Journal of Water Resource and Protection
Vol.6 No.2(2014), Article ID:43180,6 pages DOI:10.4236/jwarp.2014.62016
Non-Fermentative Gram-Negative Bacteria in Drinking Water
Vytautus Magnus University, Kaunas, Lithuania
Email: a.paulauskas@gmf.vdu.lt
Copyright © 2014 Daiva Staradumskyte, Algimantas Paulauskas. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Daiva Staradumskyte, Algimantas Paulauskas. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received June 27, 2013; revised July 29, 2013; accepted August 24, 2013
Keywords:Drinking Water; Microbiology; CDC; Pseudomonas
ABSTRACT
The description of a microbiological community of drinking water distribution systems is intended for a safe and proper use of drinking water. These studies were aimed at characterising the microbial condition of the drinking water supply system in Lithuania by means of culture-based methods and biochemical techniques. Samples of drinking water (in cities supplied in centralised way from taps, wells, and boreholes) were collected in different locations in Lithuania. Seeking to grow microflora present in water the membrane filtration method was applied to study the samples. Most often water samples were studied to identify coliform bacteria (Citrobacter spp., Enterobacter spp., Klebsiella spp., Serratia spp.), Escherichia coli (or faecal coliform), Enterococci (or fecal Streptococcus), Pseudomonas aeruginosa (other non-fermentative gram-negative bacteria) according to the international ISO standards. Large amounts of non-fermenting gram-negative bacteria were found in centralised urban water. The investigation showed that more than 50% of the wells under study had increased microbial contamination with faecal coliform bacteria and faecal Streptococcus. Bacteria of the Pseudomonas group, CDC group and other non-fermentative gram-negative bacteria were detected in more than 30% samples of centralised urban water studied.
1. Introduction
Drinking water is suitable for drinking if it is clean and does not contain any micro-organisms, parasites and other substances, which can pose danger to human health. To ensure the quality of drinking water, microbiological investigations of each tap are carried out. The greatest risk from water contaminated with microbes is related to consumption of drinking-water that is contaminated with human and animal excreta, although other sources and routes of contamination can also be of significance [1,2].
Groundwater is a source of drinking water in Lithuania. Publicly supplied water usually comes from deep-water aquifers which are not affected by environmental pollution very much. Microbiological properties of this water are usually good too. There are, however, exceptions, when surface water is artificially added to groundwater [3].
Almost one million people use well water in Lithuania [3]. This is the only source of drinking water for many people in Lithuania. Unfortunately, groundwater is not actually not safe to use. Investigations show that more than 50% of well water samples studied are noted for an increased level of microbial contamination, approximately 30% of all the wells and 90% of the wells located in towns or villages are contaminated with nitrates.
According to the international ISO standards, water samples are most often investigated to detect coliform bacteria (Citrobacter spp., Enterobacter spp., Klebsiella spp. Serratia spp., Enterobacter spp.), Escherichia coli (or faecal coliform) and faecal Enterococcus, but it was noticed that water samples contained large amounts of non-fermentative gram-negative bacteria.
Non fermentative gram-negative bacilli (non-fermenters) are ubiquitous in the environment. Non fermenters can cause a vast variety of infections [4-6]. Characteristically, these organisms fail to acidify the butt of Kligler or triple sugar iron agar or oxidative-fermentative media and grow significantly better under aerobic conditions than they do under aerobic conditions; many strains even fail to grow anaerobically [7].
Biochemically inactive bacteria of group NO-1 reproduce in drinking water and Pseudomonas spp. bacteria with the positive citochrom oxidase activity. Group NO-1 bacteria are oxidase negative, asaccharolytic, immobile, medium-sized gram-negative rods forming small colonies on sheep blood agar (SBA) [7]. In humans, P. aeruginosa is the second most frequent gram-negative nosocomial pathogen in hospitals and has the highest case-fatality rate of all hospital acquired bacteremia’s [8]. Pseudomonas genera represents opportunistic pathogens, which disperse and adhere easily to surfaces forming a biofilm, which interferes with the cleaning and disinfection procedures in hospitals and industrial environments [9].
The main objective of these investigations was to assess the total microbial community in the samples of different drinking water (centralised tap water, well water, borehole water).
