Open Access Library Journal
Vol.03 No.10(2016), Article ID:71512,16 pages
10.4236/oalib.1103108
Facilitating the Automatic Characterisation, Classification and Description of Biological Images with the VisionBioShape Package for R
Biel Stela1, Antonio Monleón-Getino1,2*
1Section of Statistics, Faculty of Biology, University of Barcelona, Barcelona, Spain
2Statistics and Bioinformatics Research Group (GRBIO), Barcelona, Spain

Copyright © 2016 by authors and Open Access Library Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: September 28, 2016; Accepted: October 22, 2016; Published: October 26, 2016
ABSTRACT
Here, we present the VisioBioshapeR package for R [R Core, 2014]. This new library is a comprehensive, multifunctional toolbox designed to automatically analyse biological images. The package extends other common libraries (Momocs, ShapeR) used for biological shape analysis by allowing the user to extract closed contour outlines automatically from reading binary images. Current functionalities of VisioBioshapeR include: random extraction of image coordinates, analysis of the shape of a biological image by the elliptic Fourier descriptor (EFD) method, extraction of an image characteristic vector using multivariate principal component analysis (PCA) and geometrical analysis. The image vector of characteristics can be directly exported to a wide range of statistical packages in R and can be used to perform classification or other types of analysis in order to sort new images into classes. The package could prove useful in studies of any two-dimensional images and is presented with three examples of its application in ecology. The library is useful when multiple images are processed at a time and we wish to automate their analysis for example for recognition of images from patterns.
Subject Areas:
Applied Statistical Mathematics, Computer Vision, Ecology, Ecosystem Science, Environmental Sciences
Keywords:
Computer Vision, Biological Images, Ecology, Statistics, Elliptic Fourier Descriptor, Principal Component Analysis

1. Introduction
Automatic identification of images, and biological images in particular (Figure 1), is a major area of current research where images are transformed into information that can be used later for image characterisation, classification or description. Currently it is a prominent subject with dozens of tools already developed in the field of biology, such as the study “ShapeR: An R Package to Study Otolith Shape Variation among Fish Populations” [1] where the authors develop an R package to analyse otoliths morphometrically as a method to delineate fish stocks, characterise population movements and detect the natal origin of fish. Another example is “A tool for developing an automatic insect identification system based on wing outlines” where [2] developed a Matlab library to identify insects automatically based on wing outlines. Others techniques used for processing biological images can be found at [3] - [8] .
The principal image processing tasks involve several processes such as grey-level transformation, binarisation, image filtering, image segmentation, visual object tracking, optical flow and image registration. After acquiring image information, statistical techniques must be used for characterisation, classification or description, depending on the target. Image pattern recognition is the technique used to classify an input image into one of a set of predefined classes [1] . So the tasks of image processing can be divided into a feature extraction module and a statistical analysis module. We should emphasise here that a bio-image is a very difficult target even for state-of-the-art image processing and pattern recognition techniques, due to noise, deformations, etc., but unfortunately it is vital to have a suitable feature extraction module to permit good statistical analysis later on.
Here, we introduce VisioBioshapeR, an R package designed to analyse biological images automatically with a small number of easy-to-use functions (Table 1). There are built-in functions which allow users to perform automatic processes such as extracting the outlines from images, visualising the mean shape and transforming the outlines into independent coefficients using the result of principal component analysis (PCA) of the elliptic Fourier descriptor (EFD). The image vector of characteristics can be directly exported to a wide range of statistical packages in R and can be used to perform classification analysis or other types of analysis (cluster) in order to classify new images into classes.
The R library presented here is a general purpose library and resolves the problem of analysing numerous specimens at a time to recognise automatically biological images efficiently and within a reasonable time. As with the applications for identification of organisms, despite the existence of many image analysis applications, most free applications (SHERPA, ImageJ, Google’s Image Recognition Software) do not completely resolve the difficulty posed by biological imaging or processing, partly due to the variability and the large number of images needed to implement future automatic identification systems in biological imaging.
