobs, J. W. Janetka, F. Maltais, G. Martinez-Botella, M. N. Namchuk, J. Straub, Q. Tang and X. Xie, “Flipped Out: Structure-Guided Design of Selective Pyrazolylpyrrole Erk Inhibitors,” Journal of Medicinal Chemistry, Vol. 50, No. 5, 2007, pp. 1280-1287. 2doi:org/10.1021/jm061381f [54] K. S. Smalley, “A Pivotal Role for Erk in the Oncogenic Behaviour of Malignant Melanoma?” International  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 550 Journal of Cancer, Vol. 104, No. 5, 2003, pp. 527-532. 2doi:org/10.1002/ijc.10978 [55] H. Gear, H. Williams, E. G. Kemp and F. Roberts, “Braf Mutations in Conjunctival Melanoma,” Invest Ophthal- mology Visual Science, Vol. 45, No. 8, 2004, pp. 2484- 2488. 2doi:org/10.1167/iovs.04-0093 [56] V. K. Goel, A. J. Lazar, C. L. Warneke, M. S. Redston and F. G. Haluska, “Examination of Mutations in Braf, Nras, and Pten in Primary Cutaneous Melanoma,” Jour- nal of Investigative Dermatology, Vol. 126, 2006, pp. 154-160. [57] H. Davies, G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Mar- shall, R. Wooster, M. R. Stratton and P. A. Futreal, “Mu- tations of the Braf Gene in Human Cancer,” Nature, Vol. 417, No. 6892, 2002, pp. 949-954. [58] R. S. Lo and O. N. Witte, “Transforming Growth Fac- tor-Beta Activation Promotes Genetic Context-Dependent Invasion of Immortalized Melanocytes,” Cancer Re- search, Vol. 68, No. 4248, 2008, pp. 4248-4257. 2doi:org/10.1158/0008-5472.CAN-07-5671 Supporting Information Available There are 467 proteins were up-regulated (Supplementary Materials, Table S1) and 535 proteins were down-regulated (Supplementary Materials, Table S2) in the Dnmt3a-D cell line as compared with the NC cell line. Supplementary ma- terials Table S3 including the abbreviated words and their full name. Table S1 protein Protein probability num unique peps precursor ion charge peptide sequence sp|Q8CGP0|H2B3B_MOUSE 0.9997 2 2 AMGIMNSFVNDIFER sp|Q8QZT1|THIL_MOUSE 1 18 2 ASKPTLNEVVIVSAIR sp|Q9QZQ8|H2AY_MOUSE 1 9 2 AASADSTTEGTPTDGFTVLSTK sp|O88544|CSN4_MOUSE 1 11 2 AIQLSGTEQLEALK sp|Q9CR16|PPID_MOUSE 1 15 2 DGSGDSHPDFPEDADIDLK sp|Q9WVJ2|PSD13_MOUSE 1 10 2 LYENFISEFEHR sp|P40336|VP26A_MOUSE 1 11 2 ELALPGELTQSR sp|Q921H8|THIKA_MOUSE 1 12 2 AEELGLPILGVLR sp|P63085|MK01_MOUSE 1 12 2 FDM[147]ELDDLPK sp|Q9QWL7|K1C17_MOUSE 1 5 2 LLEGEDAHLTQYK sp|Q99J62|RFC4_MOUSE 1 10 2 AITFLQSATR sp|Q60737|CSK21_MOUSE 1 11 2 GGPNIITLADIVK sp|Q91WK2|EIF3H_MOUSE 1 11 2 LFM[147]AQALQEYNN sp|O54984|ARSA1_MOUSE 1 8 2 GM[147]NFSVVVFDTAPTGH sp|Q07417|ACADS_MOUSE 1 10 2 GISAFLVPM[147]PTPGLTLGK sp|Q99KV1|DJB11_MOUSE 1 7 2 FQDLGAAYEVLSDSEK sp|Q9DAR7|DCPS_MOUSE 1 9 2 IVFENPDPSDGFVLIPDLK sp|Q9R1T2|SAE1_MOUSE 1 9 2 AQNLNPM[147]VDVK sp|Q9D0M1|KPRA_MOUSE 1 10 2 GQDIFIIQTIPR sp|Q9JHJ0|TMOD3_MOUSE 1 7 2 DLGDYKDLDEDELLGK sp|Q9Z2K1|K1C16_MOUSE 1 4 2 TRLEQEIATYR sp|Q9Z1G3|VATC1_MOUSE 1 11 2 VAQYM[147]ADVLEDSKDK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 551 sp|P70362|UFD1_MOUSE 1 7 2 FQPQSPDFLDITNPK sp|P47811|MK14_MOUSE 1 9 2 HENVIGLLDVFTPAR sp|Q9R0U0|FUSIP_MOUSE 1 6 2 GFAYVQFEDVR sp|P63276|RS17_MOUSE 1 3 2 IAGYVTHLM[147]K sp|Q9CXW2|RT22_MOUSE 1 8 2 LM[147]TQAQLEEATR sp|Q6Y7W8|PERQ2_MOUSE 1 9 2 EVESPYEVHDYTR sp|Q9CPX6|ATG3_MOUSE 1 5 2 LWLFGYDEQR sp|P70318|TIAR_MOUSE 1 4 2 FEDVVNQSSPK sp|P05132|KAPCA_MOUSE 1 9 2 ILQAVNFPFLVK sp|Q9DBS1|TMM43_MOUSE 1 9 2 LLSDPNYGVHLPAVK sp|P54923|ADPRH_MOUSE 1 7 3 DGETIHQQLAQM[147]GDLEAIDVAR sp|Q9D7G0|PRPS1_MOUSE 1 6 2 IFSGSSHQDLSQK sp|Q9ER00|STX12_MOUSE 1 4 2 ELGSLPLPLSASEQR sp|Q9ER88|RT29_MOUSE 1 6 2 FDQPLEASTWLK sp|Q9Z0G0|GIPC1_MOUSE 1 6 2 APPLVENEEAEPSR sp|Q99LG2|TNPO2_MOUSE 1 4 2 ALVM[147]LLEVR sp|Q9CQT1|EI2BL_MOUSE 1 6 2 AGAGGPGLAALVAFVR sp|Q9WUL7|ARL3_MOUSE 1 6 2 LNVWDIGGQR sp|P47964|RL36_MOUSE 0.9975 2 2 YPM[147]AVGLNK sp|P26516|PSD7_MOUSE 1 5 2 TNDQM[147]VVVYLASLIR sp|Q8K157|GALM_MOUSE 1 4 2 VSPDGEEGYPGELK sp|P58774|TPM2_MOUSE 0.9994 1 2 ATDAEADVASLNR sp|P61963|WDR68_MOUSE 1 3 2 DM[147]FASVGADGSVR sp|Q64442|DHSO_MOUSE 1 4 2 AM[147]GAAQVVVTDLSASR sp|Q99J09|MEP50_MOUSE 1 4 2 IWDLAQQVSLNSYR sp|Q9CX34|SUGT1_MOUSE 1 6 2 DYASALETFAEGQK sp|Q9QYA2|TOM40_MOUSE 1 6 2 FVNWQVDGEYR sp|Q9QYJ3|DNJB1_MOUSE 1 6 2 DGSDVIYPAR sp|P68181|KAPCB_MOUSE 0.9987 1 2 ILQAVEFPFLVR sp|Q60766|IRGM_MOUSE 1 5 2 DLSTSVLSEVR sp|Q64213|SF01_MOUSE 1 4 3 ILRPWQSSETR sp|Q8R323|RFC3_MOUSE 1 7 2 AIYHLEAFVAK sp|Q8R574|KPRB_MOUSE 1 5 2 IFVM[147]ATHGLLSSDAPR sp|Q99JB2|STML2_MOUSE 1 4 2 AEQINQAAGEASAVLAK sp|Q9EQ80|NIF3L_MOUSE 1 5 2 TLM[147]QVLAFLSQDR sp|Q9JLC8|SACS_MOUSE 1 9 2 LLLVLNK sp|O09174|AMACR_MOUSE 1 5 2 GQNILDGGAPFYTTYK sp|O35435|PYRD_MOUSE 1 7 2 QTQLTTDGLPLGINLGK sp|P24288|BCAT1_MOUSE 1 5 2 HLTM[147]DDLATALEGNR sp|P70697|DCUP_MOUSE 1 5 2 LVQQM[147]LDDFGPQR sp|Q61187|TS101_MOUSE 1 5 2 ELVNLTGTIPVR sp|Q6PD19|CJ076_MOUSE 1 6 2 LQDGLDQYER sp|Q91WM2|CECR5_MOUSE 1 5 2 QM[147]LVSGQGPLVENAR sp|P18872|GNAO_MOUSE 0.9998 2 2 TTGIVETHFTFK sp|P47941|CRKL_MOUSE 1 4 2 IHYLDTTTLIEPAPR sp|Q8VDT9|RM50_MOUSE 1 5 2 DVLDFYNVPVQDK sp|Q99M71|EPDR1_MOUSE 1 3 2 ALVSYDGLNQR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 552 sp|Q9WTQ8|TIM23_MOUSE 1 3 2 NVQILNM[147]VTR sp|Q80WQ2|VAC14_MOUSE 1 4 2 DFVAQNNTM[147]QIK sp|Q9CQ92|FIS1_MOUSE 1 2 2 GLLQTEPQNNQAK sp|Q9CXE7|TMED5_MOUSE 1 4 2 LEDILESINSIK sp|Q9DC50|OCTC_MOUSE 1 5 2 AASDLQIAASTFTSFGK sp|Q9JK48|SHLB1_MOUSE 1 4 2 LAADAGTFLSR sp|Q8K0D0|PCTK2_MOUSE 1 4 2 LDSEGIELITK sp|Q99J95|CDK9_MOUSE 0.9997 3 2 LADFGLAR sp|P21278|GNA11_MOUSE 1 6 2 IATVGYLPTQQDVLR sp|Q8BMS9|RASF2_MOUSE 1 5 2 TSVFTPAYGSVTNVR sp|Q99JT2|MST4_MOUSE 1 3 2 LADFGVAGQLTDTQIK sp|O70493|SNX12_MOUSE 0.9966 2 2 TNLPIFK sp|O08915|AIP_MOUSE 1 3 2 VLELDPALAPVVSR sp|P59708|PM14_MOUSE 1 3 2 GTAYVVYEDIFDAK sp|P70404|IDH3G_MOUSE 1 6 2 ENTEGEYSSLEHESVAGVVESLK sp|Q80V26|IMPA3_MOUSE 1 3 2 EM[147]LAVAVLAAER sp|Q8BGR9|UBCP1_MOUSE 1 4 2 LDDFLELNHK sp|Q8BTZ7|GMPPB_MOUSE 1 5 2 ALILVGGYGTR sp|Q8N9S3|AHSA2_MOUSE 1 4 2 LQASPVALGVR sp|Q91YM2|GRLF1_MOUSE 1 6 2 EQLTEGEEIAQEIDGR sp|Q99JF8|PSIP1_MOUSE 1 4 2 QVDTEEAGM[147]VTAATASNVK sp|Q9CQR4|THEM2_MOUSE 1 2 2 TLAFASVDLTNK sp|Q9CXY9|GPI8_MOUSE 1 3 2 NVLITDFFGSVR sp|Q9JHI5|IVD_MOUSE 1 4 2 IGQFQLM[147]QGK sp|Q9WVL0|MAAI_MOUSE 1 2 2 VITSGFNALEK sp|O08583|THOC4_MOUSE 1 3 2 QQLSAEELDAQLDAYNAR sp|Q63844|MK03_MOUSE 1 3 2 FDM[147]ELDDLPK sp|Q00899|TYY1_MOUSE 1 3 2 