2. Materials and Methods
Samples were taken from the following sources: tap water, well water, borehole water. All the samples were collected in Kaunas and Marijampolė regions. After collection the samples were transported to the laboratory as soon as possible. Different dilutions and volumes were filtered through membrane filter (0.45 µm pore size), which is used on a vacuum pump. The filter is placed on a petri dish containing selective agar. Membrane filters are transferred onto Tergitol 7 agar (T7A), which is used to detect coliform bacteria, when incubated at elevated temperatures (36˚C ± 2˚C, 44˚C ± 0.5˚C, 24 h). To confirm the identity of coliform bacteria, all red, yellow, orange and other colonies are cultivated in Tryptone water for the indole test [10]. Phenol Red Lactose Peptone Broth (PRLPB) for lactose degradation and gas formation test and Tryptone soy agar for oxidase test. Bacteria colonies whose oxidase test is positive and whole indole is negative are confirmed to be coliform bacteria. If Triple Sugar Iron agar and the column are not decomposed and no gas is produced, these colonies or cultivars are gram-negative rods, non-fermenters. Bacteria whose oxidase test is positive and indole is negative, Triple Sugar Iron agar and the column are not decomposed and no gas is produced, are non-fermenting gram-negative rods with positive citochrom oxidase activity. To identify the species biochemical tests (Triple Sugar Iron Agar, Lysine Iron Agar, Urease Test Broth, RapID ONE or RapID NF systems were used (Figure 1).
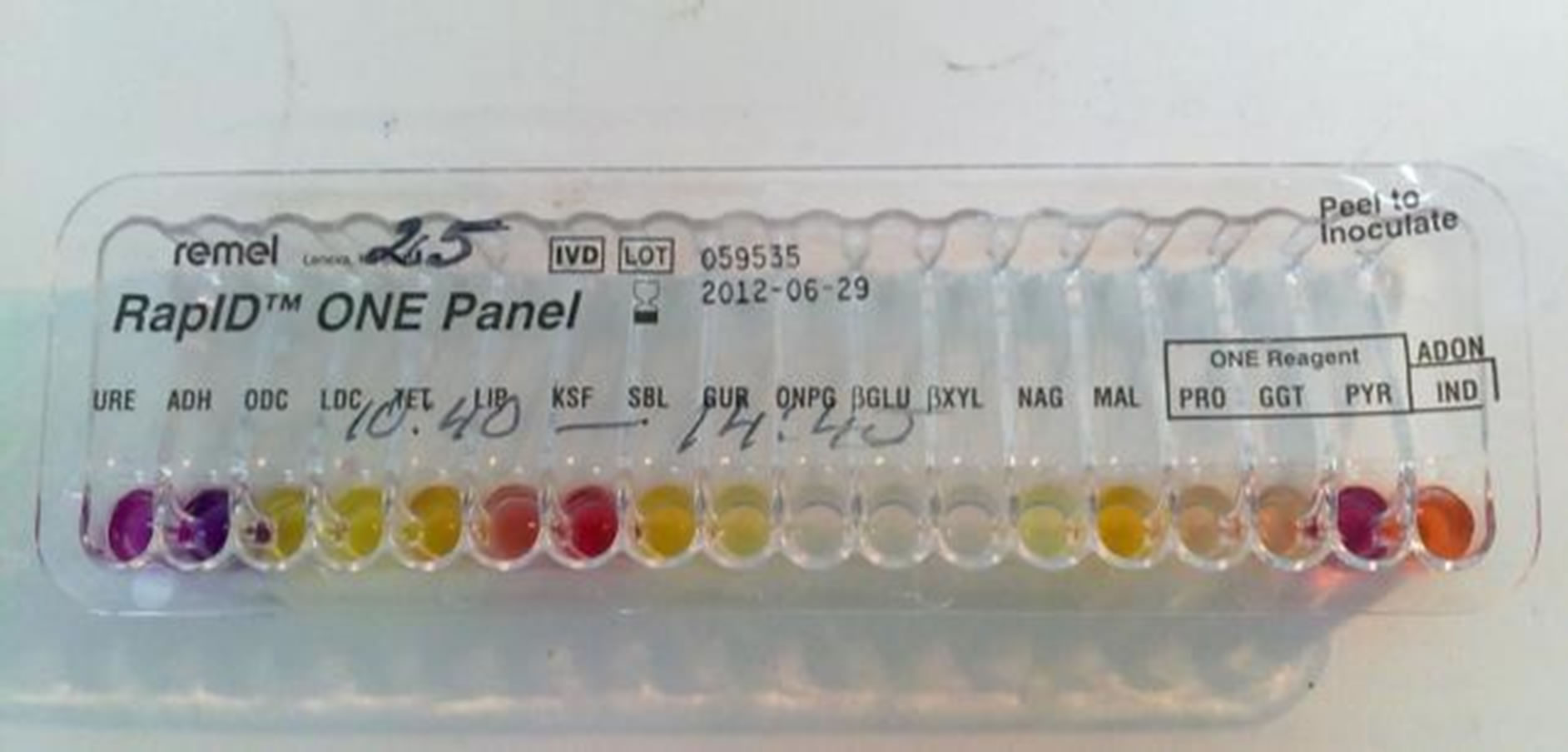
Figure 1. Rapid One System.