So, in this text, we present the mathematical basis for understanding the functions contained in the VisioBioshape library.
Figure 1. Phase contrast image of diatoms from the inventory of the River Ebro (Spain).
Table 1. R functions available in VisioBioshapeR.
1.1. Elliptic Fourier Descriptors
The functions e.fourier.graph() and automatic.PCAfourier.analysis1()are supported by the Elliptic Fourier Descriptor (EFD) method. It is a mathematical way to describe closed contours of images using coordinates previously extracted by the function coordtiff() or image.to.coords. It was first proposed as an application of the Fourier transform method [9] . The EFD performs the Fourier expansion of the x and y coordinates of the outline and outputs 4 coefficients for each harmonic. The first harmonic describes the best fit ellipse for the closed contour. The higher harmonics add more complex contour features; thus, the higher the harmonic, the finer the detail it describes. This method increases the number of coefficients in accordance with the harmonic.
We implemented the EFD algorithm in the functions e.fourier.graph() and automatic.PCAfourier.analysis1() laid out in [10] , as the base of the e.fourier.graph() function available in VisioBioshapeR.
Let T be the perimeter of the outline, and this perimeter then becomes the period of the signal. One sets 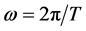 to be the pulse. Then the curvilinear abscissa t varies from 0 to T. The outline contains k, a finite number, points. One can therefore calculate discrete estimators for every coefficient of the nth harmonics:
to be the pulse. Then the curvilinear abscissa t varies from 0 to T. The outline contains k, a finite number, points. One can therefore calculate discrete estimators for every coefficient of the nth harmonics:
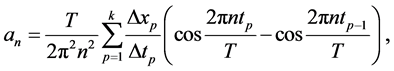 (1)
(1)
where:
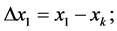 (2)
(2)
and:
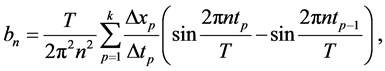 (3)
(3)
where n is the index of the nth harmonic.
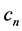 and
and 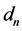 are calculated similarly.
are calculated similarly.
The first harmonic defines the ellipse that best fits the outline. One can then use the parameter of the first harmonic to normalise the data so that they can be invariant to size, rotation, and the starting position of the outline trace. The function then calculates a new set of Fourier coefficients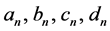 . The invariant properties of the new normalised coefficients allow us to use the new coefficients for further multivariate analysis and comparison.
. The invariant properties of the new normalised coefficients allow us to use the new coefficients for further multivariate analysis and comparison.
One obtains the set of normalised coefficients following the equations:
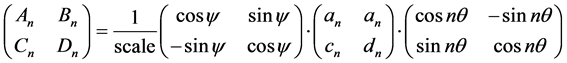 (4)
(4)
where “scale” is the magnitude of the semi-major axis of the ellipse defined by the first harmonic, 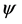 is the rotation angle of the orientation of the first ellipse,
is the rotation angle of the orientation of the first ellipse, 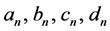 are the original harmonic coefficients and the last term,
are the original harmonic coefficients and the last term, 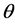 , is the rotation angle of the starting point to the end of the ellipse.
, is the rotation angle of the starting point to the end of the ellipse.
To compute these parameters one can use the formulae supplied in [11] :
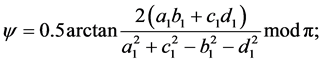
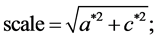
with:
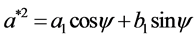
and:
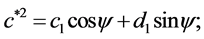
and:
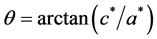
Using this method, the function e.fourier.graph() extracts valuable information from the image. Figure 2 presents an example of the use of the EFD to analyse an image of a dragonfly using the first nine harmonics and the function e.fourier.graph().