TLEGEFSVTM[147]WSSDEKK sp|Q3URS9|CCD51_MOUSE 1 4 2 EDNQYLELATLEHR sp|Q8R2U6|NUDT4_MOUSE 1 2 2 LLGIFENQDR sp|P13011|ACOD2_MOUSE 0.9913 1 2 DDLYDPTYQDDEGPPPK sp|O70439|STX7_MOUSE 1 2 2 TLNQLGTPQDSPELR sp|O88543|CSN3_MOUSE 1 2 2 AM[147]DQEITVNPQFVQK sp|P47199|QOR_MOUSE 1 3 2 VFEFGGPEVLK sp|Q8BG94|COMD7_MOUSE 1 3 2 FGVTSGSSELEK sp|Q8BH69|SPS1_MOUSE 1 4 2 IIEVAPQVATQNVNPTPGATS sp|Q8CAY6|THIC_MOUSE 1 5 2 ILVTLLHTLER sp|Q8K2Z2|PRP39_MOUSE 1 4 2 DLLTGEQFIQLR sp|Q8R349|CDC16_MOUSE 1 4 2 YAEALDYHR sp|Q8VCG3|WDR74_MOUSE 1 3 2 VWDLQGSEEPVFR sp|Q921X9|PDIA5_MOUSE 1 4 2 FHISAFPTLK sp|Q99K23|UFSP2_MOUSE 1 4 2 INAYHFPDELYK sp|Q99LD9|EI2BB_MOUSE 1 4 2 FVAPEEVLPFTEGDILEK sp|Q9D2R6|CCD56_MOUSE 1 3 2 FLDELEDEAK sp|Q9D8X5|CNOT8_MOUSE 1 3 2 GGLQEVADQLDLQR sp|Q9DCA5|BXDC2_MOUSE 1 4 2 EFLIQIFSTPR sp|P21279|GNAQ_MOUSE 1 2 2 M[147]FVDLNPDSDK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 553 sp|Q8VE80|THOC3_MOUSE 0.9992 2 2 YVLGM[147]QELFR sp|Q62422|OSTF1_MOUSE 0.9989 2 2 ALYTFEPR sp|P61294|RAB6B_MOUSE 0.9979 1 2 GSDVIIMLVGNK sp|Q8R2E9|ERO1B_MOUSE 0.9969 1 2 TLLLSIFQDTK sp|O09110|MP2K3_MOUSE 1 2 2 FPYESWGTPFQQLK sp|O35658|C1QBP_MOUSE 1 2 2 AFVEFLTDEIKEEK sp|P62488|RPB7_MOUSE 1 3 2 GEVVDAVVTQVNK sp|Q59J78|MIMIT_MOUSE 1 3 2 ETSEELLPSPTATQVK sp|Q60866|PTER_MOUSE 1 3 2 GGGALVENTTTGLSR sp|Q61249|IGBP1_MOUSE 1 3 2 AAGM[147]LSQLDLFSR sp|Q62203|SF3A2_MOUSE 1 4 2 M[147]EKPPAPPSLPAGPPGVK sp|Q78HU3|F125A_MOUSE 1 2 2 GPLPSGFSAVNDPQDIK sp|Q78JW9|UBFD1_MOUSE 1 4 2 IM[147]VVGSTINDVLAVNTPK sp|Q8BFQ8|PDDC1_MOUSE 1 2 2 AIDFVDVTESNAR sp|Q8CFE2|CD027_MOUSE 1 5 2 DSPDELPVYVGTNEAK sp|Q8R1J3|ZCHC9_MOUSE 1 2 2 GM[147]SADYEDVLDVPK sp|Q8VDG7|PAFA2_MOUSE 1 4 2 TVVNVFPGGLDLM[147]TLK sp|Q8VDQ1|PTGR2_MOUSE 1 4 2 GLENM[147]GVAFQSM[147]M[147]TGG NVGK sp|Q91VE6|MK67I_MOUSE 1 3 2 GIDYSFPSLVLPK sp|Q91WE2|NIP30_MOUSE 1 3 2 GLDEDETNFLDEVSR sp|Q99LC2|CSTF1_MOUSE 1 3 2 LGM[147]ENDDTAVQYAIGR sp|Q9CQI9|MED30_MOUSE 1 2 2 IGQETVQDIVYR sp|Q9D753|EXOS8_MOUSE 1 3 2 ATTVNIGSISTADGSALVK sp|Q9D832|DNJB4_MOUSE 1 5 2 VIGYGLPFPK sp|Q9DBL1|ACDSB_MOUSE 1 5 2 SGNYYVLNGSK sp|Q9EPJ9|ARFG1_MOUSE 1 3 2 IFDDVSSGVSQLASK sp|Q9JK38|GNA1_MOUSE 1 2 2 VLGQLTETGVVSPEQFM[147]K sp|Q9WUN2|TBK1_MOUSE 1 5 2 LSSSQGTIESSLQDISSR sp|Q9Z2D8|MBD3_MOUSE 1 2 2 AFM[147]VTDDDIR sp|P18653|KS6A1_MOUSE 0.9993 1 3 DLKPENILLDEEGHIK sp|P97820|M4K4_MOUSE 1 4 2 GQNVLLTENAEVK sp|Q91YS8|KCC1A_MOUSE 0.9999 2 2 LIFQVLDAVK sp|Q921E2|RAB31_MOUSE 0.9999 2 2 GSAAAVIVYDITK sp|P0C7N9|PSMG4_MOUSE 0.9998 2 2 AAADADVSLHNFSAR sp|P59438|HPS5_MOUSE 0.9998 4 2 LLDPLVLFEPK sp|Q8C5Q4|GRSF1_MOUSE 0.9998 2 2 LGDEVDDVYLIR sp|Q9Z1B5|MD2L1_MOUSE 0.9998 2 2 YGLTLLTTTDPELIK sp|P47226|TES_MOUSE 0.9997 2 2 NVM[147]ILTNPVAAK sp|O09117|SYPL1_MOUSE 0.9992 2 2 SAFQINLNPLK sp|Q9D7H3|RTC1_MOUSE 0.999 3 2 AFVAGVLPLK sp|Q91X78|ERLN1_MOUSE 0.9969 1 2 ISEIEDAAFLAR sp|A6PWY4|WDR76_MOUSE 1 3 2 VFDSSSISSQLPLLSTIR sp|O08579|EMD_MOUSE 1 3 2 DYNDDYYEESYLTTK sp|O54784|DAPK3_MOUSE 1 3 2 ESLTEDEATQFLK sp|P33611|DPOA2_MOUSE 1 3 2 QLLSPSSFSPSATPSQK sp|P97789|XRN1_MOUSE 1 4 2 APELFSYIAK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 554 sp|Q3U1V6|UEVLD_MOUSE 1 5 2 DGVLSPSSQAQLSSR sp|Q64669|NQO1_MOUSE 1 3 2 ALIVLAHSEK sp|Q78JE5|FBX22_MOUSE 1 2 2 STYVLSNLAEVVER sp|Q810A3|TTC9C_MOUSE 1 3 2 AGVAFFHLQDYDR sp|Q8BKX1|BAIP2_MOUSE 1 2 2 EGDLITLLVPEAR sp|Q8BWR2|CA128_MOUSE 1 2 2 GLAYGLYLR sp|Q8C407|YIPF4_MOUSE 1 3 2 SAASLLVGEEFK sp|Q8CEC0|NUP88_MOUSE 1 2 2 GPSGGGVEPPLSQYQR sp|Q8CF89|TAB1_MOUSE 1 3 2 VLLQAFDVVER sp|Q8K194|SNR27_MOUSE 1 3 2 VDGSVNAYAINVSQK sp|Q8K409|DPOLB_MOUSE 1 3 2 EEM[147]LQM[147]QDIVLNEIK sp|Q8VDS4|RPR1A_MOUSE 1 2 2 SVYENDVLEQLK sp|Q8VDS8|STX18_MOUSE 1 3 2 TAVLDFVDDYLK sp|Q91WK1|SPRY4_MOUSE 1 3 2 VGLLLDYEAK sp|Q91XI1|DUS3L_MOUSE 1 4 2 ISEM[147]LLGPVPPGFVFLPK sp|Q9CR95|NECP1_MOUSE 1 3 2 ASGTGGLSLLPPPPGGK sp|Q9CY28|GTPB8_MOUSE 1 3 2 LFDPSLEDIGR sp|Q9CZX9|TMM85_MOUSE 1 2 2 GSGQGDSLYPVGYLDK sp|Q9D8M4|RL7L_MOUSE 1 4 2 TVPLTDNTVIEEHLGR sp|Q9DBX2|PHLP_MOUSE 1 3 2 EVLVLTSVR sp|Q9ER73|ELP4_MOUSE 1 4 2 NLSDTVVGLESFIGSER sp|Q9JLB2|MPP5_MOUSE 1 4 2 EGDEDNQPLAGLVPGK sp|Q9R099|TBL2_MOUSE 1 3 2 APIINIGIADTGK sp|Q9WVB0|RBPMS_MOUSE 1 3 2 ENTPSEANLQEEEVR sp|Q6ZPF4|FMNL3_MOUSE 0.9983 1 2 AAAVSLENVLLDVK sp|Q921Y2|IMP3_MOUSE 0.9999 2 2 VGPDVVTDPAFLVTR sp|Q3V009|TMED1_MOUSE 0.9998 2 2 SIQM[147]LTLLR sp|Q9CQ71|RFA3_MOUSE 0.9991 2 2 M[147]FILSDGEGK sp|Q9CQG2|MET10_MOUSE 0.9983 2 2 APEDVILALEER sp|O55125|NIPS1_MOUSE 0.9904 1 2 AGPNIYELR sp|O09114|PTGDS_MOUSE 1 3 2 AQGLTEEDIVFLPQPDK sp|P27601|GNA13_MOUSE 1 4 2 ALWEDSGIQNAYDR sp|P42669|PURA_MOUSE 1 2 2 GPGLGSTQGQTIALPAQGLIEFR sp|P62748|HPCL1_MOUSE 1 3 2 IYANFFPYGDASK sp|P97493|THIOM_MOUSE 1 2 2 TTFNVQDGPDFQDR sp|Q3U0V2|TRADD_MOUSE 1 3 2 DPALDSLAYEYER sp|Q3UE37|UBE2Z_MOUSE 1 3 2 GHFDYQSLLM[147]R sp|Q5SSK3|CQ042_MOUSE 1 2 2 LENLIDVPLIQYK sp|Q62086|PON2_MOUSE 1 4 2 FQEEENSLLHLK sp|Q8BJZ4|RT35_MOUSE 1 3 2 IPNFLHLTPVAIK sp|Q8BKF1|RPOM_MOUSE 1 4 2 QLAELLVQAVQM[147]PR sp|Q8BMZ5|SEN34_MOUSE 1 3 2 FGGDFLVYPGDPLR sp|Q8BNV1|TRM2A_MOUSE 1 4 2 DDLFTSEIFK sp|Q8BP40|PPA6_MOUSE 1 4 2 AAISQPGISEDLEK sp|Q8BVI5|STX16_MOUSE 1 2 2 QIVQSISDLNEIFR sp|Q8K2M0|RM38_MOUSE 1 4 2 TPPLGPM[147]PNEDIDVSNLER sp|Q8R322|GLE1_MOUSE 1 4 2 IEAITSSGQM[147]GSFIR 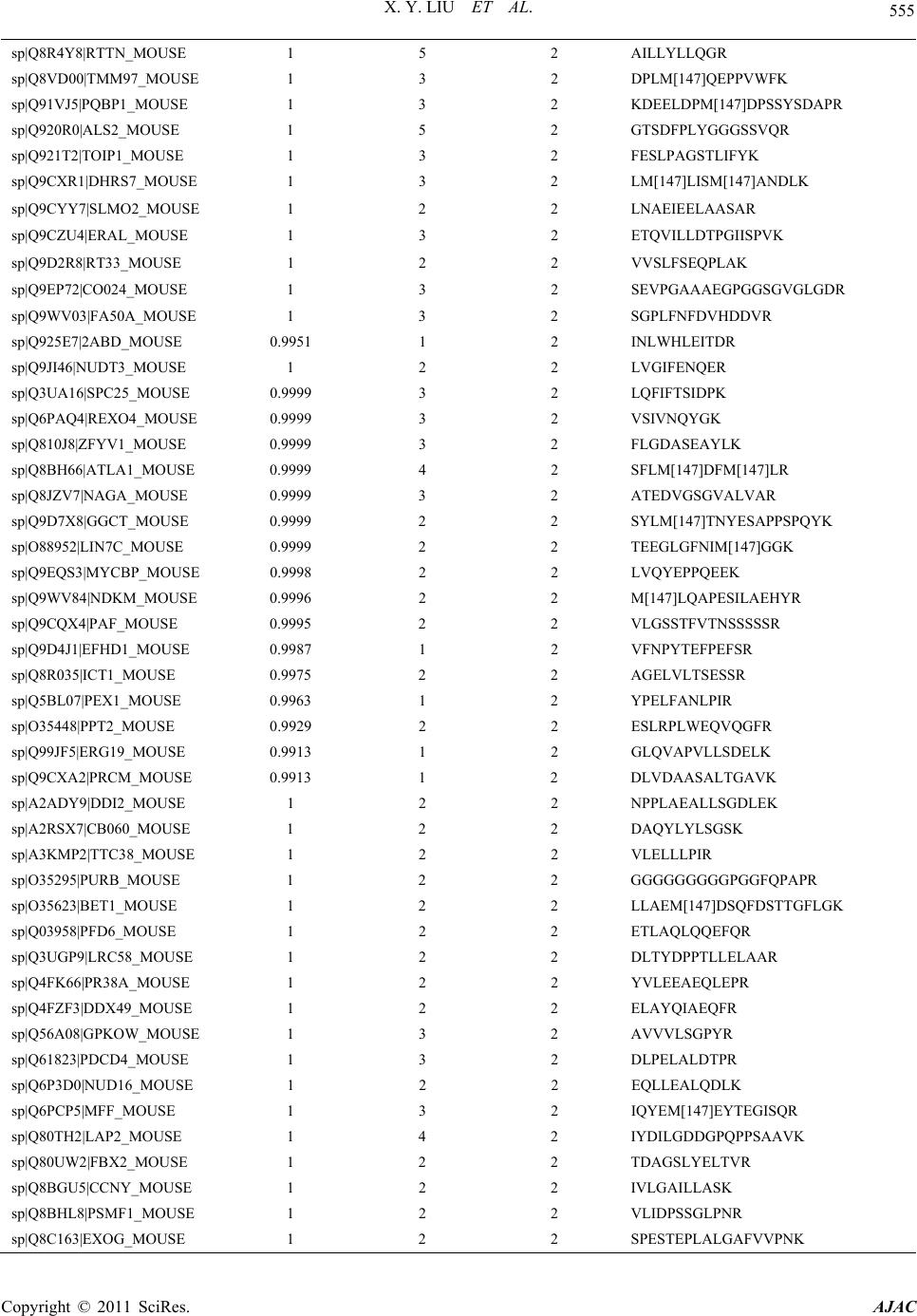 X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 555 sp|Q8R4Y8|RTTN_MOUSE 1 5 2 AILLYLLQGR sp|Q8VD00|TMM97_MOUSE 1 3 2 DPLM[147]QEPPVWFK sp|Q91VJ5|PQBP1_MOUSE 1 3 2 KDEELDPM[147]DPSSYSDAPR sp|Q920R0|ALS2_MOUSE 1 5 2 GTSDFPLYGGGSSVQR sp|Q921T2|TOIP1_MOUSE 1 3 2 FESLPAGSTLIFYK sp|Q9CXR1|DHRS7_MOUSE 1 3 2 LM[147]LISM[147]ANDLK sp|Q9CYY7|SLMO2_MOUSE 1 2 2 LNAEIEELAASAR sp|Q9CZU4|ERAL_MOUSE 1 3 2 ETQVILLDTPGIISPVK sp|Q9D2R8|RT33_MOUSE 1 2 2 VVSLFSEQPLAK sp|Q9EP72|CO024_MOUSE 1 3 2 SEVPGAAAEGPGGSGVGLGDR sp|Q9WV03|FA50A_MOUSE 1 3 2 SGPLFNFDVHDDVR sp|Q925E7|2ABD_MOUSE 0.9951 1 2 INLWHLEITDR sp|Q9JI46|NUDT3_MOUSE 1 2 2 LVGIFENQER sp|Q3UA16|SPC25_MOUSE 0.9999 3 2 LQFIFTSIDPK sp|Q6PAQ4|REXO4_MOUSE 0.9999 3 2 VSIVNQYGK sp|Q810J8|ZFYV1_MOUSE 0.9999 3 2 FLGDASEAYLK sp|Q8BH66|ATLA1_MOUSE 0.9999 4 2 SFLM[147]DFM[147]LR sp|Q8JZV7|NAGA_MOUSE 0.9999 3 2 ATEDVGSGVALVAR sp|Q9D7X8|GGCT_MOUSE 0.9999 2 2 SYLM[147]TNYESAPPSPQYK sp|O88952|LIN7C_MOUSE 0.9999 2 2 TEEGLGFNIM[147]GGK sp|Q9EQS3|MYCBP_MOUSE 0.9998 2 2 LVQYEPPQEEK sp|Q9WV84|NDKM_MOUSE 0.9996 2 2 