To detect and count Pseudomonas aeruginosa and other bacteria of Pseudomonas group membrane filters were placed on Pseudomonas detection agar (PIA). Agar is incubated at elevated temperatures (36˚C ± 2˚C, 24 - 48 h). All brown, rose-coloured or white colonies are counted. The colonies are lit in darkness with ultraviolet light to check if they fluorescent blue/green pigment. All fluorescent colonies are confirmed as Pseudomonas aeruginosa. The investigation is carried out according to LST EN ISO 16266:2008, the detection scheme is represented in Figure 2 [11].
The organism’s phenotypic characteristics were similar to the asaccharolytic strains of Acinetobacter (Table 1) but differed in their cellular morphology and the cellular fatty acid profile.
Pseudomonas (Figure 3) are aerobic, non-sporeforming, gram-negative rods that are straight or slightly curved. These bacteria are catalase positive and most of them are oxidase positive. With the exception of certain strains of P. vesicularis these organisms grow on Mac Conkey agar and appear as non-fermenters. Most strains fail to decompose glucose and most species degrade nitrate to either nitrite or nitrogen gas. Certain species have distinctive colony morphologies or pigmentation [7].
Pseudomonas aeruginosa when grown in the suitable media produces non-fluorescent bluish pigment pyocianin. Many strains also produce fluorescent green pigment pyoverdin. Pseudomonas aeruginosa, like other fluorescent pseudomonas, produces catalase, oxidase and amnion from arginine and can grow on citrate as the sole source of carbon [11].
3. Results and Discussion
From 2010 to 2012 drinking water from different water groups—tap water, well water, borehole water—was analysed (Figure 4).
In 2010, a total of 2607 water samples were analysed to identify microbial water contamination. The heaviest bacterial contamination was detected in well water, 55.8% of intestinal enterococci in all the samples tested. In 2011, a total 2174 samples were analysed to identify microbial water contamination. The highest bacterial contamination was detected in well water: 56.1% of Escherichia coli in
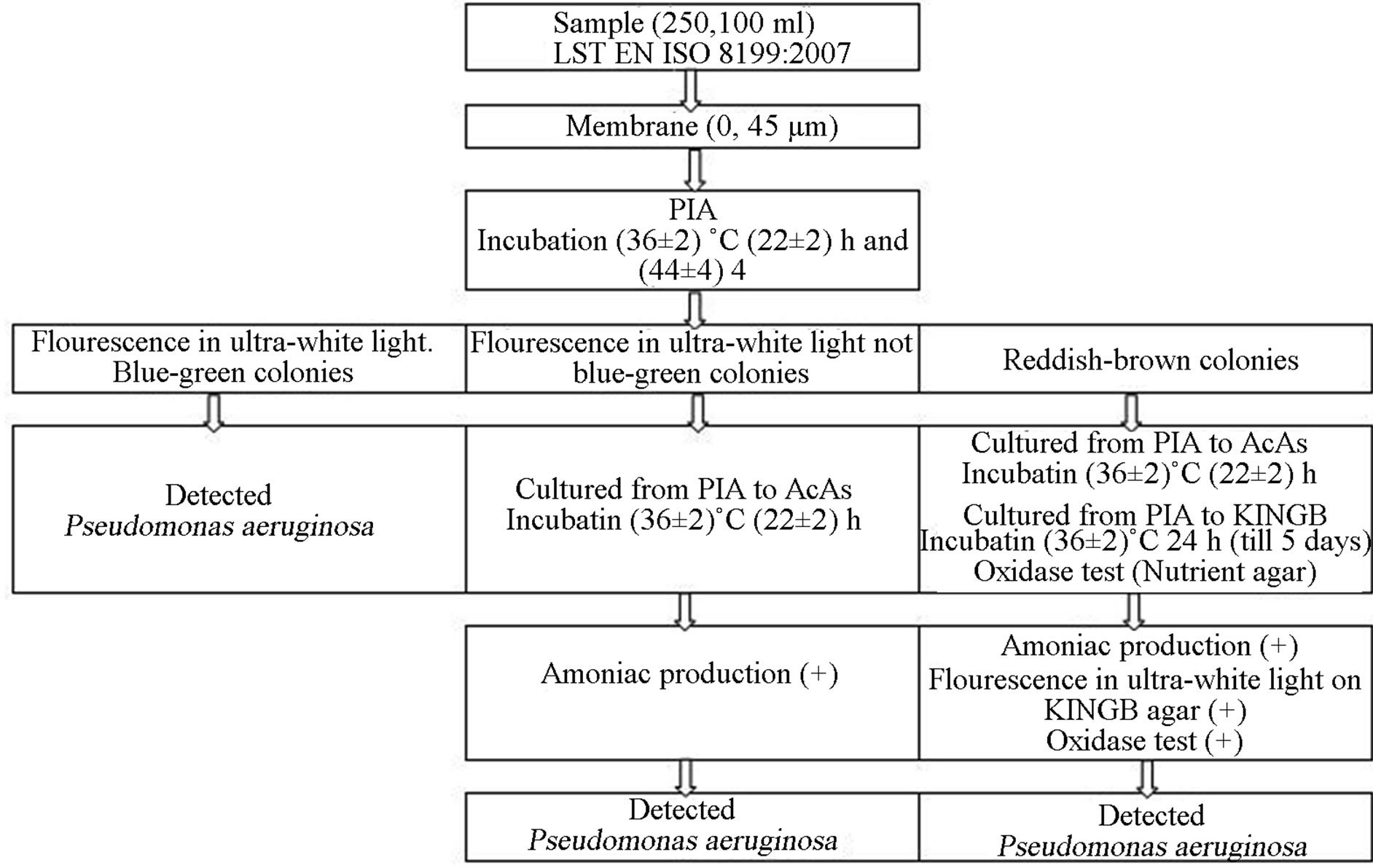
Figure 2. Detection and enumeration of Pseudomonas aeruginosa according to LST EN ISO 16266:2008 [11].