1.2. Principal Component Analysis
The automatic.PCAfourier.analysis1() and automatic.PCAfourier.analysis2() functions are assigned the task of obtaining a vector containing relevant features which are extracted from each image resulting from the geometric analysis of its form. The problem of feature selection is undoubtedly the most critical part of the framework, as the sensitivity and robustness of the identification algorithm depend closely on responsiveness to the information that has been obtained automatically. In this case, the characteristics that are extracted from the shape of the image are the main components of all the Fourier harmonics of an image using previously the EFD, the geometric centre of the image and the Euclidean distance of the coordinates.
PCA is a mathematical tool for analysing data in a way that allows us to identify patterns in the data, and for expressing the data in such a way as to highlight their similarities and differences. Patterns can be hard to find in data of a high dimension, as is the
Figure 2. Reconstruction contour with a number of harmonics.
case of the data contained and compiled within an image, where graphical representation is not available.
Conceptually, the goal of PCA is to reduce the number of variables of interest to a smaller set of components; in this case, the coefficients related to images. PCA analyses all the variance in the variables and reorganises it into a new set of components equal in number to the number of original variables. The new components are independent and they decrease with respect to the amount of variance in the originals they account for. The first component captures most of the variance; the 2nd, second most; and so on, until all the variance is accounted for. However, only some components will be retained for further study (dimension reduction). Since the first few capture most of the variance, further analysis typically focuses on them.
So, we apply PCA to the data contained in the matrix of EFD coefficients obtained in the elliptic Fourier analysis, either for the EFD coefficients obtained up to a certain number of harmonics or for all the harmonics obtained in the EFD analysis.
We use the function princomp() of R [12] , which uses the spectral decomposition approach, to perform the PCA analysis.
Thus for each image, each of the four Fourier coefficients (
The difference between automatic.PCAfourier.analysis1 and automatic.PCAfourier. analysis2 is that the first serves for the analysis of a single image at a time, either from the matrix of coordinates or a coordinate file, on the other hand automatic.PCAfourier. analysis2 has its origins in a list containing matrices of coordinates from multiple images or a single image. automatic.PCAfourier.analysis2 is therefore useful when multiple images are processed.
1.3. Image Geometry: Medoids and Euclidean Maximum Distance
The automatic.PCAfourier.analysis1() function can also analyse the image geometrically to obtain the centroid(medoid) and the Euclidean distances between the coordinates.
One of the interesting points of the image is the geometric centre; but there are different ways to calculate it. We selected the medoid as a good estimator of the image centroid. Medoids are representative objects of the coordinates X,Y whose average dissimilarity to all the coordinates is minimal. The medoid is not equivalent to a median or a geometric median. A median is only defined on a 1-dimensional coordinate, and it only minimises dissimilarity to other coordinates X,Y for a specific distance metric. A geometric median is defined in any dimension, but is not necessarily a point from within all the coordinates.
To calculate the medoid, we selected in R [12] the function pam() from the cluster library (Kaufman and Rousseeuw, 1990). pam Compared to the k-means approach in kmeans, the function pam has the following features: (a) it also accepts a dissimilarity matrix; (b) it is more robust because it minimises a sum of dissimilarities instead of a sum of squared Euclidean distances; and (c) it provides a novel graphic display.
Another important pieces of information obtained for automatic.PCAfourier. analysis1() is the distance between the coordinates X,Y of the image, which can contain information about relevant characteristic such as in the distance between tree rings or comparing some images with others. In order to calculate the distance, we used the Euclidean distance between all pairs of coordinates and extracted the maximum of them.
Many other metrics can be considered for data analysis purposes in the future.
2. Examples Section
In order to demonstrate the utility of the VisioBioshapeR package, here we present 3 examples of its use, one of which is real, including the automatic identification of 35 Tiff images of 3 genera of diatoms from an inventory of the River Ebro (Spain).
The first example consists of shape analysis of 35 diatoms using three genera: Navicula (n = 9), Cocconeis (n = 18) and Diatoma (n = 8), from the River Ebro inventory. Images can be retrieved from three folders (class 1, class 2 and class 3 respectively) in the examples directory (see “Documentation and availability” section). We then present two more simple examples with a dragonfly from a binarised digital camera image and tree rings that are designed and plotted using Paint (Microsoft Paint).
