Journal of Computer and Communications
Vol.04 No.15(2016), Article ID:72314,8 pages
10.4236/jcc.2016.415004
A GASVM Algorithm for Predicting Protein Structure Classes
Longlong Liu, Mingjiao Ma*, Tingting Zhao
Department of Mathematical Sciences, Ocean University of China, Qingdao, China



Received: September 26, 2016; Accepted: November 25, 2016; Published: November 28, 2016
ABSTRACT
The research methods of protein structure prediction mainly focus on finding effective features of protein sequences and developing suitable machine learning algorithms. But few people consider the importance of weights of features in classification. We propose the GASVM algorithm (classification accuracy of support vector machine is regarded as the fitness value of genetic algorithm) to optimize the coefficients of these 16 features (5 features are proposed first time) in the classification, and further develop a new feature vector. Finally, based on the new feature vector, this paper uses support vector machine and 10-fold cross-validation to classify the protein structure of 3 low similarity datasets (25PDB, 1189, FC699). Experimental results show that the overall classification accuracy of the new method is better than other methods.
Keywords:
Protein Structural Classes, Protein Secondary Structure, Genetic Algorithm, Support Vector Machine

1. Introduction
For today’s advances in bioinformatics, one of the main tasks is the prediction of protein structure in post-genome era of genomic research [1]. Improving the classification accuracy of the spatial structure of proteins not only helps to understand protein function but also helps to understand how proteins perform biological functions [2]. Depending on the difference of secondary structure alignment and topology fragment in protein sequence, Levitt and Chothia divided a protein sequence into four structural classes: all-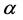 , all-
, all-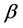 ,
, 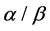 and
and 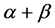 [3]. The current classification prediction algorithms are mostly concentrated on these four structural classes prediction.
[3]. The current classification prediction algorithms are mostly concentrated on these four structural classes prediction.
Current methods for protein structure prediction are mainly focus on finding effective features of protein sequences and developing suitable machine learning algorithms. The former kind of research is mostly based on the amino acid composition [4] and pseudo-amino acid composition [5], which considered that similar sequences have similar protein structures. But the prediction results are easily affected by the sequence similarity. For example, the prediction accuracy of a high similarity dataset is 95% while the prediction accuracy of a low similarity dataset may be only 40% - 60%. Because the relationship between protein structures is most associated with protein secondary structure, someone proposed methods based on protein secondary structure and protein functional domain to predict protein structure [6]. Experiments show that for low similarity datasets this method also has a high prediction accuracy. After extracting effective features, you can use a variety of classification algorithms to classify the extracted feature vector, such as Neural networks [7], Support vector machines [8], Bayesian classification [9], rough set theory [10], Fuzzy classification [11], Logit Boost classifier [12], Information about the differences method [13], etc. Thus, an appropriate machine learning algorithm is very important to the prediction.
2. Materials and Methods
In this section, first, give the methods for extracting 16 features from the protein secondary structure to compose a 16-dimensional feature vector; second, change every protein sequence of 3 low similarity datasets (25PDB, 1189 and FC699 dataset) to a 16-dimensional feature vector; finally, classify the 16-dimensional feature vectors by GASVM algorithm.
2.1. Materials
To evaluate the proposed method and facilitate its comparison with other existing methods, 3 widely used benchmark datasets 25PDB [13], 1189 [9] and FC699 [16] with sequence similarity lower than 25%, 40% and 40% respectively were selected. The compositions of 3 datasets were shown in Table 1.
2.2. 16-Dimensional Feature Vector
Through PSIPRED [14] software, each amino acid residue of protein sequences can be mapped to one kind of the following three secondary structural elements: H (Helix), E (Strand), and C (Coil). In this paper, let SSS denote secondary structure sequence and
Table 1. Compositions of 3 datasets.
no-C-SSS denote the sequence that was removed coil structure from secondary structure sequence. Let 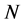 and
and 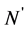 denote the length of SSS and the length of no-C-SSS respectively. For convenience, the 16-dimensional feature vector which is extracted from protein secondary structure is denoted by
denote the length of SSS and the length of no-C-SSS respectively. For convenience, the 16-dimensional feature vector which is extracted from protein secondary structure is denoted by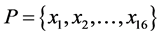 . The method to extract the feature vector will be described in more detail.
. The method to extract the feature vector will be described in more detail.
1) The first two features represent the proportion of H and E in SSS respectively, which have been proved significantly helpful in improving accuracy of protein structural classes[15], The features are as follows:
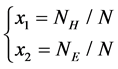
where 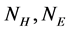 is the number of
is the number of 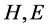 in the SSS respectively. Since
in the SSS respectively. Since 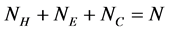 (where
(where 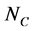 is the number of
is the number of 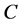 in the SSS), we only need to extract
in the SSS), we only need to extract 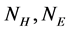 two features to represent the SSS.
two features to represent the SSS.
2) To classify the protein structures, the maximum length and the average length of 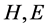 and
and  segments (the successive same letter) are also important factors. Six features are described as follows:
segments (the successive same letter) are also important factors. Six features are described as follows:
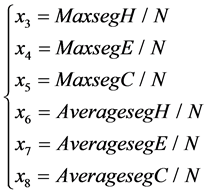
The