Figure 3. a) Pseudomonas aeruginosa on PIA agar from drinking water b) Pseudomonas aeruginosa on PIA agar fluorescence in ultra-violet light.
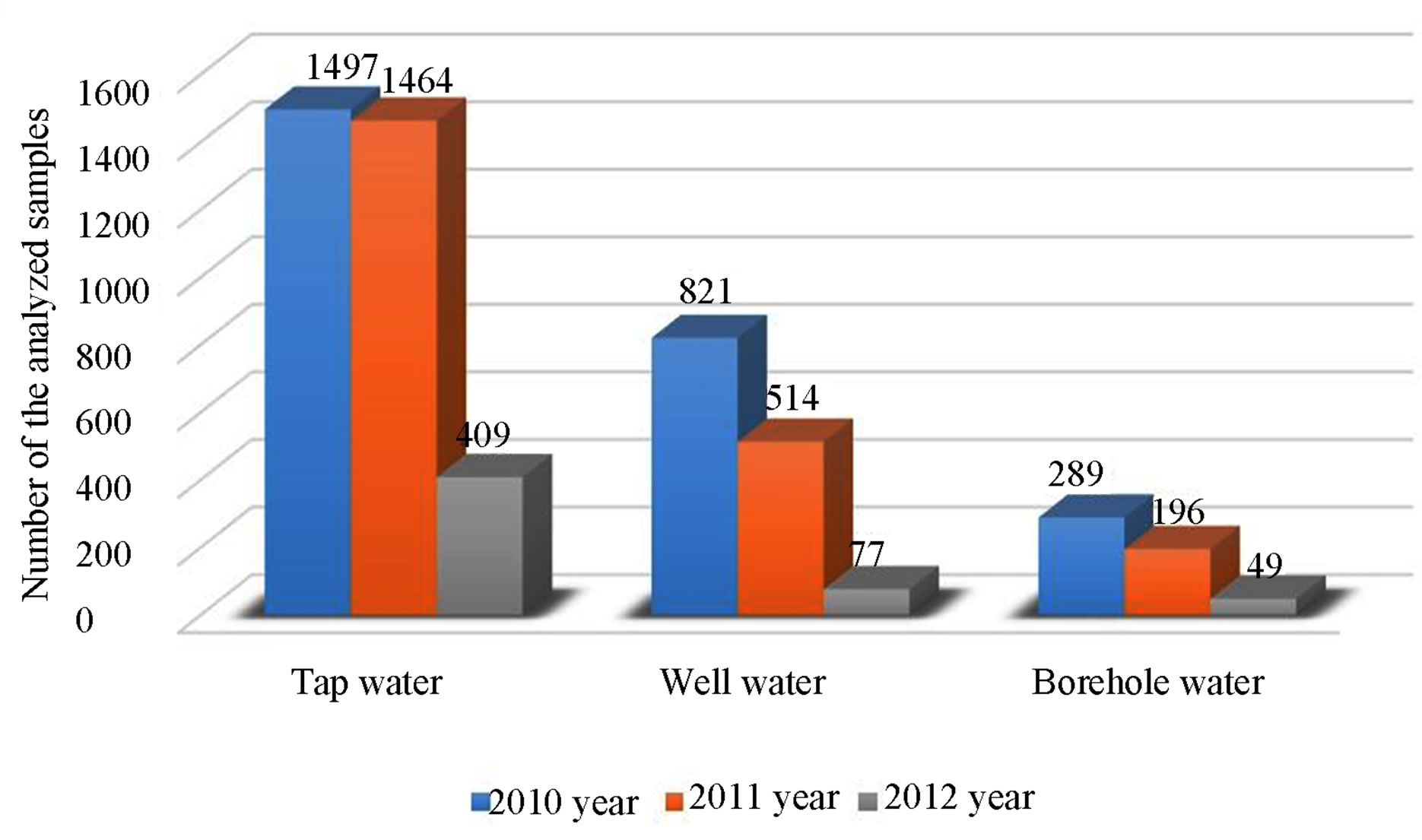
Figure 4. Number of the analyzed samples by water groups, from 2010 to 2012.