2.1. Sample Image Gallery
The VisioBioshapeR library, image samples and *.r script with some examples and analysis can be found at GitHub.
You will need to load VisioBioshapeR and retrieve in R the example data file with the commands library (VisioBioshapeR). To start the analysis, the path to the gallery of example images needs to be set to the folder “Templates” which contains three folders “class 1”, “class 2” and “class 3”, with the binarised diatom images. In the root folder, we can also find the dfly.tif (dragonfly) and tree.tif (ring tree) images.
The tree ring image was designed and plotted using Paint software.
Figure 3 represents the flow of processes performed by the VisioBioshapeR library, where you can see the image to be analysed and the result as a vector of image information. This process can be automated easily.
2.2. Automatic Identification of Diatoms
The first example of use refers to the automatic identification of diatoms; a requirement since 1999 [13] . Both the Windows software SHERPA [14] and the application developed by [15] automatically recognise diatoms, although neither of these allow use from R or general use for biological imaging.
Figure 3. Scheme using the library to read and analyze images automatically.
The samples of diatoms presented in the example of the use of VisioBioshapeR (see Figure 4) were obtained through the standard diatom sampling method and collected from a previous study on characterisation of the ecological state of the Ebro basin [16] .
We selected the three genera Navicula, Cocconeis and Diatoma because of their relative abundance in the samples and thus the higher probability of finding the target diatom valves in the samples. Moreover, these species are common in European rivers.
The taxonomic rank in this study was downgraded to genus so we can obtain more image samples from the ADIAC database [13] .
The observation and identification was performed using a Carl Zeiss Jenaval bright field microscope with phase contrast lenses Planachromat Phv 100×/1.3 and normal lenses 100×/1.3. The images were taken with an Olympus DP25 FW digital camera and the associated software Olympus Cell^ B. The images have 2560 × 1920 pixels, where each pixel equals 0.03376 mm2.
In each photograph session we obtained background images to correct irregularities in the light source of the microscope. The background was corrected by subtracting the background image from the normal image and thus obtaining a standardised image with regular background and colour correction.
We used the SHERPA software [14] for the segmentation of the standardised images. Then we obtained the final binary images by binarising the results of the SHERPA analysis in Tiff format.
In the following sections we demonstrate how to use the different functions in the three examples (diatoms, dragonfly and tree rings).
2.3. Coordinate Extraction from the Images
To obtain the outline of each image of interest we propose two solutions, one for the analysis of individual images, very versatile and allows the analysis of sub-images within a single image, it is the function Coordtiff() and another is the function image.to.coords(), more efficient computationally and that is destined to rapid removal of the images of a directory, all at once. The last must not have more than a single Tiff image to extract the coordinates correctly. Coordtiff() can interactively display the image once extracted coordinates and image.to.coords() does not.
For the Coordtiff() function we used a modification of the Conte function [10] : a classical R function used in the outline detection of a new image. This modification allows us to generate randomly all the automatic outlines for an image. The outlines are detected by first transforming the images into grey-scale images using the option “pixmapGrey”. The images are then binarised using a threshold pixel value (intensity threshold) which can be defined by the user. The outlines are then collected automatically in a list of X,Y coordinates to uniquely determine the position of a point of the image on a manifold of the graph space. The order of the coordinates is significant and is known as “the x-coordinate” and “the y-coordinate”.
The modified Conte function was named Coordtiff() and is defined as:
>Coordtiff(file, harmonic = F)
The file argument is used to read the images in Tiff format using the function readTiff [17] . The harmonic argument is used to perform a previous EFD analysis by means of the function efourier [10] and represent the reconstruction of the original image graphically using a number of the first 9 harmonics. The function returns a data frame which contains the X, Y coordinates of the outline of each image. The main advantage of the function Coordtiff() with respect to Conte is that if the image has different sub-images (for example tree rings), it can separate them all out.