M[147]LQAPESILAEHYR sp|Q9CQX4|PAF_MOUSE 0.9995 2 2 VLGSSTFVTNSSSSSR sp|Q9D4J1|EFHD1_MOUSE 0.9987 1 2 VFNPYTEFPEFSR sp|Q8R035|ICT1_MOUSE 0.9975 2 2 AGELVLTSESSR sp|Q5BL07|PEX1_MOUSE 0.9963 1 2 YPELFANLPIR sp|O35448|PPT2_MOUSE 0.9929 2 2 ESLRPLWEQVQGFR sp|Q99JF5|ERG19_MOUSE 0.9913 1 2 GLQVAPVLLSDELK sp|Q9CXA2|PRCM_MOUSE 0.9913 1 2 DLVDAASALTGAVK sp|A2ADY9|DDI2_MOUSE 1 2 2 NPPLAEALLSGDLEK sp|A2RSX7|CB060_MOUSE 1 2 2 DAQYLYLSGSK sp|A3KMP2|TTC38_MOUSE 1 2 2 VLELLLPIR sp|O35295|PURB_MOUSE 1 2 2 GGGGGGGGGPGGFQPAPR sp|O35623|BET1_MOUSE 1 2 2 LLAEM[147]DSQFDSTTGFLGK sp|Q03958|PFD6_MOUSE 1 2 2 ETLAQLQQEFQR sp|Q3UGP9|LRC58_MOUSE 1 2 2 DLTYDPPTLLELAAR sp|Q4FK66|PR38A_MOUSE 1 2 2 YVLEEAEQLEPR sp|Q4FZF3|DDX49_MOUSE 1 2 2 ELAYQIAEQFR sp|Q56A08|GPKOW_MOUSE 1 3 2 AVVVLSGPYR sp|Q61823|PDCD4_MOUSE 1 3 2 DLPELALDTPR sp|Q6P3D0|NUD16_MOUSE 1 2 2 EQLLEALQDLK sp|Q6PCP5|MFF_MOUSE 1 3 2 IQYEM[147]EYTEGISQR sp|Q80TH2|LAP2_MOUSE 1 4 2 IYDILGDDGPQPPSAAVK sp|Q80UW2|FBX2_MOUSE 1 2 2 TDAGSLYELTVR sp|Q8BGU5|CCNY_MOUSE 1 2 2 IVLGAILLASK sp|Q8BHL8|PSMF1_MOUSE 1 2 2 VLIDPSSGLPNR sp|Q8C163|EXOG_MOUSE 1 2 2 SPESTEPLALGAFVVPNK 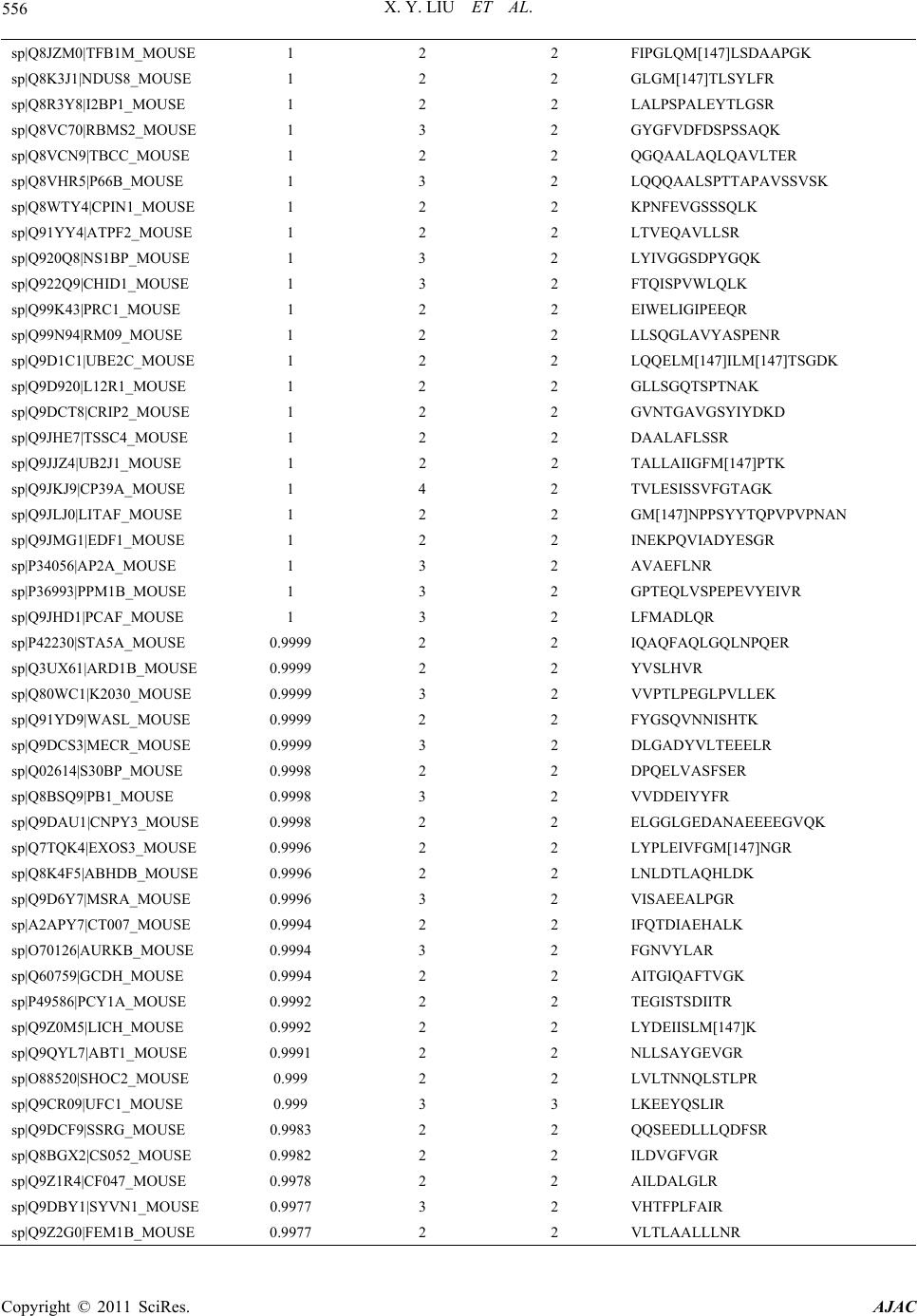 X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 556 sp|Q8JZM0|TFB1M_MOUSE 1 2 2 FIPGLQM[147]LSDAAPGK sp|Q8K3J1|NDUS8_MOUSE 1 2 2 GLGM[147]TLSYLFR sp|Q8R3Y8|I2BP1_MOUSE 1 2 2 LALPSPALEYTLGSR sp|Q8VC70|RBMS2_MOUSE 1 3 2 GYGFVDFDSPSSAQK sp|Q8VCN9|TBCC_MOUSE 1 2 2 QGQAALAQLQAVLTER sp|Q8VHR5|P66B_MOUSE 1 3 2 LQQQAALSPTTAPAVSSVSK sp|Q8WTY4|CPIN1_MOUSE 1 2 2 KPNFEVGSSSQLK sp|Q91YY4|ATPF2_MOUSE 1 2 2 LTVEQAVLLSR sp|Q920Q8|NS1BP_MOUSE 1 3 2 LYIVGGSDPYGQK sp|Q922Q9|CHID1_MOUSE 1 3 2 FTQISPVWLQLK sp|Q99K43|PRC1_MOUSE 1 2 2 EIWELIGIPEEQR sp|Q99N94|RM09_MOUSE 1 2 2 LLSQGLAVYASPENR sp|Q9D1C1|UBE2C_MOUSE 1 2 2 LQQELM[147]ILM[147]TSGDK sp|Q9D920|L12R1_MOUSE 1 2 2 GLLSGQTSPTNAK sp|Q9DCT8|CRIP2_MOUSE 1 2 2 GVNTGAVGSYIYDKD sp|Q9JHE7|TSSC4_MOUSE 1 2 2 DAALAFLSSR sp|Q9JJZ4|UB2J1_MOUSE 1 2 2 TALLAIIGFM[147]PTK sp|Q9JKJ9|CP39A_MOUSE 1 4 2 TVLESISSVFGTAGK sp|Q9JLJ0|LITAF_MOUSE 1 2 2 GM[147]NPPSYYTQPVPVPNAN sp|Q9JMG1|EDF1_MOUSE 1 2 2 INEKPQVIADYESGR sp|P34056|AP2A_MOUSE 1 3 2 AVAEFLNR sp|P36993|PPM1B_MOUSE 1 3 2 GPTEQLVSPEPEVYEIVR sp|Q9JHD1|PCAF_MOUSE 1 3 2 LFMADLQR sp|P42230|STA5A_MOUSE 0.9999 2 2 IQAQFAQLGQLNPQER sp|Q3UX61|ARD1B_MOUSE 0.9999 2 2 YVSLHVR sp|Q80WC1|K2030_MOUSE 0.9999 3 2 VVPTLPEGLPVLLEK sp|Q91YD9|WASL_MOUSE 0.9999 2 2 FYGSQVNNISHTK sp|Q9DCS3|MECR_MOUSE 0.9999 3 2 DLGADYVLTEEELR sp|Q02614|S30BP_MOUSE 0.9998 2 2 DPQELVASFSER sp|Q8BSQ9|PB1_MOUSE 0.9998 3 2 VVDDEIYYFR sp|Q9DAU1|CNPY3_MOUSE 0.9998 2 2 ELGGLGEDANAEEEEGVQK sp|Q7TQK4|EXOS3_MOUSE 0.9996 2 2 LYPLEIVFGM[147]NGR sp|Q8K4F5|ABHDB_MOUSE 0.9996 2 2 LNLDTLAQHLDK sp|Q9D6Y7|MSRA_MOUSE 0.9996 3 2 VISAEEALPGR sp|A2APY7|CT007_MOUSE 0.9994 2 2 IFQTDIAEHALK sp|O70126|AURKB_MOUSE 0.9994 3 2 FGNVYLAR sp|Q60759|GCDH_MOUSE 0.9994 2 2 AITGIQAFTVGK sp|P49586|PCY1A_MOUSE 0.9992 2 2 TEGISTSDIITR sp|Q9Z0M5|LICH_MOUSE 0.9992 2 2 LYDEIISLM[147]K sp|Q9QYL7|ABT1_MOUSE 0.9991 2 2 NLLSAYGEVGR sp|O88520|SHOC2_MOUSE 0.999 2 2 LVLTNNQLSTLPR sp|Q9CR09|UFC1_MOUSE 0.999 3 3 LKEEYQSLIR sp|Q9DCF9|SSRG_MOUSE 0.9983 2 2 QQSEEDLLLQDFSR sp|Q8BGX2|CS052_MOUSE 0.9982 2 2 ILDVGFVGR sp|Q9Z1R4|CF047_MOUSE 0.9978 2 2 AILDALGLR sp|Q9DBY1|SYVN1_MOUSE 0.9977 3 2 VHTFPLFAIR sp|Q9Z2G0|FEM1B_MOUSE 0.9977 2 2 VLTLAALLLNR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 557 sp|P62484|ABI2_MOUSE 0.996 1 2 ALFDSYTNLER sp|A2A4P0|DHX8_MOUSE 0.9957 1 2 FSQYFYEAPIFTIPGR sp|Q9D735|CS043_MOUSE 0.9913 1 2 GGPGPTLSFVGK sp|Q3UBX0|TM109_MOUSE 0.9904 1 2 ETSADILTQIGR sp|O35972|RM23_MOUSE 1 2 2 NYLEQIYNVPVAAVR sp|Q2TPA8|HSDL2_MOUSE 1 2 2 ADVVM[147]SM[147]ATDDFVK sp|Q80VL1|TDRKH_MOUSE 1 2 2 DM[147]ATETDDSLASILTETK sp|Q8C156|CND2_MOUSE 1 2 2 TIEQNLSNINVSEADGK sp|Q8CHT0|AL4A1_MOUSE 1 3 2 TVIQAEIDAAAELIDFFR sp|Q8K2L8|TTC15_MOUSE 1 2 2 QVLNASSVEQSFVGLK sp|Q8R2N2|CIR1A_MOUSE 1 2 2 TDGTVEIYNLSANYFQEK sp|Q8VE18|CQ071_MOUSE 1 2 2 AANYDFYQLLEEK sp|Q91ZR2|SNX18_MOUSE 1 2 2 APEPGPPADGGPGAPAR sp|Q921W4|QORL1_MOUSE 1 2 3 LSAGVFRPLLDEPIPLYEAK sp|Q99MZ7|PECR_MOUSE 1 2 2 NQVAVVTGGGTGIGK sp|Q9D7A8|ARMC1_MOUSE 1 2 2 AEALASAIASTK sp|Q9DC71|RT15_MOUSE 1 3 2 TLEAQIIALTVR sp|Q9ESP1|SDF2L_MOUSE 1 2 2 AM[147]EGIFIKPGADLSTGHDEL sp|Q9WV85|NDK3_MOUSE 1 2 2 ALIGATDPGDAM[147]PGTIR sp|Q3V300|KIF22_MOUSE 0.9999 3 2 ALM[147]DLLQLAR sp|Q8BHE8|CB047_MOUSE 0.9999 2 2 QLLSASYEFQR sp|Q8BVU5|NUDT9_MOUSE 0.9999 3 2 EFGEEALNSLQK sp|Q8R107|PRLD1_MOUSE 0.9999 2 2 AVQEFGLAR sp|Q9CRA5|GOLP3_MOUSE 0.9999 2 2 SDAPTGDVLLDEALK sp|Q9D125|RT25_MOUSE 0.9999 2 2 IM[147]TVNYNTYGELGEGAR sp|Q9D2E2|TOE1_MOUSE 0.9999 3 2 VPVVDVQSDNFK sp|Q9DB90|CS061_MOUSE 0.9999 2 2 GGNQTSGIDFFITQER sp|Q9WVM3|APC7_MOUSE 0.9999 2 2 AIQLNSNSVQALLLK sp|Q9Z2E1|MBD2_MOUSE 0.9999 2 2 LQGLSASDVTEQIIK sp|Q8K0C9|GMDS_MOUSE 0.9998 2 2 FYQASTSELYGK sp|Q9ESW8|PGPI_MOUSE 0.9998 2 2 SAFVHVPPLGK sp|Q62036|AZI1_MOUSE 0.9997 2 2 PAEPTDFLM[147]LFEGSTSGR sp|Q99N87|RT05_MOUSE 0.9997 3 2 EPEPEPEVPDTK sp|Q9CYI4|LUC7L_MOUSE 0.9997 2 2 ALLDQLM[147]GTAR sp|Q9R1Z7|PTPS_MOUSE 0.9997 2 3 LHSPSLSDEENLR sp|Q64437|ADH7_MOUSE 0.9997 3 2 M[147]LTYDPM[147]LLFTGR sp|Q924W5|SMC6_MOUSE 0.9996 2 2 SAVLTALIVGLGGK sp|Q9R0X0|MED20_MOUSE 0.9996 2 2 SLQQTVELLTK sp|Q9D8V7|SC11C_MOUSE 0.9993 2 2 GDLLFLTNFR sp|Q9DB40|MED27_MOUSE 0.9993 2 2 TPLYSQLLQAYK sp|Q80W93|HYDIN_MOUSE 0.9992 3 3 QKLT[181]LLAQGQGLEPR sp|Q91Y86|MK08_MOUSE 0.9988 3 2 NIIGLLNVFTPQK sp|Q63810|CANB1_MOUSE 0.9977 2 3 M[147]M[147]VGNNLKDTQLQQIVDK sp|Q91ZF0|DJC24_MOUSE 0.9973 2 2 LILLYHPDK sp|Q8VC65|NRM_MOUSE 0.996 2 2 YFGVLQR sp|Q99KK9|SYHM_MOUSE 0.9945 1 2 DQGGELLSLR sp|Q9D6M3|GHC1_MOUSE 0.9933 2 2 SEGYFGM[147]YR 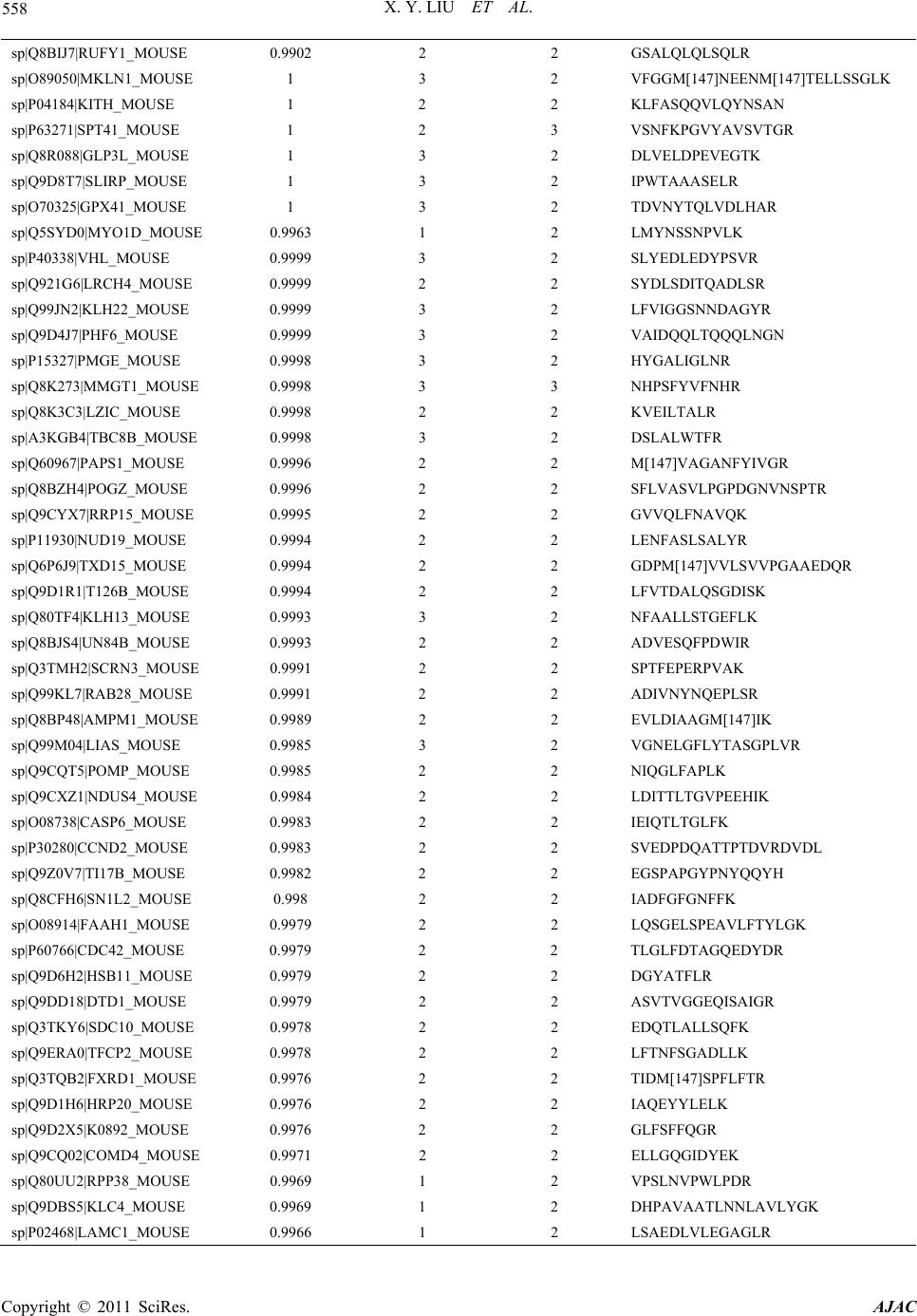 X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 558 sp|Q8BIJ7|RUFY1_MOUSE 0.9902 2 2 GSALQLQLSQLR sp|O89050|MKLN1_MOUSE 1 3 2 VFGGM[147]NEENM[147]TELLSSGLK sp|P04184|KITH_MOUSE 1 2 2 KLFASQQVLQYNSAN sp|P63271|SPT41_MOUSE 1 2 3 VSNFKPGVYAVSVTGR sp|Q8R088|GLP3L_MOUSE 1 3 2 DLVELDPEVEGTK sp|Q9D8T7|SLIRP_MOUSE 1 3 2 IPWTAAASELR sp|O70325|GPX41_MOUSE 1 3 2 TDVNYTQLVDLHAR sp|Q5SYD0|MYO1D_MOUSE 0.9963 1 2 LMYNSSNPVLK sp|P40338|VHL_MOUSE 0.9999 3 2 SLYEDLEDYPSVR sp|Q921G6|LRCH4_MOUSE 0.9999 2 2 SYDLSDITQADLSR sp|Q99JN2|KLH22_MOUSE 0.9999 3 2 LFVIGGSNNDAGYR