all the samples. In 2012, as many as 1678 samples were analysed to identify microbial water contamination. The highest bacterial contamination was detected in well water 93.3% of coliform bacteria in all the samples tested.
The results of the investigated water samples showed that water contained large amounts of non-fermentative gram-negative bacteria. Our results show (Figure 4) that the majority of the inhabitants think that the quality of drinking water is good because of the use of water filters alone, therefore the number of people who wish the
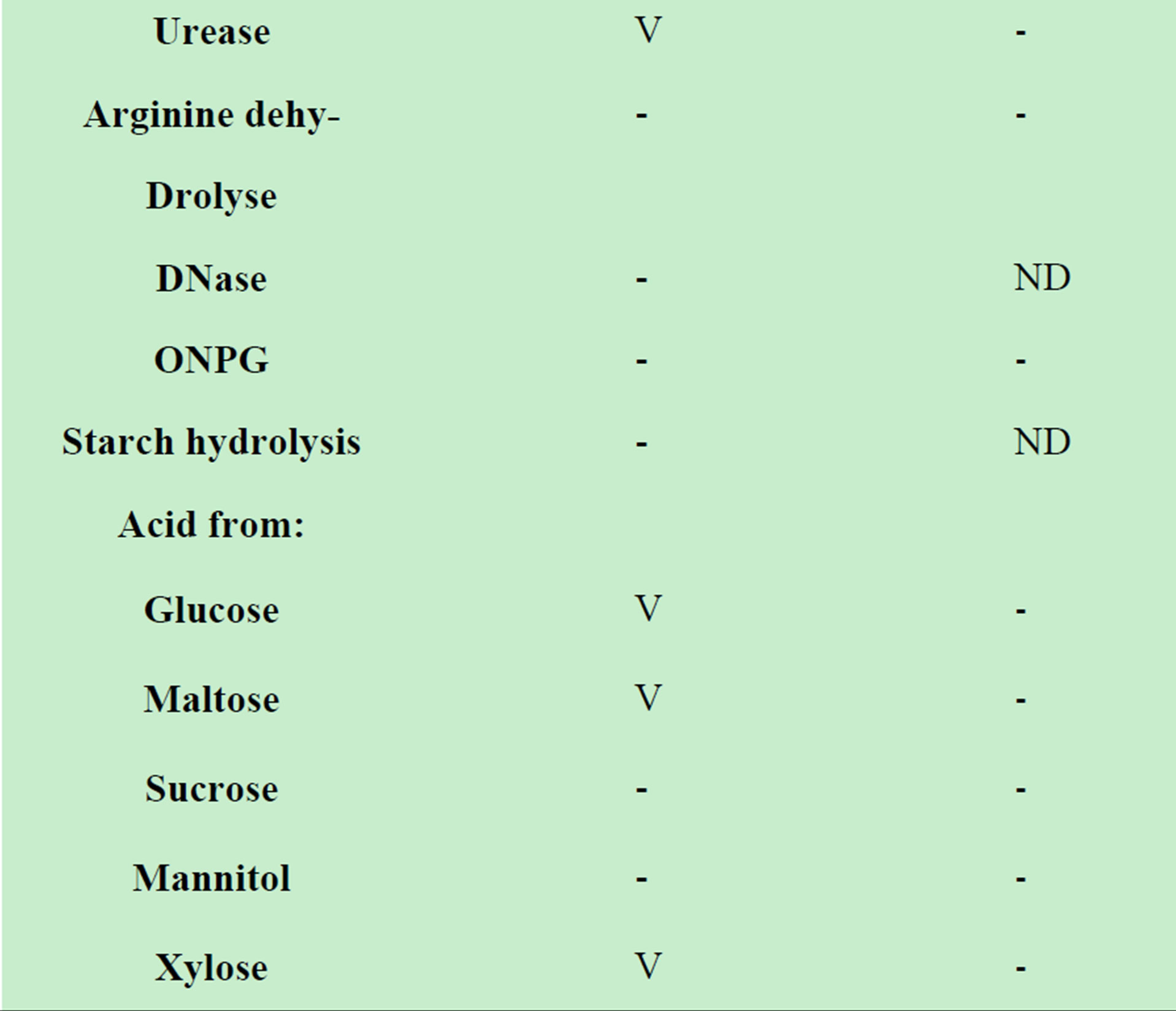