An example of use of the function Coordtiff() is presented in Figure 3 using a binarised image of a dragonfly, where we can see a red line that represents the coordinates extracted overlaid on the image.
image.to.coords() is defined as:
>image.to.coords(filename, foldername, folder = F)
The filename argument is used to read a single image in Tiff format. The folder argument is a Boolean value that tells the function if we are reading a set of images in a folder (T) or a single image (F). foldername argument is the relative path to the folder. The algorithm for the outline extraction is from the raster library [17] originally used for extract the coordinates of a contour map. So the benefits of the function are a faster execution time and a more scalable data set reading tool.
2.4. EFD Analysis and Reconstruction of the Image
When all the outlines have been captured and stored in the form of X,Y coordinates, the shape coefficients of the image can be extracted using the function e.fourier.graph(). The EFD technique is performed using the efourier function [10] and the original im-
Figure 4. Example of diatoms used for automatic identification from the River Ebro (Spain) inventory.
age is represented with the reconstructed image using its harmonics (by default: 9 harmonics) by means of the iefourier function. The function e.fourier.graph is a modification of efourier which adds the possibility of normalising the images with regard to size and rotation using the function NEFnormalization and groups the coefficients with regards to size and rotation, then displays the coefficients.
The function is defined as:
>e.fourier.graph(M,n.harmonics = 9, coef = F, normalized = T)
The M argument is used to read the coordinates X and Y from the images using the function Coordtiff The n.harmonic argument is used to perform EFD analysis by means of the function efourier and to represent the reconstruction of the original image graphically using the function iefourier using a first n.harmonic harmonics.
The function returns the four coefficients 

The normalised argument is used to normalise (or not) the images with regards to size and rotation, and group the coefficients with regards to size and rotation, then to display the coefficients
Figure 2 and Figure 3 show examples of the use of e.fourier.graph() using an image of a dragonfly, reading the X,Y coordinates (Figure 3) and performing EFD analysis and reconstruction the image with the first 9 harmonics.
One of the purposes of this functions is adjust the number of harmonics required to a suitable reconstruction of the images in the extraction of the vector containing image information using the function automatic.PCAfourier.analysis1().
2.5. Shape Coefficients
When all the outlines have been captured in a image, the number of harmonics required is adjusted and normalisation is performed if necessary, the shape coefficients can be extracted and used for PCA analysis using the function automatic.PCAfouri- er.analysis1().
15 harmonics by default are used to give 4 elliptic Fourier coefficients (if the normalised option is used, the first three coefficients are omitted due to standardisation in relation to size, rotation and starting point).
The function is defined as:
>automatic.PCAfourier.analysis1(file, M, id.num, nom.label, tipus.input = 2, normalised = FALSE, num.harmonic = 15)
The file and M arguments are used to read the coordinates X, Y from the images using the function Coordtiff() in the case of using M or directly if we read a file containing the X,Y coordinates of the image.
The id.num argument is used to assign a number to the image analysed. The nom.label argument is used to assign a label (character) to the new image analysed. The argument tipus.input = 2 indicates to the function that we are reading a matrix, M, with the coordinates X and Y, and if we use tipus.input = 1 we are reading a file with the coordinates.
The normalised argument is used to normalise (or not) the images with regards to size and rotation and groups the coefficients with regards to size and rotation, and then displays the coefficients.
After the EFD analysis, in a second step, the function automatic.PCAfourier. analysis1() performs PCA analysis of the four first coefficients 

The function returns 21 variables containing information about the geometry of the image using the EFD, PCA and Euclidean analysis of the image:
・ Column 1: numerical ID of the image
・ Column 2: image label
・ Columns 3 - 10: PCA coefficients of all the EFD coefficients
・ columns 11 - 18: PCA coefficients of selected (nharmonics) elliptic Fourier coefficients
・ Columns 20 - 21: X and Y centroids of the image using the medoids
・ Column 21: Euclidean maximum distance between the X,Y coordinates.