sp|Q9D4J7|PHF6_MOUSE 0.9999 3 2 VAIDQQLTQQQLNGN sp|P15327|PMGE_MOUSE 0.9998 3 2 HYGALIGLNR sp|Q8K273|MMGT1_MOUSE 0.9998 3 3 NHPSFYVFNHR sp|Q8K3C3|LZIC_MOUSE 0.9998 2 2 KVEILTALR sp|A3KGB4|TBC8B_MOUSE 0.9998 3 2 DSLALWTFR sp|Q60967|PAPS1_MOUSE 0.9996 2 2 M[147]VAGANFYIVGR sp|Q8BZH4|POGZ_MOUSE 0.9996 2 2 SFLVASVLPGPDGNVNSPTR sp|Q9CYX7|RRP15_MOUSE 0.9995 2 2 GVVQLFNAVQK sp|P11930|NUD19_MOUSE 0.9994 2 2 LENFASLSALYR sp|Q6P6J9|TXD15_MOUSE 0.9994 2 2 GDPM[147]VVLSVVPGAAEDQR sp|Q9D1R1|T126B_MOUSE 0.9994 2 2 LFVTDALQSGDISK sp|Q80TF4|KLH13_MOUSE 0.9993 3 2 NFAALLSTGEFLK sp|Q8BJS4|UN84B_MOUSE 0.9993 2 2 ADVESQFPDWIR sp|Q3TMH2|SCRN3_MOUSE 0.9991 2 2 SPTFEPERPVAK sp|Q99KL7|RAB28_MOUSE 0.9991 2 2 ADIVNYNQEPLSR sp|Q8BP48|AMPM1_MOUSE 0.9989 2 2 EVLDIAAGM[147]IK sp|Q99M04|LIAS_MOUSE 0.9985 3 2 VGNELGFLYTASGPLVR sp|Q9CQT5|POMP_MOUSE 0.9985 2 2 NIQGLFAPLK sp|Q9CXZ1|NDUS4_MOUSE 0.9984 2 2 LDITTLTGVPEEHIK sp|O08738|CASP6_MOUSE 0.9983 2 2 IEIQTLTGLFK sp|P30280|CCND2_MOUSE 0.9983 2 2 SVEDPDQATTPTDVRDVDL sp|Q9Z0V7|TI17B_MOUSE 0.9982 2 2 EGSPAPGYPNYQQYH sp|Q8CFH6|SN1L2_MOUSE 0.998 2 2 IADFGFGNFFK sp|O08914|FAAH1_MOUSE 0.9979 2 2 LQSGELSPEAVLFTYLGK sp|P60766|CDC42_MOUSE 0.9979 2 2 TLGLFDTAGQEDYDR sp|Q9D6H2|HSB11_MOUSE 0.9979 2 2 DGYATFLR sp|Q9DD18|DTD1_MOUSE 0.9979 2 2 ASVTVGGEQISAIGR sp|Q3TKY6|SDC10_MOUSE 0.9978 2 2 EDQTLALLSQFK sp|Q9ERA0|TFCP2_MOUSE 0.9978 2 2 LFTNFSGADLLK sp|Q3TQB2|FXRD1_MOUSE 0.9976 2 2 TIDM[147]SPFLFTR sp|Q9D1H6|HRP20_MOUSE 0.9976 2 2 IAQEYYLELK sp|Q9D2X5|K0892_MOUSE 0.9976 2 2 GLFSFFQGR sp|Q9CQ02|COMD4_MOUSE 0.9971 2 2 ELLGQGIDYEK sp|Q80UU2|RPP38_MOUSE 0.9969 1 2 VPSLNVPWLPDR sp|Q9DBS5|KLC4_MOUSE 0.9969 1 2 DHPAVAATLNNLAVLYGK sp|P02468|LAMC1_MOUSE 0.9966 1 2 LSAEDLVLEGAGLR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 559 sp|Q64152|BTF3_MOUSE 0.9963 2 2 TATADDKKLQFSLK sp|Q8BRN9|C2D1B_MOUSE 0.9963 1 2 GM[147]NLPAPPGVTPDDLDAFVR sp|P19182|IFRD1_MOUSE 0.9961 2 2 VLYEFVLER sp|Q91Z38|TTC1_MOUSE 0.9958 2 2 AIQLNPTYIR sp|Q9D8T0|FAM3A_MOUSE 0.9958 2 2 IVSGAANVIGPK sp|O09111|NDUBB_MOUSE 0.9954 2 2 EVNGLPIM[147]ESNYFDPSK sp|Q9CYA0|CREL2_MOUSE 0.9949 2 2 YEFSEIR sp|Q6ZQF0|TOPB1_MOUSE 0.9942 2 2 NLTVALANSSR sp|Q8CI70|LRC20_MOUSE 0.9933 2 2 FM[147]TTFNQLR sp|Q7TSG2|CTDP1_MOUSE 0.9931 2 2 VLTQLVLSPDAPDR sp|Q8BRK8|AAPK2_MOUSE 0.9923 1 2 IADFGLSNM[147]M[147]SDGEFLR sp|O35459|ECH1_MOUSE 0.9913 1 2 EVDM[147]GLAADVGTLQR sp|P17095|HMGA1_MOUSE 0.9913 1 2 KQPPVSPGTALVGSQK sp|Q9CQ26|STABP_MOUSE 0.9913 1 2 NEFTITHVLIPR sp|Q9CRB2|NHP2_MOUSE 0.9913 1 2 ELLVNLNPIAQPLASR sp|Q9D0I4|STX17_MOUSE 0.9913 1 2 VAGIAAALGGGVLGFTGGK sp|Q8BW94|DYH3_MOUSE 0.9908 2 2 VFLEALNNNIR sp|P70353|NFYC_MOUSE 0.9904 1 2 M[147]ISAEAPVLFAK sp|P60762|MO4L1_MOUSE 1 2 2 VDPTVENEETFM[147]NR sp|Q8BLY7|HPS6_MOUSE 1 2 2 LLSDLSNFTGAAR sp|Q9DBZ1|IKIP_MOUSE 1 2 2 LLQTESSEFQGLQSK sp|Q8C863|ITCH_MOUSE 0.9904 1 2 FIDTGFSLPFYK sp|Q91W34|CP058_MOUSE 0.9999 2 2 ALVLETLNESR sp|Q4VAA7|SNX33_MOUSE 0.9998 2 2 IAETYSIEM[147]GPR sp|Q9DCT5|SDF2_MOUSE 0.9997 2 2 AM[147]EGIFM[147]KPSELLR sp|Q9D287|SPF27_MOUSE 0.9996 2 2 EAAAALVEEETR sp|Q9JKK1|STX6_MOUSE 0.9995 2 2 DQM[147]SASSVQALAER sp|P97434|MPRIP_MOUSE 0.9993 2 2 LLAEETAATISAIEAMK sp|Q3TRM4|PLPL6_MOUSE 0.9993 3 2 GDLIGVVEALTR sp|Q9D1I5|MCEE_MOUSE 0.9988 2 3 M[147]ELLHPLGSDSPITGFLQK sp|O55060|TPMT_MOUSE 0.9983 2 2 GALVAINPGDHDR sp|Q64362|AKTIP_MOUSE 0.9983 2 2 IDTTSPLNPEAAVLYEK sp|Q9Z2Q5|RM40_MOUSE 0.9983 2 2 VYTQVEFKR sp|Q3UH68|LIMC1_MOUSE 0.9978 2 2 TSVPESIASAGTGSPSK sp|Q8CE50|SNX30_MOUSE 0.9976 2 2 VEFDLPEYSVR sp|Q8BQZ4|K1219_MOUSE 0.9973 2 2 EVPVIFIHPLNTGLFR sp|Q99JY4|TRABD_MOUSE 0.9971 2 2 TVTQLVAEDGSR sp|Q8VCR3|CF035_MOUSE 0.9966 1 2 GTM[147]ATAALPESGSSLALR sp|Q9D892|ITPA_MOUSE 0.9931 3 2 IDLPEYQGEPDEISIQK sp|A2RTL5|RSRC2_MOUSE 0.9913 1 2 NTAM[147]DAQEALAR sp|O54916|REPS1_MOUSE 0.9913 1 2 LVAVAQSGFPLR sp|Q8BKW4|ZCHC4_MOUSE 0.9913 1 2 YLSFIQLPLAQR sp|Q8BU04|UBR7_MOUSE 0.9913 1 2 EDIQQFFEEFQSK sp|Q9CXK8|NIP7_MOUSE 0.9913 1 2 LHVTALDYLAPYAK sp|Q9D8Z1|ASCC1_MOUSE 0.9913 1 2 TFENFYFGSLR sp|Q9ET47|ESPN_MOUSE 0.9913 1 2 SFNM[147]M[147]SPTGDNSELLAEIK sp|Q9JI44|DMAP1_MOUSE 0.9913 1 2 TPEQVAEEEYLLQELR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 560 sp|Q9WU81|SPX2_MOUSE 0.9913 1 2 EQGPEPEAISFLGALR sp|P55302|AMRP_MOUSE 0.9913 1 2 IQEYNVLLDTLSR sp|Q5SS80|DHR13_MOUSE 0.991 3 2 LLT[181]HLLLPR sp|Q8BGI5|PEX26_MOUSE 0.9904 1 2 LSEAEELAVR sp|O35144|TERF2_MOUSE 0.9977 2 2 ALSTAQDSEAAFAK sp|Q80X71|T106B_MOUSE 0.997 2 2 GQENQLVALIPYSDQR sp|Q8C0M9|ASGL1_MOUSE 0.9967 2 2 GLGGLILVNK sp|Q69ZW3|EHBP1_MOUSE 0.9954 2 3 PK[142]PTSPNNLVNTVQEGETER sp|P70428|EXT2_MOUSE 0.9914 1 2 YVDDAGVPVSSAISR sp|P70445|4EBP2_MOUSE 0.9913 1 2 HAVGDEAQFEM[147]DI sp|Q61827|MAFK_MOUSE 0.9913 1 2 EAGENAPVLSDDELVSM[147]SVR sp|Q9WTK2|CDYL_MOUSE 0.9913 1 2 EVQSALSTAAADDSK sp|P55937|GOGA3_MOUSE 0.9913 1 2 VADAAASLEQQLEQVK sp|O35516|NOTC2_MOUSE 0.9904 1 2 EPLPPIVTFQLIPK sp|P61205|ARF3_MOUSE 1 2 2 DAVLLVFANK sp|Q61781|K1C14_MOUSE 1 2 2 VLDELTLAR sp|Q9Z1K7|APC2_MOUSE 0.9993 2 2 LPVS[167]IPAPQR sp|P09041|PGK2_MOUSE 0.9961 1 3 VSHVSTGGGASLELLEGK sp|P62309|RUXG_MOUSE 0.9961 1 2 GNSIIMLEALER sp|Q66JX5|FR1OP_MOUSE 0.9961 1 2 DLLVQTLENSGVLNR sp|P62746|RHOB_MOUSE 0.9942 1 2 QVELALWDTAGQEDYDR sp|Q9D1M7|FKB11_MOUSE 0.9942 1 2 DPLVIELGQK sp|Q61474|MSI1H_MOUSE 0.9914 1 2 GFGFVTFMDQAGVDK Table S2 protein protein probability Num unique peps precursor ion charge peptide sequence sp|P68033|ACTC_MOUSE 1 9 2 AGFAGDDAPR sp|Q64524|H2B2E_MOUSE 1 10 2 AMGIMNSFVNDIFER sp|Q7TPR4|ACTN1_MOUSE 1 35 2 AGTQIENIEEDFR sp|Q9WU78|PDC6I_MOUSE 1 33 2 ATLVKPTPVNVPVSQK sp|Q62448|IF4G2_MOUSE 1 23 2 EWLTELFQQSK sp|Q6PIC6|AT1A3_MOUSE 0.9951 4 2 GVGIISEGNETVEDIAAR sp|P16110|LEG3_MOUSE 1 13 2 IQVLVEADHFK sp|Q61206|PA1B2_MOUSE 1 2 2 IIVLGLLPR sp|Q61941|NNTM_MOUSE 1 11 2 AVVLAANHFGR sp|Q8BH74|NU107_MOUSE 1 13 2 AIYAALSGNLK sp|Q9Z2I0|LETM1_MOUSE 1 9 2 ADDKLISEEGVDSLTVK sp|Q921N6|DDX27_MOUSE 1 13 2 ALQEFDLALR sp|Q6PAR5|GAPD1_MOUSE 1 12 2 AVETPPM[147]SSVNLLEGLSR sp|Q9DBY8|NVL_MOUSE 1 13 2 AVANESGLNFISVK sp|Q9WVL3|S12A7_MOUSE 1 13 2 DLQMFLYHLR sp|P28271|ACOC_MOUSE 1 10 2 TSLSPGSGVVTYYLR sp|Q9QZQ1|AFAD_MOUSE 1 12 2 LAAGDQLLSVDGR sp|Q80TP3|UBR5_MOUSE 1 12 2 LLTATNLVTLPNSR 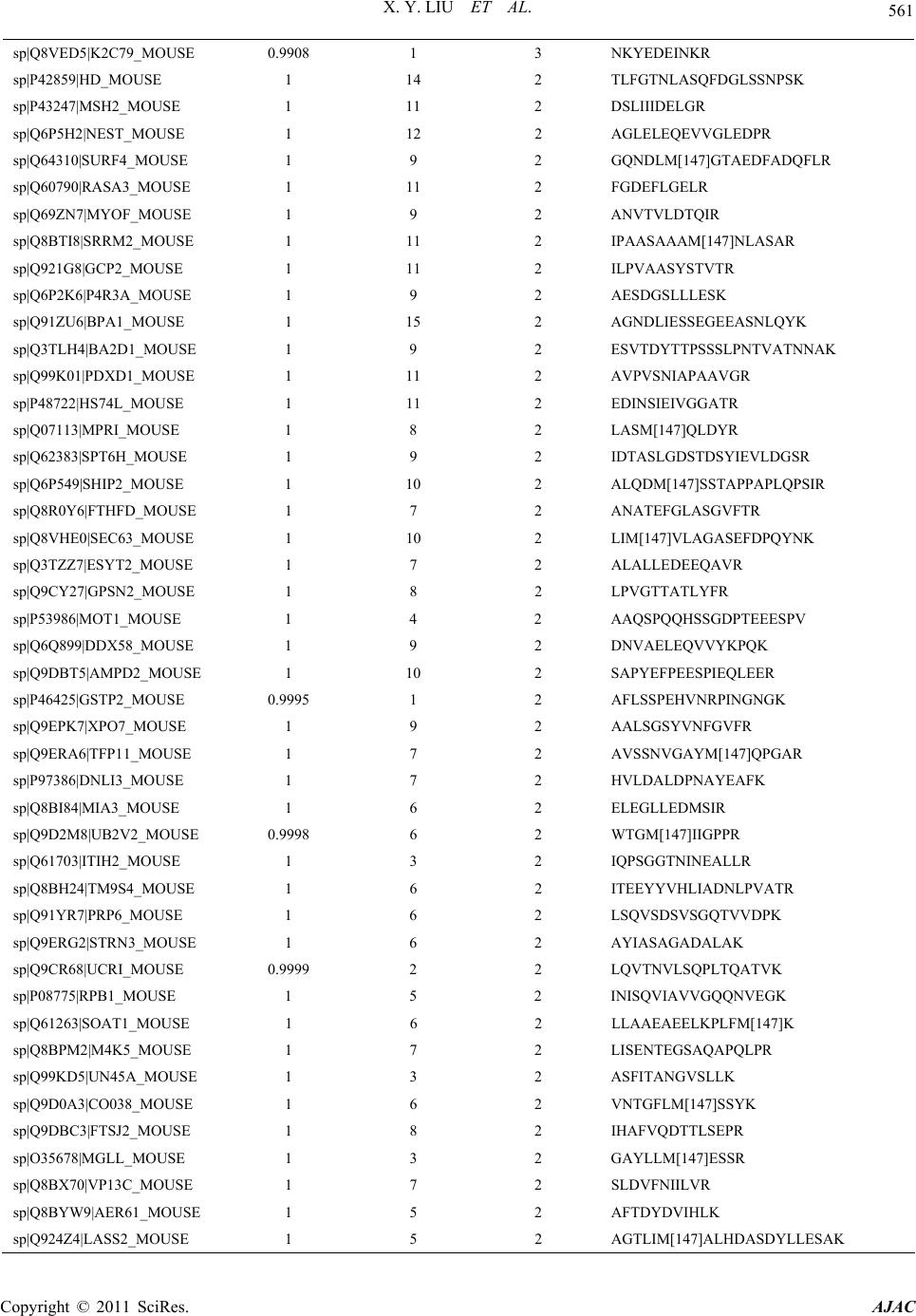 X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 561 sp|Q8VED5|K2C79_MOUSE 