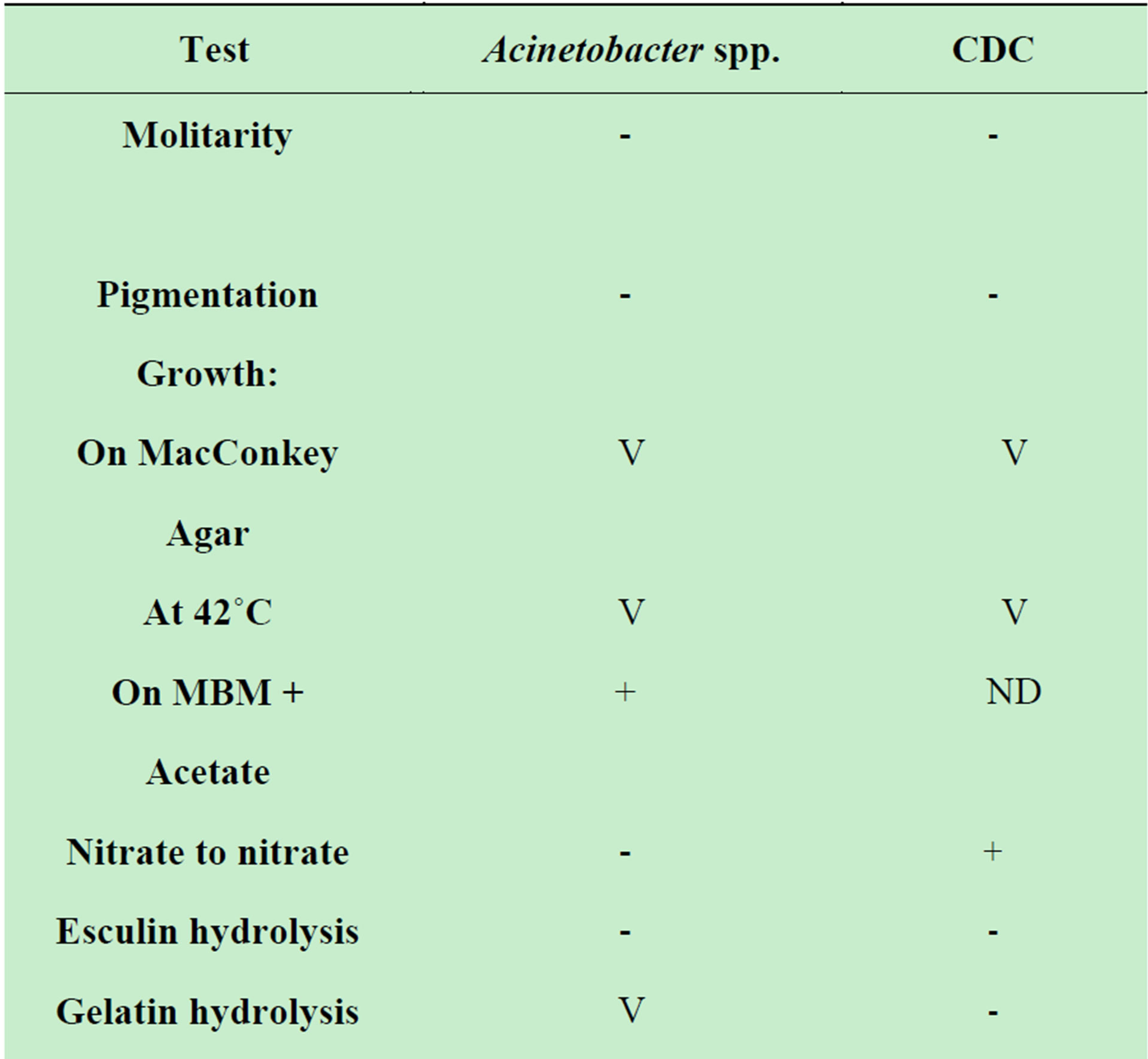
Table 1. Oxidase-negative, indole negative, non-fermentative gram–negative rods [7].
microbiological quality of their drinking water to be tested is on the decrease.
The investigation results showed (Figure 5) that centralised urban tap water is most often analysed by water control laboratories in Lithuania. This is because it is obligatory for public institutions to have the quality of water tested once a year, but, unfortunately, the tests are carried out to detect only those micro-organisms, which are indicated in the hygiene standards (coliform bacteria and faecal enterococci) [12].