2.6. Automatic Classification Analysis
Finally, in the example of R script we present a complete example of classification of biological images using 35 Tif images of 3 frequent genera of diatoms (navicula (n = 9), cocconeis (n = 18) and diatoma (n = 8)) from the River Ebro (Spain) that are shown in Figure 4. The objective was to obtain an automatic classification of the biological images for use with new images, using VisioBioshapeR.
The first step (see Figure 3) was to read the coordinates of all the Tiff images automatically (Figure 4) using the function Coordtiff() and store them in a vector of coordinates X,Y. The second step was to create a matrix to collect all the PCA-EFD coefficients obtained during the previous analysis using the automatic.PCAfourier.analysis1() function and group them in a matrix with 35 rows and more than 21 columns, indicating an ID and a label for each image. At the end, we created a new row with the class of diatoms (1 for navicula, 2 for cocconeis, 3 for diatoma). In the R script we present the use of some classification multivariate methods to obtain a discriminant function that allows us to separate different genera (Table 2).
In Table 2 we can see the percentage (%) of good classification using different methods:
We used the following discriminant methods to separate the three genera of diatoms using the variables calculated in the EFD-PCA:
Table 2. In brackets: the number of variables used.
・ LDA: Fisher linear discriminant analysis (with CV: Cross validation) using the MASS library,
・ RRLDA: Robust Regularised Linear Discriminant Analysis using the rrlda library,
・ FDA: Flexible Discriminant Analysis using the mda library,
・ NN: Neural Nets using the nnet library,
・ SVM: support vector machine using the kernlab library.
The percentage of good classification varies between 50% and 100%, and specially some methods can be considered useful to classify organism using the procedures by means of the library.
2.7. Other Examples
Another two examples are offered in the R script. First was a dragonfly (Figure 2, Figure 5) and the second, more complex was to analyse automatically an image of tree rings designed in order to apply it to tree growth analysis (see Figure 6 and Figure 7). Thus, we can obtain the distance between rings represented in Figure 7, where the X coordinates are the tree rings and Y is the distance between rings. So this could be useful to study tree growth or for climate reconstruction.
3. Conclusions and Prospects
The VisioBioshapeR library has demonstrated its usefulness in the three examples presented in this work. All the examples used in this paper were collected in R script to illustrate the utilities.
As we have seen in the above examples (dragonfly, tree rings and diatoms) this VisioBioshapeR library is acceptable for the specific purpose and has a wide variety of uses in automatic identification of biological images.
Acknowledgements
We thank the research group in diatoms led by Professor Jaume Cambra, Nuria Flor and Andrea Burfeid of the Botanical Department at the UB for their help and support, analysis of samples and providing images of diatoms.
Figure 5. Extraction of the X,Y coordinates from the image of a dragonfly.
Figure 6. Extraction of the coordinates X,Y in a tree rings analysis using the Coordtiff() function.
Figure 7. Analysis of the growth of a tree from automatic image analysis of its rings.
We also thank the Faculty of Biology at the UB for a grant it awarded in 2015 to support this project.
Documentation and Resources
The website for VisioBioshapeR can be found at GITHUB: https://github.com/BielStela/VisioBioShapeR.
It contains the library, an “R” script with the examples and a “zip”’ folder with all the images commented on and used in the examples.
Cite this paper
Stela, B. and Monleón-Getino, A. (2016) Facilitating the Automatic Characterisation, Classification and Description of Biological Images with the VisionBioShape Package for R. Open Access Library Journal, 3: e3108. http://dx.doi.org/10.4236/oalib.1103108
References
- 1. Libungan, L.A. and Pálsson, S. (2015) Shaper: An r Package to Study Otolith Shape Variation among Fish Populations. PloS ONE, 10, e0121102.
http://dx.doi.org/10.1371/journal.pone.0121102 - 2. Yang, H.-P., Ma, C.-S., Wen, H., Zhan, Q.-B. and Wang, X.-L. (2015) A Tool for Developing an Automatic Insect Identification System Based on Wing Outlines. Scientific Reports, 5, 12786.