3) The more segments whose length reaches a certain value, the more likely to determine the structure of a protein. We respectively selected the segment 

where, 






4) While proteins in the 
























2.3. Construction of Classification Algorithm
2.3.1. Support Vector Machine
There are many algorithms to solve the protein multi-classification problem, such as neural net-work classification, support vector machine (SVM), Bayesian classification and so on. In this paper, support vector machine is selected for protein classification. The basic idea of SVM is map the data to a high dimensional space, and then find the data partition hyper plane in the high dimensional space. SVM has been widely used in protein secondary structure classification for its high prediction accuracy [17]. In this paper, we use “one-to-one” multi-classification method, and then combine 6 two-clas- sifiers to achieve multi-classification. Compared with other kernel function, the radial basis kernel function is better when deal with nonlinear problem [18]. So we select the radial basis kernel function 
2.3.2. GASVM Algorithm
Genetic algorithm (GA) is a method based on the principle of natural selection and genetic optimization search. It includes several steps, such as chromosome coding, population initialization, fitness function calculating, basic genetic operation and so on. Here, GASVM algorithm is proposed to optimize the coefficients of these 16 features in the classification. The classification accuracy of SVM is regarded as the fitness function value of GASVM algorithm. The steps of GASVM algorithm are described as follows:
1) Let the coefficient vector be
2) The new feature vector 


3) Calculate the new feature vector 
4) The first 160 individuals with big fitness function values are selected as parents in the next generation. In order to obtain the global optimum solution and improve the convergence rate, sorting selection method is adopted, the top 80% chromosomes with higher fitness from population are selected and copied into the mating pool.
5) A new generation is produced by the genetic and crossover operation of paternal generation chromosomes. The multi-point crossover is adopted.
6) In the new generation, 40 population samples are selected randomly, and then mutation is performed. It means the values of certain genes of a chromosome are replaced with other values to generate a new individual. Here, 5% of the chromosomes are mutated by point mutation method.
7) Repeat steps (2) to (6) until the fitness function values satisfy the requirement or the maximum number of cycles is reached.
3. Results and Discussion
The protein sequences in 25PDB, 1189, FC699 3 datasets were classified by GASVM algorithm and 10-fold cross-validation was used. The classification accuracy can be seen in Table 2, the overall accuracy of the 25PDB, 1189, FC699 dataset is 83.32%, 85.44% and 93.36% respectively, the accuracy of all-