0.9908 1 3 NKYEDEINKR sp|P42859|HD_MOUSE 1 14 2 TLFGTNLASQFDGLSSNPSK sp|P43247|MSH2_MOUSE 1 11 2 DSLIIIDELGR sp|Q6P5H2|NEST_MOUSE 1 12 2 AGLELEQEVVGLEDPR sp|Q64310|SURF4_MOUSE 1 9 2 GQNDLM[147]GTAEDFADQFLR sp|Q60790|RASA3_MOUSE 1 11 2 FGDEFLGELR sp|Q69ZN7|MYOF_MOUSE 1 9 2 ANVTVLDTQIR sp|Q8BTI8|SRRM2_MOUSE 1 11 2 IPAASAAAM[147]NLASAR sp|Q921G8|GCP2_MOUSE 1 11 2 ILPVAASYSTVTR sp|Q6P2K6|P4R3A_MOUSE 1 9 2 AESDGSLLLESK sp|Q91ZU6|BPA1_MOUSE 1 15 2 AGNDLIESSEGEEASNLQYK sp|Q3TLH4|BA2D1_MOUSE 1 9 2 ESVTDYTTPSSSLPNTVATNNAK sp|Q99K01|PDXD1_MOUSE 1 11 2 AVPVSNIAPAAVGR sp|P48722|HS74L_MOUSE 1 11 2 EDINSIEIVGGATR sp|Q07113|MPRI_MOUSE 1 8 2 LASM[147]QLDYR sp|Q62383|SPT6H_MOUSE 1 9 2 IDTASLGDSTDSYIEVLDGSR sp|Q6P549|SHIP2_MOUSE 1 10 2 ALQDM[147]SSTAPPAPLQPSIR sp|Q8R0Y6|FTHFD_MOUSE 1 7 2 ANATEFGLASGVFTR sp|Q8VHE0|SEC63_MOUSE 1 10 2 LIM[147]VLAGASEFDPQYNK sp|Q3TZZ7|ESYT2_MOUSE 1 7 2 ALALLEDEEQAVR sp|Q9CY27|GPSN2_MOUSE 1 8 2 LPVGTTATLYFR sp|P53986|MOT1_MOUSE 1 4 2 AAQSPQQHSSGDPTEEESPV sp|Q6Q899|DDX58_MOUSE 1 9 2 DNVAELEQVVYKPQK sp|Q9DBT5|AMPD2_MOUSE 1 10 2 SAPYEFPEESPIEQLEER sp|P46425|GSTP2_MOUSE 0.9995 1 2 AFLSSPEHVNRPINGNGK sp|Q9EPK7|XPO7_MOUSE 1 9 2 AALSGSYVNFGVFR sp|Q9ERA6|TFP11_MOUSE 1 7 2 AVSSNVGAYM[147]QPGAR sp|P97386|DNLI3_MOUSE 1 7 2 HVLDALDPNAYEAFK sp|Q8BI84|MIA3_MOUSE 1 6 2 ELEGLLEDMSIR sp|Q9D2M8|UB2V2_MOUSE 0.9998 6 2 WTGM[147]IIGPPR sp|Q61703|ITIH2_MOUSE 1 3 2 IQPSGGTNINEALLR sp|Q8BH24|TM9S4_MOUSE 1 6 2 ITEEYYVHLIADNLPVATR sp|Q91YR7|PRP6_MOUSE 1 6 2 LSQVSDSVSGQTVVDPK sp|Q9ERG2|STRN3_MOUSE 1 6 2 AYIASAGADALAK sp|Q9CR68|UCRI_MOUSE 0.9999 2 2 LQVTNVLSQPLTQATVK sp|P08775|RPB1_MOUSE 1 5 2 INISQVIAVVGQQNVEGK sp|Q61263|SOAT1_MOUSE 1 6 2 LLAAEAEELKPLFM[147]K sp|Q8BPM2|M4K5_MOUSE 1 7 2 LISENTEGSAQAPQLPR sp|Q99KD5|UN45A_MOUSE 1 3 2 ASFITANGVSLLK sp|Q9D0A3|CO038_MOUSE 1 6 2 VNTGFLM[147]SSYK sp|Q9DBC3|FTSJ2_MOUSE 1 8 2 IHAFVQDTTLSEPR sp|O35678|MGLL_MOUSE 1 3 2 GAYLLM[147]ESSR sp|Q8BX70|VP13C_MOUSE 1 7 2 SLDVFNIILVR sp|Q8BYW9|AER61_MOUSE 1 5 2 AFTDYDVIHLK sp|Q924Z4|LASS2_MOUSE 1 5 2 AGTLIM[147]ALHDASDYLLESAK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 562 sp|Q9Z160|COG1_MOUSE 1 8 2 ALQLLYDLR sp|O08759|UBE3A_MOUSE 1 6 2 M[147]M[147]ETFQQLITYK sp|P22892|AP1G1_MOUSE 1 6 2 AM[147]ELSFALVNGNNIR sp|Q52KI8|SRRM1_MOUSE 1 6 2 DSSVQEATSTSDILK sp|Q6PGH2|HN1L_MOUSE 1 4 2 GSGIFDESTPVQTR sp|Q80U72|LAP4_MOUSE 1 7 2 SLEELLLDANQLR sp|Q8BVG4|DPP9_MOUSE 1 4 2 ELVQPFSSLFPK sp|Q8R3S6|EXOC1_MOUSE 1 6 2 ALQEGDLVSSR sp|Q9D4H1|EXOC2_MOUSE 1 6 2 ASNTADTLFQEVLGR sp|O54988|SLK_MOUSE 1 6 2 AGNILFTLDGDIK sp|P16056|MET_MOUSE 1 5 2 GDLTIANLGTSEGR sp|O35382|EXOC4_MOUSE 1 5 2 FIQEIEHALGLGPAK sp|P39053|DYN1_MOUSE 1 4 2 SSVLENFVGR sp|P39447|ZO1_MOUSE 1 5 2 LAGGNDVGIFVAGVLEDSPAAK sp|Q7TSC1|BAT2_MOUSE 1 5 2 AVGTPGANAGGAGPGISAM[147]SR sp|Q8BU03|PWP2_MOUSE 1 3 2 AGQLLPVVQFLQK sp|Q8BYH7|TBC17_MOUSE 1 5 2 IFSGGLSPGLR sp|Q8CCB4|VPS53_MOUSE 1 6 2 M[147]VLLDLPSIGSQVVR sp|Q8K368|FANCI_MOUSE 1 6 2 FVSDLLTALFR sp|Q8R5L3|VPS39_MOUSE 1 8 2 AINLLPANTQINDIR sp|Q99NH0|ANR17_MOUSE 1 6 2 NVSDYTPLSLAASGGYVNIIK sp|Q9DC40|TELO2_MOUSE 1 6 2 LLGDLPDELLEAR sp|Q9QX47|SON_MOUSE 1 5 2 AGIEGPLLASEVER sp|Q9R0L6|PCM1_MOUSE 1 7 2 ALYALQDIVSR sp|P83741|WNK1_MOUSE 1 5 2 IGDLGLATLK sp|Q8CJG0|I2C2_MOUSE 1 5 2 YAQGADSVEPM[147]FR sp|P17897|LYZ1_MOUSE 0.9976 2 2 STDYGIFQINSR sp|P03911|NU4M_MOUSE 1 5 2 IILPSLM[147]LLPLTWLSSPK sp|P45377|ALD2_MOUSE 1 4 3 AVQREDLFIVSK sp|P97452|BOP1_MOUSE 1 4 2 VNVDPEDLIPK sp|Q3U487|HECD3_MOUSE 1 4 2 AGLPLPAALAFVPR sp|Q6PF93|PK3C3_MOUSE 1 5 2 DGDESSPILTSFELVK sp|Q6PR54|RIF1_MOUSE 1 7 2 TIGDLSTLTASEIK sp|Q8BIG7|CMTD1_MOUSE 1 2 2 PGGVLAVLR sp|Q8BUV3|GEPH_MOUSE 1 5 2 DLVQDPSLLGGTISAYK sp|Q8VBZ3|CLPT1_MOUSE 1 4 2 NLLTGETEADPEM[147]IK sp|Q9D8V0|HM13_MOUSE 1 3 2 QYQLLFTQGSGENK sp|Q9WV70|NOC2L_MOUSE 1 3 2 SIAFPELVLPTVLQLK sp|Q99KH8|STK24_MOUSE 1 7 2 KTSYLTELIDR sp|P01027|CO3_MOUSE 0.9999 3 2 SSVAVPYVIVPLK sp|Q8BIV3|RNBP6_MOUSE 0.9985 2 2 EGFVEYTEQVVK sp|O54827|AT10A_MOUSE 0.9919 2 2 ASPSPSLVIDGR sp|O35604|NPC1_MOUSE 1 5 2 LQEETLDQQLGR sp|O70579|PM34_MOUSE 1 4 2 LSSLDVFIIGAIAK sp|P58021|TM9S2_MOUSE 1 5 2 RPSENLGQVLFGER  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 563 sp|Q3UDW8|HGNAT_MOUSE 1 2 2 ADPLSADYQPETR sp|Q61464|ZN638_MOUSE 1 6 2 DVTVLSM[147]AEEQDLQQER sp|Q6PIP5|NUDC1_MOUSE 1 6 2 LPTDVTAYDNR sp|Q8VDF2|UHRF1_MOUSE 1 6 2 YDDYPEHGVDIVK sp|Q91V04|TRAM1_MOUSE 1 3 2 LDFSTGNFNVLAVR sp|Q9DBF1|AL7A1_MOUSE 1 5 2 GAPTTSLVSVAVTK sp|Q9QY36|ARD1A_MOUSE 1 4 2 YYADGEDAYAM[147]K sp|Q91W97|HKDC1_MOUSE 0.9956 2 2 M[147]ISGLYM[147]GELVR sp|P31786|ACBP_MOUSE 1 5 2 TYVEKVDELKK sp|P41216|ACSL1_MOUSE 1 2 2 LM[147]ITGAAPVSATVLTFLR sp|P58404|STRN4_MOUSE 1 5 2 ALIASAGADALAK sp|Q0VGY8|TANC1_MOUSE 1 7 2 ANFQEIISALPFVK sp|Q3TCJ1|F175B_MOUSE 1 3 2 AIYQVYNALQEK sp|Q3UVK0|ERMP1_MOUSE 1 5 2 AFINLEAAGVGGK sp|Q5H8C4|VP13A_MOUSE 1 4 2 ALVGGAVGGLAGAASK sp|Q60953|PML_MOUSE 1 4 2 DNSVSSFLDSTR sp|Q6P9Q6|FKB15_MOUSE 1 5 2 EVATDGLLQGNSR sp|Q80Y81|RNZ2_MOUSE 1 4 2 LDNIFLTR sp|Q8C8R3|ANK2_MOUSE 1 3 2 VVTEEVTTTTTTITEK sp|Q8K1X1|BRWD2_MOUSE 1 3 2 LLLDPDFSLLQR sp|Q8R4G6|MGT5A_MOUSE 1 4 2 TLAVLLDNILQR sp|Q8VDC0|SYLM_MOUSE 1 4 2 AM[147]QDALADLPEWYGIK sp|Q91W86|VPS11_MOUSE 1 3 2 GNYPVTGLAFR sp|Q99PG2|OGFR_MOUSE 1 3 2 QSALDYFLFAVR sp|Q9EPU4|CPSF1_MOUSE 1 4 2 VLVDSSFGQPTTQGEVR sp|Q9ERG0|LIMA1_MOUSE 1 5 2 SQDVGFWEGEVVR sp|Q9ET30|TM9S3_MOUSE 1 4 2 DAFVYAIK sp|Q9JIX8|ACINU_MOUSE 1 4 2 KVTLGDTLTR sp|P46467|VPS4B_MOUSE 1 4 2 GILLFGPPGTGK sp|Q922W5|P5CR1_MOUSE 0.9999 2 2 LGAQALLGAAK sp|O35609|SCAM3_MOUSE 1 3 2 AQQEFAAGVFSNPAVR sp|P11688|ITA5_MOUSE 1 3 2 VTAPLEAEYSGLVR sp|P81117|NUCB2_MOUSE 1 3 2 AATADLEQYDR sp|Q0KL02|TRIO_MOUSE 1 5 2 DNFDAFYSEVAELGR sp|Q3TMX7|QSOX2_MOUSE 1 5 2 EGSDAVWLLDSGSVR sp|Q3U186|SYRM_MOUSE 1 2 2 GGVTFLEDVLNEVQSR sp|Q3UMF0|COBL1_MOUSE 1 3 2 DYQAQEPLDLTK sp|Q6ZQ93|UBP34_MOUSE 1 10 2 SFLLLAASTLLK sp|Q8BTY8|SCFD2_MOUSE 1 3 2 SQIAVNDVFM[147]ALR sp|Q8BXN9|TM87A_MOUSE 1 3 2 ADEIESYLENLK sp|Q8C0L8|COG5_MOUSE 1 3 2 GALEAYVQSVR sp|Q8C754|VPS52_MOUSE 1 3 2 FLEQLQELDAK sp|Q8CBQ5|P4K2B_MOUSE 1 3 2 IAAIDNGLAFPFK sp|Q8K2C9|PTAD1_MOUSE 1 3 3 VELSDVQNPAISITDNVLHFK sp|Q8R0A0|T2FB_MOUSE 1 4 3 VVTTNYKPVANHQYNIEYER  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 564 sp|Q8R5J9|PRAF3_MOUSE 1 2 2 TPM[147]GIILDALEQQEDNINK sp|Q8VEL2|MTMRE_MOUSE 1 2 2 AATPSPSGAIGGLLEQFAR