http://dx.doi.org/10.1038/srep12786 - 3. Uchida, S. (2013) Image Processing and Recognition for Biological Images. Development, Growth & Differentiation, 55, 523-549.
http://dx.doi.org/10.1111/dgd.12054 - 4. Ginoris, Y.P., Amaral, A.L., Nicolau, A., Coelho, M.A.Z. and Ferreira, E.C. (2007) Recognition of Protozoa and Metazoa Using Image Analysis Tools, Discriminant Analysis, Neural Networks and Decision Trees. Analytica Chimica Acta, 595, 160-169.
http://dx.doi.org/10.1016/j.aca.2006.12.055 - 5. Ghazali, K.H., Alsameraai, R.S.H. and Mohamed, Z. (2013) Automated System for Diagnosis Intestinal Parasites by Computerized Image Analysis. Modern Applied Science, 7, 98- 114. http://dx.doi.org/10.5539/mas.v7n5p98
- 6. Shamir, L., Delaney, J.D., Orlov, N., Mark Eckley, D. and Goldberg, I.G. (2010) Pattern Recognition Software and Techniques for Biological Image Analysis. PLoS Computational Biology, 6, e1000974.
http://dx.doi.org/10.1371/journal.pcbi.1000974 - 7. Tan, C.S., Lau, P.Y., Phang, S.-M. and Low, T.J. (2014) A Framework for the Automatic Identification of Algae (Neomeris vanbosseae M.A. Howe): U3S. 2014 International Conference on Computer and Information Sciences (ICCOINS), Kuala Lumpur, 3-5 June 2014, 1-6.
- 8. Avci, D. and Varol, A. (2009) An Expert Diagnosis System for Classification of Human Parasite Eggs Based on Multi-Class SVM. Expert Systems with Applications, 36, 43-48.
http://dx.doi.org/10.1016/j.eswa.2007.09.012 - 9. Kuhl, F. and Giardina, C. (1982) Elliptical Fourier Features of a Closed Contour. Computer Graphics Image Processing, 18, 236-258.
http://dx.doi.org/10.1016/0146-664X(82)90034-X - 10. Claude, J. (2008) Morphometrics with R. Springer, Berlin.
- 11. Rohlf, F.J., Ferson, S. and Koehn, R.K. (1985) Measuring Shape Variation of Two-Dimensional Outlines. Systematic Zoology, 34, 59-68.
http://dx.doi.org/10.2307/2413345 - 12. R Core Team (2015) R: A Language and Environment for Statistical Computing.
- 13. Buf, H.D., Bayer, M., Droop, S., Botanic, R., Edinburgh, G., Head, R., Juggins, S., Fischer, S. and Bunke, H. (1999) Diatom Identification: A Double Challenge Called ADIAC. International Conference on Image Analysis and Processing, Italy, 27-29 September 1999.
http://dx.doi.org/10.1109/ICIAP.1999.797682 - 14. Kloster, M., Kauer, G. and Beszteri, B. (2014) SHERPA: An Image Segmentation and Outline Feature Extraction Tool for Diatoms and Other Objects. BMC Bioinformatics, 15, 218.
http://dx.doi.org/10.1186/1471-2105-15-218 - 15. Jalba, A.C., Wilkinson, M.H.F., Roerdink, J.B.T.M., Bayer, M.M. and Juggins, S. (2005) Automatic Diatom Identification Using Contour Analysis by Morphological Curvature Scale Spaces. Machine Vision and Applications, 16, 217-228.
http://dx.doi.org/10.1007/s00138-005-0175-8 - 16. Flor-Arnau, N., Cambra, J. and Burfeid Castellanos, A.M. (2012) Referencia y red básica de diatomeas en la cuenca del ebro. evaluación del estado ecológico de las masas de agua superficiales de la cuenca del ebro utilizando las diatomeas bentónicas como bioindicadores resultados anos 2011-2012. Confederación Hidrográfica del Ebro.
- 17. Hijmans, R.J. (2015) Raster: Geographic Data Analysis and Modeling. R Package Version 2.5-2.