The microbiological analysis of drinking water from individual boreholes showed (Table 2, Figure 6) that in 2012 non-fermentative gram-negative bacteria were found in eight water samples.
The results showed (Figure 5) that in water samples people look for micro-organisms, which are indicated in
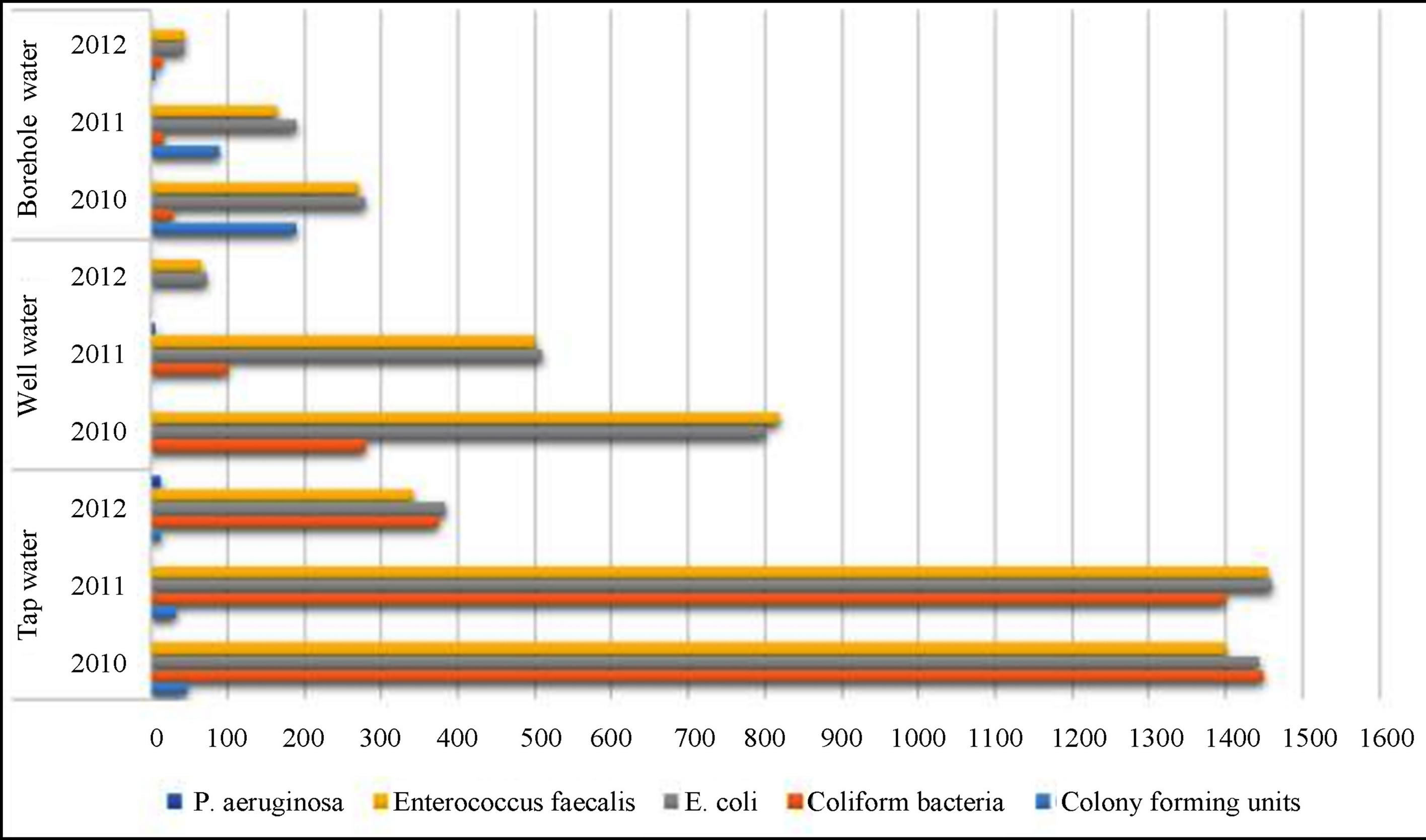
Figure 5. The number of samples analyzed by microbiological indicators between 2010 and 2012 by water source groups.

Figure 6. Non-fermentative bacteria in drinking water by group, from 2010 to 2012.