Table 2. The results for the 3 datasets with 10-fold cross-validation.
Figure 1. The optimal coefficients of 3 datasets.
4. Comparison with Other Methods
The SCPRED, MODAS and RKS-PPSC methods are widely accepted in protein structure classification and the 25PDB, 1189, FC699 3 datasets are adopted to validate the effects. Here, the results of GASVM algorithm were compared with SCPRED, MODAS, RKS-PPSC and reference [20] (see Table 3). The data in Table 3 show that the overall accuracies obtained by our method are higher than other methods on 25PDB, 1189 and FC699 datasets, which increase 0.42%, 1.90% and 1.59% respectively.
Our method obtains the highest prediction accuracies for the classes among all the tested methods on 3 datasets. As for the class, the accuracy is 83.53% on 25PDB dataset and 86.69% on 1189 dataset, which is 0.17% and 0.41% lower than that of the famous MODAS method [21] respectively, but is 3.43% higher than SCPRED [17] and 2.29% higher than kongs’ method [20]. About the class, the accuracy is 81.02% on 25PDB dataset, which is 4.78% lower than that of the RKS-PPSC [22], but is 7.02% higher than SCPRED [17]; the accuracy is 87.11% on 1189dataset, which is 2.49% lower than SCPRED, but is 4.51% higher than RKS-PPSC. It is also noticed that the significant improvement is made in particular for the class, which is the difficult class to predict.
5. Conclusion
In the paper, the importance of the weights of different features in protein structure classification are considered, so GASVM algorithm is proposed to optimize the coeffi-
Table 3. The comparison of different methods.
cients of these 16 features in the classification. Finally, 10-fold cross-validation is used to classify the protein structures of3 low similarity datasets (25PDB, 1189, FC699) and experimental results show that the overall classification accuracy of the new method is better than other methods. GASVM algorithm is very effective in protein structure classification. Weights of different features are considered is very necessary.
Acknowledgements
The authors would like to thank all of the researchers who made publicly available data used in this study and thank the National Natural Science Foundation of China (No: 61303145) for the support to this work.
Cite this paper
Liu, L.L., Ma, M.J. and Zhao, T.T. (2016) A GASVM Algorithm for Predicting Protein Structure Classes. Journal of Computer and Communications, 4, 46-53. http://dx.doi.org/10.4236/jcc.2016.415004
References
- 1. Black, D.L. (2000) Protein Diversity from Alternative Splicing: A Challenge for Bioinformatics and Post-Genome Biology. Cell, 103, 367-370. http://dx.doi.org/10.1016/S0092-8674(00)00128-8
- 2. Anfinsen, C.B. (1973) Principles That Govern the Folding of Protein Chains. Science, 181, 223-230. http://dx.doi.org/10.1126/science.181.4096.223
- 3. Levitt, M. and Chothia, C. (1976) Structural Pattern in Globular Proteins. Nature, 261, 552-558. http://dx.doi.org/10.1038/261552a0
- 4. Chou, K.C. (1999) A Key Driving Force in Determination of Protein Structural Classes. Biochemical & Biophysical Research Communications, 264, 216-224. http://dx.doi.org/10.1006/bbrc.1999.1325
- 5. Chou, K.C. (2011) Some Remarks on Protein Attribute Prediction and Pseudo Amino Acid Composition. Journal of Theoretical Biology, 273, 236-247. http://dx.doi.org/10.1016/j.jtbi.2010.12.024
- 6. Chou, K.C. (2004) Structural Bioinformatics and Its Impact to Biomedical Science. Current Medicinal Chemistry, 11, 2105-2134. http://dx.doi.org/10.2174/0929867043364667
- 7. Volpato, V., Adelfio, A. and Pollastri, G. (2013) Accurate Prediction of Protein Enzymatic Class by n-to-1 Neural Networks. Bmc Bioinformatics, 14, 1-7. http://dx.doi.org/10.1186/1471-2105-14-s1-s11