sp|Q922U1|PRPF3_MOUSE 1 5 2 LFEAVEEGR sp|Q99LB7|SARDH_MOUSE 1 3 2 DPSGGPVSLDFVK sp|Q9CQJ6|DENR_MOUSE 1 3 2 QETGITEGQGPVGEEEEK sp|Q9D853|METLA_MOUSE 1 3 2 SGNTVAALVFQK sp|Q9WV68|DECR2_MOUSE 1 5 2 VAFITGGGSGIGFR sp|Q61411|RASH_MOUSE 1 3 2 LVVVGAGGVGK sp|Q3UV17|K22O_MOUSE 0.99 1 2 AQYEDIAQK sp|Q8K0V4|CNOT3_MOUSE 1 3 2 M[147]LDNDSILVDAIR sp|Q921C5|BICD2_MOUSE 1 4 2 VGLLATLQDTQK sp|Q811J3|IREB2_MOUSE 0.9999 3 2 EGIPLIILAGK sp|P00397|COX1_MOUSE 0.9983 2 2 M[147]IGAPDM[147]AFPR sp|Q8BZ98|DYN3_MOUSE 0.9964 2 2 SSVLENFVGR sp|Q9ERY9|ERG28_MOUSE 0.9955 2 2 YLEAEPVSR sp|O08784|TCOF_MOUSE 1 3 2 AGAVTSSASLSSPALAK sp|O35657|NEUR1_MOUSE 1 3 2 GTLLAFAEAR sp|P03888|NU1M_MOUSE 1 3 2 GPNIVGPYGILQPFADAM[147]K sp|P70206|PLXA1_MOUSE 1 4 2 FVDDLFETIFSTAHR sp|Q3UMC0|SPAT5_MOUSE 1 6 2 ALANESGLNFLAIK sp|Q5DTT3|CJ018_MOUSE 1 4 2 VIPILPALSYALLEAK sp|Q5RJG1|NOL10_MOUSE 1 3 2 DLENLGLTHLIGSPFLR sp|Q68FF6|GIT1_MOUSE 1 3 2 SLSSPTDNLELSAR sp|Q80YV3|TRRAP_MOUSE 1 5 2 NFIQTILTSLIEK sp|Q8BM72|HSP13_MOUSE 1 4 2 IFTPEELEAEVGR sp|Q8R151|ZNFX1_MOUSE 1 5 2 INVFDFGQWPSK sp|Q8VCR7|ABHEB_MOUSE 1 3 2 FSVLLLHGIR sp|Q91X52|DCXR_MOUSE 1 4 2 GVPGAIVNVSSQASQR sp|Q9D8N2|FAM45_MOUSE 1 3 2 M[147]M[147]ESYIAVLTK sp|Q9QWT9|KIFC1_MOUSE 1 4 2 LTYLLQNSLGGSAK sp|Q9Z2G6|SE1L1_MOUSE 1 3 2 AADM[147]GNPVGQSGLGM[147]AYLYGR sp|P10648|GSTA2_MOUSE 1 3 2 SHGQDYLVGNR sp|Q3B7Z2|OSBP1_MOUSE 1 4 2 IPM[147]PVNFNEPLSM[147]LQR sp|Q8BLR2|CPNE4_MOUSE 0.9999 2 2 DIVQFVPFR sp|P28028|BRAF1_MOUSE 0.9999 3 2 M[147]LNVTAPTPQQLQAFK sp|Q8K4L0|DDX54_MOUSE 0.9997 3 2 AGLTEPVLIR sp|Q6GQT1|A2MP_MOUSE 0.9992 3 2 M[147]VSGFIPLKPTVK sp|Q8BFR1|ZCCHL_MOUSE 0.9989 2 2 FLLQEVELR sp|O35621|PMM1_MOUSE 0.994 1 2 NGM[147]LNVSPIGR sp|Q9CZX7|TM55A_MOUSE 0.9917 2 2 ISSVGSALPR sp|O88487|DC1I2_MOUSE 1 3 2 EAAVSVQEESDLEK sp|P52875|TM165_MOUSE 1 3 2 M[147]SPDEGQEELEEVQAELK sp|P97478|COQ7_MOUSE 1 2 2 IYAGQM[147]AVLGR sp|Q2HXL6|EDEM3_MOUSE 1 3 2 FTGATIFEEYAR sp|Q3THK3|T2FA_MOUSE 1 3 2 IYQEEEM[147]PESGAGSEFNR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 565 sp|Q3UHJ0|AAK1_MOUSE 1 4 2 AGQTQPNPGILPIQPALTPR sp|Q60596|XRCC1_MOUSE 1 3 2 HFFLYGEFPGDER sp|Q60855|RIPK1_MOUSE 1 3 2 AEYNEVLLEEGK sp|Q80TE0|RPAP1_MOUSE 1 4 2 VSSLLLPVPK sp|Q80U70|SUZ12_MOUSE 1 5 2 ETLTTELQTR sp|Q811D0|DLG1_MOUSE 1 3 2 QVTPDGESDEVGVIPSK sp|Q8BI72|CARF_MOUSE 1 4 2 GSASFVSSLLK sp|Q8BM55|TM214_MOUSE 1 4 2 SQSVFTGNPSVWLK sp|Q8BT07|CEP55_MOUSE 1 5 2 YSSSSLFEQLEEK sp|Q8BZ20|PAR12_MOUSE 1 4 2 LGLSSDLVSR sp|Q8CGU1|CACO1_MOUSE 1 3 2 AALLGEELASAAGAR sp|Q8K0Q5|RHG18_MOUSE 1 4 2 IEEGSLETEGLLR sp|Q8R2M2|TDIF2_MOUSE 1 3 2 LTSSSIDPGLNIK sp|Q8VIM9|IRGQ_MOUSE 1 3 2 PLPQGDVTALFLGPPGSGK sp|Q91VW5|GOGA4_MOUSE 1 3 2 SLLEELASQLDSR sp|Q91WR3|ASCC2_MOUSE 1 3 2 HNIFQNDEFDVFSR sp|Q91YK2|RRP1B_MOUSE 1 3 2 LGALPDSSSDLPVQK sp|Q99J27|ACATN_MOUSE 1 4 2 YTAGPQPLNIFYK sp|Q99KB8|GLO2_MOUSE 1 3 2 HVEPGNAAIQEK sp|Q99LB2|DHRS4_MOUSE 1 4 2 LAEDGAHVVVSSR sp|Q99M28|RNPS1_MOUSE 1 3 2 DHIMEIFSTYGK sp|Q9D2N9|VP33A_MOUSE 1 4 2 IISAAFEER sp|Q9DBR0|AKAP8_MOUSE 1 3 2 TVEFLQEYIINR sp|Q9JJA2|COG8_MOUSE 1 3 2 ISQFLQVLETDLYR sp|Q9WTR1|TRPV2_MOUSE 1 3 2 GVPEELTGLLEYLR sp|Q9Z2V5|HDAC6_MOUSE 1 3 2 LVDALM[147]GAEIR sp|Q64261|CDK6_MOUSE 0.9917 1 2 VQTSEEGM[147]PLSTIR sp|P63011|RAB3A_MOUSE 1 2 2 M[147]SESLDTADPAVTGAK sp|Q04692|SMRCD_MOUSE 0.9999 2 2 QEQLYSGLFNR sp|Q3U308|CP084_MOUSE 0.9999 3 2 DLPSLDPLPPYVLAEAQLR sp|Q8VHE6|DYH5_MOUSE 0.9999 7 2 RTDLNYIAAVDLK sp|P24638|PPAL_MOUSE 0.9998 2 2 LQGGVLLAQILK sp|Q8BZQ7|ANC2_MOUSE 0.9998 2 2 IEELFSIIR sp|Q99KR7|PPIF_MOUSE 0.9998 3 2 HVGPGVLSM[147]AN sp|P83510|TNIK_MOUSE 0.9993 2 2 NIATYYGAFIK sp|P62858|RS28_MOUSE 0.9984 2 2 EGDVLTLLESER sp|Q8R2U4|ME11A_MOUSE 0.9978 2 2 TAGLSLLAEER sp|P45878|FKBP2_MOUSE 0.9968 2 2 LVIPSELGYGER sp|P08032|SPTA1_MOUSE 0.9967 3 2 FLTLLAK sp|Q8C7B8|ZSWM4_MOUSE 0.9943 2 2 LQPALTSR sp|O35682|MYADM_MOUSE 1 2 2 TTITTTTSSSTTVGSAR sp|O70481|UBR1_MOUSE 1 2 2 INSENAEALAQLLTLAR sp|O88746|TOM1_MOUSE 1 4 2 SSPDLTGVVAVYEDLR sp|P48771|CX7A2_MOUSE 1 2 2 LFQEDNGM[147]PVHLK sp|P62313|LSM6_MOUSE 1 2 2 GNNVLYISTQK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 566 sp|P97314|CSRP2_MOUSE 1 2 2 GFGYGQGAGALVHAQ sp|Q00547|HMMR_MOUSE 1 3 2 DVTAQLESVQEK sp|Q3TVI8|PBIP1_MOUSE 1 2 2 QEGLALFGVELAPVR sp|Q3UVG3|F91A1_MOUSE 1 3 2 VQGDYFETLLYK sp|Q60772|CDN2C_MOUSE 1 3 2 DGTGFAVIHDAAR sp|Q64FW2|RETST_MOUSE 1 3 2 ATVQSVLLDSAGR sp|Q6DVA0|LEMD2_MOUSE 1 3 2 ELQALGFQPGPITDTTR sp|Q6NVE8|WDR44_MOUSE 1 2 2 LLASAGQDNIVR sp|Q6NZN0|RBM26_MOUSE 1 3 2 VIQPLVQQPILPVVK sp|Q80XU3|NUCKS_MOUSE 1 2 3 TPSPKEEDEEAESPPEKK sp|Q8BFW7|LPP_MOUSE 1 3 2 M[147]LYDM[147]ENPPADDYFGR sp|Q8BTZ5|ANR46_MOUSE 1 2 2 LLESLEEQEVK sp|Q8BUY5|CC001_MOUSE 1 2 2 AGAVAADSPGFVEDR sp|Q8C3I8|BRP16_MOUSE 1 3 2 DQGAYLILR sp|Q8C9B9|DIDO1_MOUSE 1 3 2 TASPLEHILQTLFGK sp|Q8K3X4|EAP1_MOUSE 1 2 2 YGLSAAAAAAAAAAAVEQR sp|Q8R307|VPS18_MOUSE 1 3 2 LGALQGDPDALTLYR sp|Q8R3P6|CO044_MOUSE 1 3 2 AALAFGFLDLLK sp|Q8R3V5|SHLB2_MOUSE 1 2 2 LASDAGIFFTR sp|Q8R5H6|WASF1_MOUSE 1 3 2 IENDVATILSR sp|Q8VD04|GRAP1_MOUSE 1 4 2 TQTGDSSSVSSFSYR sp|Q91VS7|MGST1_MOUSE 1 2 2 IYHTIAYLTPLPQPNR sp|Q91VX9|TM168_MOUSE 1 2 2 TVDIEEADPPQLGDFTR sp|Q91VY9|ZN622_MOUSE 1 3 3 VHSFFIPDIEYLSDLK sp|Q922S8|KIF2C_MOUSE 1 4 2 FSLVDLAGNER sp|Q922Y1|UBXN1_MOUSE 1 2 2 GEEPGQDQDPVQLLSGFPR sp|Q923D5|WBP11_MOUSE 1 2 2 AVSILPLLGHGVPR sp|Q9CQU3|RER1_MOUSE 1 2 2 LGQIYQSWLDK sp|Q9CYK1|SYWM_MOUSE 1 3 2 YGEFFPLPK sp|Q9D0M0|EXOS7_MOUSE 1 3 3 VYIVHGVQEDLR sp|Q9D1C8|VPS28_MOUSE 1 2 2 AM[147]DEIQPDLR sp|Q9DB96|NGDN_MOUSE 1 2 2 ASGASLQGHPAVLR sp|Q9DC29|ABCB6_MOUSE 1 2 2 APDIILLDEATSALDTSNER sp|Q9DCD2|SYF1_MOUSE 1 2 2 FYEDNGQLDDAR sp|Q9QUJ7|ACSL4_MOUSE 1 4 2 SDQSYVISFVVPNQK sp|Q9QWF0|CAF1A_MOUSE 1 2 2 LVGGQGPIDSFLR sp|Q9QY06|MYO9B_MOUSE 1 3 2 TPIESLFIEATER sp|Q9Z1T6|FYV1_MOUSE 1 3 2 DYFPEQIYWSPLLNK sp|Q9Z2A5|ATE1_MOUSE 1 3 2 SLEDLIFQSLPENASHK sp|Q9Z2L7|CRLF3_MOUSE 1 2 2 LIEHGVNTADDLVR sp|O35638|STAG2_MOUSE 0.9999 2 2 M[147]YSDAFLNDSYLK sp|P25425|PO2F1_MOUSE 1 3 2 LYGNDFSQTTISR sp|P42232|STA5B_MOUSE 1 4 2 IQAQFAQLGQLNPQER sp|P61620|S61A1_MOUSE 1 5 3 GM[147]EFEGAIIALFHLLATR sp|P84075|HPCA_MOUSE 0.997 1 2 IYANFFPYGDASK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 567 sp|P15116|CADH2_MOUSE 0.9999 3 2 LNGDFAQLNLK sp|P70700|RPA2_MOUSE 0.9999 3 2 ISLTIVDAVISPPSVPK sp|Q60928|GGT1_MOUSE 0.9999 2 2 FVDVSQVIR sp|Q8CI95|OSB11_MOUSE 0.9999 3 2 SVILHLLSQLK sp|Q91V01|PCAT3_MOUSE 0.9999 2 2 LATSLGASEQALR sp|Q9D0P8|RAYL_MOUSE 0.9999 2 2 SQTSGISLPGVLVGTK sp|Q9JJL8|SYSM_MOUSE 0.9999 2 2 VLIALLESNQQK sp|Q9QZ08|NAGK_MOUSE 0.9999 2 2 LGILTHLYR sp|Q02780|NFIA_MOUSE 0.9999 2 2 LDLVM[147]VILFK sp|Q91W69|EPN3_MOUSE 0.9999 2 2 NIVHNYSEAEIK sp|Q9CTG6|AT132_MOUSE 0.9996 3 2 ELSLLGLLVM[147]R sp|Q9R0Q9|MPU1_MOUSE 0.9996 2 2 GLGLGIVAGSLLVK sp|Q60848|HELLS_MOUSE 0.9995 2 2 LISQIQPEVNR sp|Q91ZN5|S35B2_MOUSE 0.9994 2 2 APDEVLLAPR sp|Q9WUU9|MCM3A_MOUSE 0.9994 3 2 LPLYLPQTLVSFPDSIK sp|P00848|ATP6_MOUSE 0.9993 2 2 LSM[147]AIPLWAGAVITGFR sp|Q8BTI7|ANR52_MOUSE 0.9993 2 2 DAVSPFSFSLLK sp|Q9JHS3|MAPIP_MOUSE 0.9993 2 2 NGNQAFNEDSLK sp|Q3TYS2|CQ062_MOUSE 0.9991 2 2 DIQDVNVEEEK sp|O88848|ARL6_MOUSE 0.999 3 2 IPILFFANK sp|Q80TZ9|RERE_MOUSE 0.999 2 2 VDSFFYILGYNPETR sp|P97412|LYST_MOUSE 0.9988 5 2 DLSGLLVSAFK sp|Q9CPP0|NPM3_MOUSE 0.9988 2 2 DHDNQEIAVPVANLR sp|Q8BFS6|CSTP1_MOUSE 0.9986 2 2 LTEQAVEAINK sp|Q8R1S0|COQ6_MOUSE 0.9981 2 2 ILLLEAGPK sp|Q8VHN7|GPR98_MOUSE 0.998 4 2 FAEPCVLR sp|Q9R0M6|RAB9A_MOUSE 0.9976 2 2 EPESFPFVILGNK sp|Q8BYU6|TOIP2_MOUSE 0.9957 5 2 FESLPAGSTLIFYK sp|Q9WTL7|LYPA2_MOUSE 0.9956 2 2 FGALTAEK sp|Q9Z2R6|U119A_MOUSE 0.9949 1 2 QPIGPEDVLGLQR sp|Q9CQT2|RBM7_MOUSE 0.9919 2 2 TLFVGNLETK sp|Q9CY57|CA077_MOUSE 0.9917 1 2 EQLDNQLDAYM[147]SK sp|P03930|ATP8_MOUSE 1 2 2 VSSQTFPLAPSPK sp|P24610|PAX3_MOUSE 1 3 2 TTFTAEQLEELER sp|P70302|STIM1_MOUSE 1 3 2 YAEEELEQVR sp|P70677|CASP3_MOUSE 1 4 2 LEFM[147]HILTR sp|Q09143|CTR1_MOUSE 1 2 2 VIYAM[147]AEDGLLFK sp|Q3UA37|QRIC1_MOUSE 1 3 2 EIQEAIAVANATTM[147]H sp|Q3UGY8|BIG3_MOUSE 1 4 2 NLIDTLSTPLTGR sp|Q3UW53|NIBAN_MOUSE 1 4 2 VLTSEEEYSLLSDK sp|Q64430|ATP7A_MOUSE 1 5 2 QIEAVGFPAFIK sp|Q8BHY8|SNX14_MOUSE 1 3 2 LVSLITLLR sp|Q8BTG3|T11L1_MOUSE 1 3 2 AIFSVLDLM[147]K sp|Q8C5W3|TBCEL_MOUSE 1 3 2 YYVDVPQEEVPFR sp|Q8CIM8|INT4_MOUSE 1 3 2 FLQEVDFFQR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 568 sp|Q91VZ6|SMAP1_MOUSE 1 3 2 LLYEANLPENFR sp|Q9JLV1|BAG3_MOUSE 1 3 2 ELLALDSVDPEGR sp|Q9QYC7|VKGC_MOUSE 1 3 2 DGLTGELGYLNPGVFTQSR sp|Q9QYH6|MAGD1_MOUSE 1 3 2 ATEAVLWEALR sp|Q9R0I7|YLPM1_MOUSE 1 3 2 VFSSEQGLGESSALSQSIIAAK sp|P61028|RAB8B_MOUSE 0.9993 2 2 SSTNVEEAFFTLAR sp|Q8JZR6|S4A8_MOUSE 0.9984 2 2 FLFILLGPVGK sp|Q9CQK7|RWDD1_MOUSE 1 4 3 AKFDAELLEIKK sp|P10922|H10_MOUSE 0.9999 2 2 YSDM[147]IVAAIQAEK sp|Q3UUQ7|PGAP1_MOUSE 0.9998 3 2 AFFDLIDADTK sp|Q8CHC4|SYNJ1_MOUSE 0.9998 3 2 VLDAYGLLGVLR sp|Q8K370|ACD10_MOUSE 0.9998 4 2 LSLQPSEAIFLDDLGSNLK sp|Q8BGZ2|F168A_MOUSE 0.9997 2 2 SIPSAIYPAPVAAPR sp|Q922H1|ANM3_MOUSE 0.9997 3 2 QTVFLLEKPFPVK sp|P58854|GCP3_MOUSE 0.9996 3 2 YLLLGQGDFIR sp|Q9CQZ6|NDUB3_MOUSE 0.9996 3 2 M[147]ELPDYR sp|P47713|PA24A_MOUSE 0.9995 3 2 DVPVVAILGSGGGFR sp|Q99JP7|GGT7_MOUSE 0.9995 3 2 LPEDEPAPAAPLR sp|O55242|OPRS1_MOUSE 0.9994 2 2 QYAGLDHELAFSR sp|Q8K5B2|MCFD2_MOUSE 0.9994 2 2 DDDKNNDGYIDYAEFAK sp|Q5U430|UBR3_MOUSE 0.9992 3 2 LDPDYFISSVFER sp|Q9QZH6|ECSIT_MOUSE 0.9992 3 2 IFVHYPR sp|Q9Z0J0|NPC2_MOUSE 0.9991 2 2 LPVKNEYPSIK sp|Q6P8H8|ALG8_MOUSE 0.999 2 2 AILLAILPM[147]SLLSVEK sp|Q8K4M5|COMD1_MOUSE 0.999 2 2 LSEVEESINR sp|Q9CQB5|CISD2_MOUSE 0.9987 3 2 QLPVPDSITGFAR sp|Q03173|ENAH_MOUSE 0.9986 2 2 VEDGSFPGGGNTGSVSLASSK sp|Q9EPN1|NBEA_MOUSE 0.9984 3 2 EISNFEYLM[147]FLNTIAGR sp|Q9R0Q4|MO4L2_MOUSE 0.9983 2 2 EYAVNEVVGGIK sp|Q8VCI5|PEX19_MOUSE 0.9975 2 2 ELAEEEPHLVEQFQK sp|Q80U78|PUM1_MOUSE 0.9973 1 2 SASSASSLFSPSSTLFSSSR sp|Q6ZPE2|MTMR5_MOUSE 0.9964 2 2 GLLALLFPLR sp|Q8CHT3|INT5_MOUSE 0.9957 2 2 EQPLLFELLK sp|Q91XL9|OSBL1_MOUSE 0.9917 1 2 ITM[147]PVIFNEPLSFLQR sp|Q80UG2|PLXA4_MOUSE 0.9911 1 2 FVDDLFETIFSTAHR sp|A2AAJ9|OBSCN_MOUSE 0.9904 3 2 EDENFVCIR sp|P97857|ATS1_MOUSE 0.99 1 2 GIGYFFVLQPK sp|O89017|LGMN_MOUSE 1 2 2 DYTGEDVTPENFLAVLR sp|Q3UFM5|NOM1_MOUSE 1 3 2 ELITEAQTQASGAGNK sp|Q5PRF0|HTR5A_MOUSE 1 2 2 VLILEQLLNSIK sp|Q5SUC9|SCO1_MOUSE 1 2 2 LVGLTGTKEEIDGVAR sp|Q64282|IFIT1_MOUSE 1 3 2 ISEQVQFLDIK sp|Q91WG2|RABE2_MOUSE 1 2 2 LQAELETSEQVQR sp|Q99MU3|DSRAD_MOUSE 1 3 2 YLNTNPVGGLLEYAR sp|Q9CR88|RT14_MOUSE 1 2 2 HLADHGLLSGVQR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 569 sp|Q9D187|FA96B_MOUSE 1 2 2 SGERPVTAGEEDEEVPDSIDAR sp|Q9Z0R9|FADS2_MOUSE 1 3 2 ALIDIVSSLK sp|Q61771|KIF3B_MOUSE 1 2 2 HLIIENFIPLEEK sp|O08601|MTP_MOUSE 0.9999 3 2 SGFTTANQVLGVSSK sp|O35350|CAN1_MOUSE 0.9999 2 2 APSDLYQIILK sp|O88879|APAF_MOUSE 0.9999 2 2 GSPLVVSLIGALLR sp|Q3SXD3|HDDC2_MOUSE 0.9999 2 2 LQDFYDSTAGK sp|Q3U5F4|YRDC_MOUSE 0.9999 2 2 LPESEPVEAASPER sp|Q80XQ2|TBCD5_MOUSE 0.9999 2 2 TFPEM[147]QFFQQENVR sp|Q8BKX6|SMG1_MOUSE 0.9999 4 2 AQDTFQTIEGIIR sp|Q9CQ75|NDUA2_MOUSE 0.9999 2 2 TVSLNNLSADEVTR sp|Q9DBA9|TF2H1_MOUSE 0.9999 2 2 M[147]LQEDPVLFQLYK sp|Q9EQG9|C43BP_MOUSE 0.9999 2 2 DVLYLSAIR sp|Q3TEL6|RN157_MOUSE 0.9999 2 2 VSYLLQEIYGIENK sp|Q66T02|PKHG5_MOUSE 0.9998 3 2 SLGEVLLPVFER sp|Q8R080|GTSE1_MOUSE 0.9998 2 2 VPQFSVGESPGGVTPK sp|Q8VHL1|SETD7_MOUSE 0.9998 2 2 VYVADSLISSAGEGLFSK sp|Q99LH1|NOG2_MOUSE 0.9998 2 2 VIDSSDVVVQVLDAR sp|Q05793|PGBM_MOUSE 0.9997 5 2 VIPYFTQTPYSFLPLPTIK sp|Q3UGP8|AG10B_MOUSE 0.9997 2 2 YFILPYIIYR sp|Q9D4H2|GCC1_MOUSE 0.9997 2 2 TQLATLTSSLATVTQEK sp|Q9Z1X9|CC45L_MOUSE 0.9997 2 2 LQEFLADM[147]GLPLK sp|Q6IQX7|CHSS2_MOUSE 0.9996 2 2 LTVLLPLAAAER sp|Q6PEV3|WIPF2_MOUSE 0.9996 2 2 GSSGGYGPGAAALQPK sp|Q6XUX1|RIPK5_MOUSE 0.9996 2 3 SPLYGQLVDLGYLSSSHR sp|Q6ZWZ2|UB2R2_MOUSE 0.9996 2 2 FPIDYPYSPPTFR sp|P40201|CHD1_MOUSE 0.9927 2 2 M[147]LDILAEYLK sp|Q5SWT3|S2535_MOUSE 0.9994 2 2 LGTYGLAESR sp|Q91ZR1|RAB4B_MOUSE 0.9994 2 2 M[147]GSGIQYGDISLR sp|P46414|CDN1B_MOUSE 0.9993 2 2 NLFGPVNHEELTR sp|Q3U5Q7|CMPK2_MOUSE 0.9993 2 2 AFYSLGNYLVASEIAK sp|Q9CQ86|CQ037_MOUSE 0.9993 2 2 EEYPGIEIESR sp|P62311|LSM3_MOUSE 0.9992 2 2 GDGVVLVAPPLR sp|Q5BLK4|ZCHC6_MOUSE 0.9992 2 2 NTEPVGQLWLGLLR sp|Q8R550|SH3K1_MOUSE 0.9992 2 2 M[147]EPAVSSQAAIEELK sp|P54116|STOM_MOUSE 0.9991 2 2 EASM[147]VITESPAALQLR sp|Q501J2|F173A_MOUSE 0.9991 2 2 LQAELPVGAR sp|Q3UDR8|YIPF3_MOUSE 0.999 2 2 DIPAVLPAAR sp|Q9EPQ7|STAR5_MOUSE 0.999 2 2 SM[147]AEFYPNLQK sp|Q9CYZ6|CS060_MOUSE 0.9989 2 2 DAPIATLVQR sp|P28741|KIF3A_MOUSE 0.9988 2 2 SAKPETVIDSLLQ sp|Q9CYA6|ZCHC8_MOUSE 0.9988 2 2 LVNYPGFNISTPR sp|Q9R1S3|PIGN_MOUSE 0.9988 2 2 EATLPFLFTPFK sp|Q9R207|NBN_MOUSE 0.9988 2 2 LLPAAGAAPGEPYR sp|Q9JKX4|AATF_MOUSE 0.9987 2 2 