- 8. Chen, Y.K. and Li, K.B. (2013) Predicting Membrane Protein Types by Incorporating Protein Topology, Domains, Signal Peptides, and Physicochemical Properties into the General form of Chou’s Pseudo Amino Acid Compo-sition. Journal of Theoretical Biology, 318, 1-12. http://dx.doi.org/10.1016/j.jtbi.2012.10.033
- 9. Wang, Z. and Zheng, Y. (2000) How Good Is Prediction of Protein Structural Class by the Component-Coupled Method? Proteins Structure Function & Bioinformatics, 38, 165-175. http://dx.doi.org/10.1002/(SICI)1097-0134(20000201)38:2<165::AID-PROT5>3.0.CO;2-V
- 10. Cao, Y., Liu, S., Zhang, L., Qin, J., Wang, J. and Tang, K. (2005) Prediction of Protein Structural Class with Rough Sets. Bmc Bioinformatics, 7, 1-6.
- 11. Shen, H.B., Yang, J., Liu, X.J. and Chou, K.C. (2005) Using Supervised Fuzzy Clustering to Predict Protein Structural Classes. Biochemical & Biophysical Research Communications, 334, 577-581. http://dx.doi.org/10.1016/j.bbrc.2005.06.128
- 12. Cai, Y.D., Feng, K.Y., Lu, W.C. and Chou, K.C. (2006) Using LogitBoost Classifier to Predict Protein Structural Classes. Journal of Theoretical Biology, 238, 172-176. http://dx.doi.org/10.1016/j.jtbi.2005.05.034
- 13. Jin, L., Fang, W. and Tang, H. (2003) Prediction of Protein Structural Classes by a New Measure of Information Discrepancy. Computational Biology & Chemistry, 27, 373-380. http://dx.doi.org/10.1016/S1476-9271(02)00087-7
- 14. Jones, D.T. (1999) Protein Secondary Structure Prediction Based on Position-Specific Scoring Matrices. Journal of Molecular Biology, 292, 195-202. http://dx.doi.org/10.1006/jmbi.1999.3091
- 15. Anand, A., Pugalenthi, G. and Suganthan, P.N. (2008) Predicting Protein Structural Class by SVM with Class-Wise Optimized Features and Decision Probabilities. Journal of Theoretical Biology, 253, 375-80. http://dx.doi.org/10.1016/j.jtbi.2008.02.031
- 16. Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped Blast and Psi-Blast: A New Generation of Protein Database Search Programs. Nucleic Acids Research, 25, 3389-3402. http://dx.doi.org/10.1093/nar/25.17.3389
- 17. Kurgan, L., Cios, K. and Chen, K. (2008) Scpred: Accurate Prediction of Protein Structural Class for Sequences of Twilight-Zone Similarity with Predicting Sequences. BMC Bioinformatics, 9, 815-818. http://dx.doi.org/10.1186/1471-2105-9-226
- 18. Zhang, S., Ding, S. and Wang, T. (2011) High-Accuracy Prediction of Protein Structural Class for Low-Similarity Sequences Based on Predicted Secondary Structure. Biochimie, 93, 710-714. http://dx.doi.org/10.1016/j.biochi.2011.01.001
- 19. Liu, T. and Jia, C. (2010) A High-Accuracy Protein Structural Class Prediction Algorithm Using Predicted Secondary Structural Information. Journal of Theoretical Biology, 267, 272-275. http://dx.doi.org/10.1016/j.jtbi.2010.09.007
- 20. Liang, K., Zhang, L. and Lv, J. (2014) Accurate Prediction of Protein Structural Classes by Incorporating Predicted Secondary Structure Information into the General Form of Chou’s Pseudo Amino Acid Composition. Journal of Theoretical Biology, 344, 12-18. http://dx.doi.org/10.1016/j.jtbi.2013.11.021
- 21. Mizianty, M.J. and Lukasz, K. (2009) Modular Prediction of Protein Structural Classes from Sequences of Twilight-Zone Identity with Predicting Sequences. BMC Bioinformatics, 10, 1-24. http://dx.doi.org/10.1186/1471-2105-10-414
- 22. Yang, J.Y., Peng, Z.L. and Xin, C. (2010) Prediction of Protein Structural Classes for Low- Homology Sequences Based on Predicted Secondary Structure. BMC Bioinformatics, 11, 1-10. http://dx.doi.org/10.1186/1471-2105-11-s1-s9