ALLTTNQLPQPDVFPVFK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 570 sp|Q5KU39|VPS41_MOUSE 0.9986 4 2 IVLLM[147]DFDSEK sp|P0C1Q2|PDE11_MOUSE 0.9985 2 2 DISNDLDLTSLSYK sp|Q76KJ5|RPA34_MOUSE 0.9985 2 2 EATLLASSSEAGGR sp|Q9D850|TMM68_MOUSE 0.9985 2 2 TFLGDPIPYDPK sp|Q9R0D8|WDR54_MOUSE 0.9985 2 2 TISALDLAPEVGK sp|Q8K2F8|LS14A_MOUSE 0.9981 2 2 YEGILYTIDTENSTVALAK sp|Q9CPV5|PMF1_MOUSE 0.9981 2 2 NQELADAVLAGR sp|Q9DCI9|RM32_MOUSE 0.998 2 2 QQIGAQEGGPFR sp|Q9CQ18|RNH2C_MOUSE 0.9978 2 2 HDADGLQASFR sp|Q9D8Y1|T126A_MOUSE 0.9978 2 2 ILNVTQAR sp|Q9WTK3|GPAA1_MOUSE 0.9976 2 2 YGVEALTLR sp|P70444|BID_MOUSE 0.9975 2 2 IEPDSESQEEIIHNIAR sp|Q07139|ECT2_MOUSE 0.9974 2 2 LPSVALLLNDLK sp|Q8BMD6|CN101_MOUSE 0.9974 3 2 LPGTGIDPEVLLSEAIR sp|Q9D600|PSF2_MOUSE 0.9974 2 2 TNLQPSESTQSQDF sp|Q8BTJ4|ENPP4_MOUSE 0.9973 2 2 EVDDLIGDIVLK sp|Q8QZX2|CD015_MOUSE 0.9973 2 2 TNPM[147]VFLSQFPLGK sp|Q924L1|LTMD1_MOUSE 0.9972 2 2 LGIGQLTAQEVK sp|Q7TSZ8|NACC1_MOUSE 0.9971 2 2 FSTPDLALNR sp|Q61207|SAP_MOUSE 0.997 2 2 TVVTEAGNLLK sp|Q9D023|BR44_MOUSE 0.9969 3 2 YSLVIIPK sp|Q8K1A6|C2D1A_MOUSE 0.9967 2 2 SFDPVLEALSR sp|Q9CPR1|RWDD4_MOUSE 0.9966 2 2 SIYEGDNSFR sp|Q3TBW2|RM10_MOUSE 0.996 2 2 VFPSQVLKPFLENSK sp|Q8BK75|CC075_MOUSE 0.9958 2 2 GQLVFLEGLK sp|P50096|IMDH1_MOUSE 0.9954 1 2 NLIDAGVDGLR sp|P63139|NFYB_MOUSE 0.9951 2 2 EQDIYLPIANVAR sp|Q9R059|FHL3_MOUSE 0.9949 2 2 TLTQGGVTYR sp|Q3ULF4|SPG7_MOUSE 0.9944 2 2 EGGFSAFNQLK sp|Q80UZ2|SDA1_MOUSE 0.9943 1 2 DLLVQYATGK sp|Q8VE19|MIO_MOUSE 0.9932 2 2 NLAIFDLR sp|Q9JKK8|ATR_MOUSE 0.9931 3 2 FLDLIPQDTLAVASFR sp|Q8CIG3|AOF1_MOUSE 0.9927 2 2 VLVTVPLAILQR sp|Q3TDD9|KLRAQ_MOUSE 0.9917 2 2 EGLAQQVQQSLEK sp|Q6PCM2|INT6_MOUSE 0.9917 1 2 NLQAEGLTTLGQSLR sp|Q8K2F0|BRD3_MOUSE 0.9917 1 2 EYPDAQGFAADIR sp|Q8R0J7|VP37B_MOUSE 0.9917 1 2 SLAEGNLLYQPQLDAQK sp|Q9QUG2|POLK_MOUSE 0.9912 2 2 ASTVPAAISTAEEIFAIAK sp|Q68EF0|RAB3I_MOUSE 0.9908 1 2 IDVLQAEVAALK sp|Q7TPM1|BAT2L_MOUSE 0.99 1 2 LLSFSPEEFPTLK sp|O35379|MRP1_MOUSE 1 2 2 LYAWELAFQDK sp|P70399|TP53B_MOUSE 1 3 2 VITDVYYVDGTEVER sp|Q4FZC9|SYNE3_MOUSE 1 2 2 AATLLEQVTSSVR sp|Q6P8M1|TATD1_MOUSE 1 2 2 HQDDLQDVIER sp|Q8BK03|FA73B_MOUSE 1 2 2 PAAAYEEALQLVK  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 571 sp|Q8BW10|NOB1_MOUSE 1 2 2 TDVFAPDYIAGVSPFAENDISSR sp|Q99K74|MED24_MOUSE 1 2 2 VESLVALLNNSSEM[147]K sp|Q9CQF4|CF203_MOUSE 1 2 2 SEQEEELESEPGVAK sp|Q9Z2A7|DGAT1_MOUSE 1 2 2 LQDSLFSSDSGFSNYR sp|P59016|VP33B_MOUSE 0.9999 2 2 AGLLTEQAPGDTLTAVESK sp|Q5DU25|IQEC2_MOUSE 0.9999 2 2 LIEAFSQR sp|Q8VDD9|PHIP_MOUSE 0.9999 2 3 YHDM[147]PDVIDFLVLR sp|A2AR02|PPIG_MOUSE 0.9998 2 2 DFM[147]VQGGDFSEGNGR sp|Q8C3X8|LMF2_MOUSE 0.9998 2 2 LFGSVEHLQLANSYGLFR sp|P70188|KIFA3_MOUSE 0.9997 2 2 SLNANTDITSLAR sp|Q9ER69|FL2D_MOUSE 0.9997 2 2 STM[147]VDPAINLFFLK sp|Q9QZ73|DCNL1_MOUSE 0.9997 2 2 QFM[147]IFTQSSEK sp|P58501|GCFC_MOUSE 0.9996 2 2 TLQELSIDGLLNR sp|P22366|MYD88_MOUSE 0.9991 2 2 FALSLSPGVQQK sp|P97480|EYA3_MOUSE 0.999 2 2 SNVGGLLSPQR sp|Q0P678|ZCH18_MOUSE 0.999 2 3 ASQQAAAPQPAVPGQPQQGSFVAHK sp|Q9CX00|K0174_MOUSE 0.999 2 2 ELDSGLAESVSTLIWAAPR sp|Q61037|TSC2_MOUSE 0.9987 3 2 LGYLPYSLLFR sp|P46938|YAP1_MOUSE 0.9985 2 2 GDSETDLEALFNAVM[147]NPK sp|P62046|LRCH1_MOUSE 0.9985 2 2 NLESIDPQFTIR sp|P97473|TRBP2_MOUSE 0.9985 2 2 TPISLLQEYGTR sp|Q5PSV9|MDC1_MOUSE 0.9985 3 2 LGLPLLSPEFLLTGVLK sp|Q9JKL4|CC060_MOUSE 0.9985 2 2 IEIVVVGTGNK sp|P97300|NPTN_MOUSE 0.9984 2 3 KRPDEVPDDDEPAGPM[147]K sp|Q99KY4|GAK_MOUSE 0.9984 2 2 IAVM[147]SFPAEGVESAIK sp|P59672|ANS1A_MOUSE 0.9981 3 2 LLLNGFDDVR sp|Q8VDR9|DOCK6_MOUSE 0.9979 2 2 VAELYLPLLSLAR sp|Q80VQ1|LRRC1_MOUSE 0.9967 1 2 SLEELLLDANQLR sp|Q9CXF7|CHD1L_MOUSE 0.9963 2 2 GIPTYIYYFPR sp|Q9D820|U566_MOUSE 0.9962 2 2 FVLDSAFLEGGHEK sp|Q8K2J4|CCD14_MOUSE 0.9952 3 2 NVSQTAEK sp|P49769|PSN1_MOUSE 0.9937 2 2 NETLFPALIYSST sp|Q3V1H1|CKAP2_MOUSE 0.9924 2 2 IEPITSPIENIISIYEK sp|P70236|MP2K6_MOUSE 0.9923 2 2 ADDLEPIVELGR sp|O35375|NRP2_MOUSE 0.9917 1 2 IFQANNDATEVVLNK sp|P50427|STS_MOUSE 0.9917 1 2 VLAALDELGLAR sp|P84089|ERH_MOUSE 0.9917 1 2 ADTQTYQPYNK sp|Q5DTK1|CHSS3_MOUSE 0.9917 1 2 DNTVQGQQVYYPIIFSQYDPK sp|Q8K4R9|DLGP5_MOUSE 0.9917 1 2 ANEILVQQGLESLTDR sp|Q91YU8|SSF1_MOUSE 0.9917 1 2 TEEELQAILAAK sp|Q9CQ39|MED21_MOUSE 0.9917 1 2 IQSALADIAQSQLK sp|Q9Z0R4|ITSN1_MOUSE 0.9917 1 2 LQEIDVFNNQLK sp|Q8VCF0|MAVS_MOUSE 0.9914 2 2 DTLWGLFNNLQR sp|Q6TEK5|VKORL_MOUSE 0.9908 1 2 GFGLLGSIFGK sp|Q9JIA7|SPHK2_MOUSE 0.9908 1 2 LLILVNPFGGR  X. Y. LIU ET AL. Copyright © 2011 SciRes. AJAC 572 sp|Q99LC9|PEX6_MOUSE 0.9902 3 2 LVFVGASEDR sp|Q3U1G5|I20L2_MOUSE 0.9985 2 2 IDLLGEFQSALPK sp|Q8BXA1|GOLI4_MOUSE 0.9981 2 2 DAGFQALEEQNQVEPR sp|P70158|ASM3A_MOUSE 0.998 2 2 NGNPLNSVFVAPAVTPVK sp|Q61624|ZN148_MOUSE 0.9979 2 2 LPQGLQYALNVPISVK sp|Q9QXE7|TBLX_MOUSE 0.9979 2 2 HQEPVYSVAFSPDGK sp|P59997|JHD1A_MOUSE 0.9974 2 2 ILLEELASSDPK sp|Q8BRG8|TM209_MOUSE 0.9918 2 2 YTVAPTSLVVSPGQQALLGLK sp|Q3URD3|SLMAP_MOUSE 0.9917 1 2 IEALQADNDFTNER sp|Q62388|ATM_MOUSE 0.9917 1 2 SVATSSIVGYILGLGDR sp|Q7TMW6|NARFL_MOUSE 0.9917 1 2 APDTEGSELLQQLER sp|Q80YR4|ZN598_MOUSE 0.9917 1 2 TPGLAPTPQAYLVPENFR sp|Q8K3K8|OPTN_MOUSE 0.9917 1 2 ADLLGIVSELQLK sp|Q9CPS7|PNO1_MOUSE 0.9917 1 2 RPVFPPLSGDQLLTGK sp|P70261|PALD_MOUSE 0.9908 1 2 ALGNILAYLSDAK sp|Q8BW49|TTC12_MOUSE 0.9908 1 2 ANTAIGILTDLALEER sp|Q91W92|BORG5_MOUSE 0.9908 1 2 LTADM[147]ISPPLGDFR sp|Q9Z0P7|SUFU_MOUSE 0.9908 1 2 VSILPDVVFDSPLH sp|P62737|ACTA_MOUSE 1 5 2 AVFPSIVGR sp|P12246|SAMP_MOUSE 0.9965 1 2 APPSIVLGQEQDNYGGGFQR sp|P62835|RAP1A_MOUSE 0.9952 1 2 INVNEIFYDLVR sp|Q80Y17|L2GL1_MOUSE 0.9948 1 2 APVVAIAVLDGR Table S3 All the abbreviated words and their full name 1DE-LC-MS/MS dimensional gel electrophoresis- liquid chromatography-mass spectrometry ACN Acetonitrile BSA Bovine Albumin Standards DMEM Dulbecco's Modified Eagle Media Dnmt3a DNA methyltransferase 3A Dnmt3a-D Dnmt3a depletion B16 melanoma ERK extracellular signal-regulated kinase FBS Fetal bovine serum GO Genome Ontology IEF isoelectro- focusing IPG immobilized pH gradient IPI International Protein Index JNK c-Jun N-terminal kinase KEGG Kyoto Encyclopedia of Genes and Genomes LTQ-Orbitrap Linear ion trap Orbitrap MAPK mitogen activated protein kinase signaling pathway NC negative control cell line PBS phosphate-buffered saline PMSF phenylmethanesulfonyl fluoride TPP Trans-Proteomic Pipeline
|