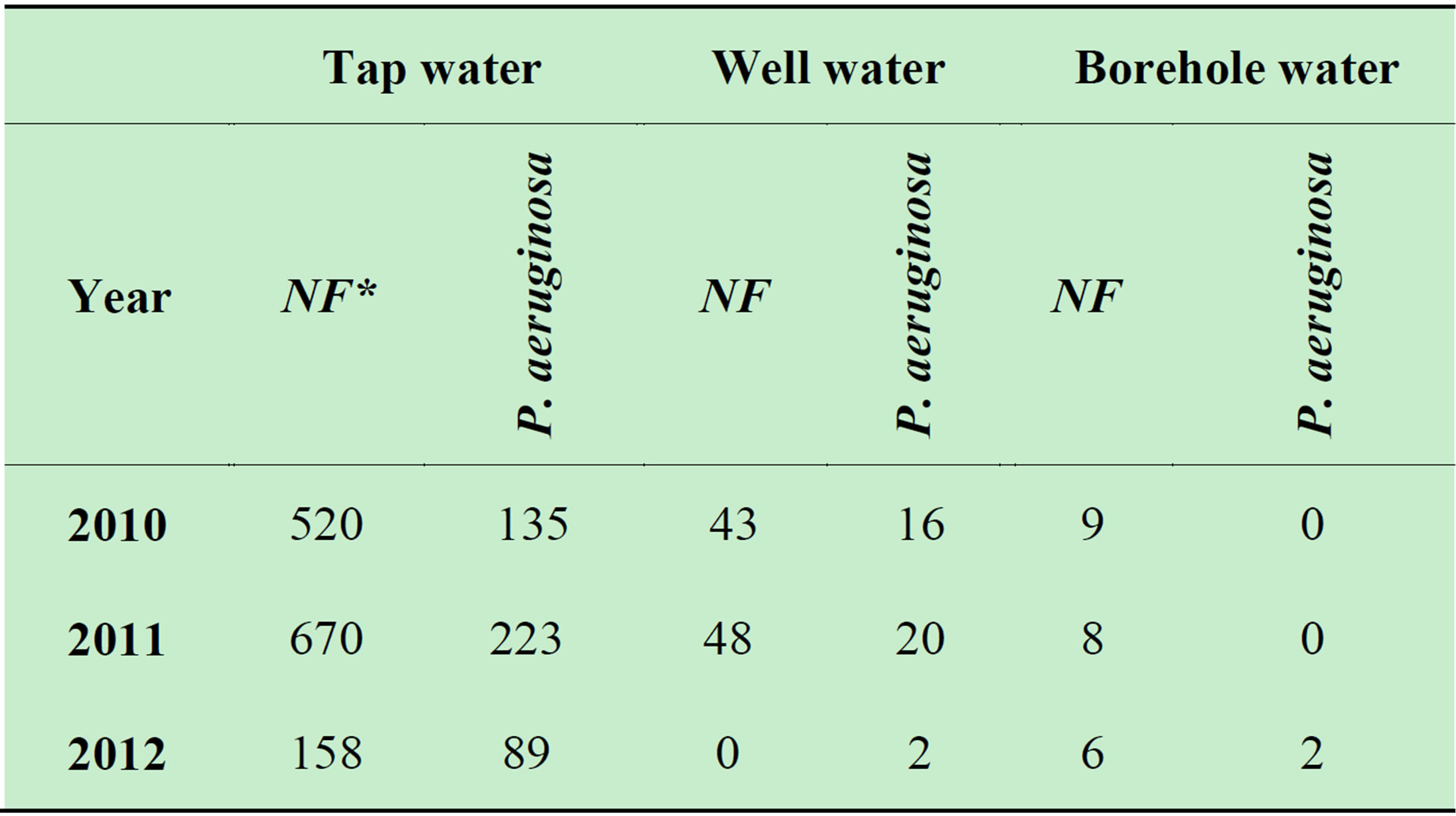
Table 2. Detected non-fermentative gram-negative bacteria in drinking water (*NF-Non-fermentative).
the hygiene standards. Our investigations, however, showed that their water systems contain non-fermentative gram-negative bacteria Acinetobacter, Pseudomonas or CDC (Table 2, Figure 6); these bacteria are not indicated in the hygiene standard as an indicated organism to assess the quality of drinking water therefore people do not ask samples of their drinking water to be tested for the said micro-organisms.
P. aeruginosa is a bacterium that does not require any nutrients. This is indicated by the ability of these bacteria to grow even in distilled water [8]. P. aeruginosa tends to grow in moist environment and this is most likely a reflection of their natural existence in soil and water. They can grow on the inside surface areas of polyvinyl chloride pipes and garden watering hoses. P. aeruginosa is a bacterium, which causes skin rash and infections acquired in swimming pools and saunas. In water these bacteria stick to the skin and penetrate into the hair follicles where they reproduce, and products of their metabolism cause inflammation. The affected spot can look like red itching rash or like a burn. The affected spot reminds of chemical rash and since its incubation period is long some cases might be wrongly diagnosed as chemical rash rather than bacterial infections. P. aeruginosa is the main cause of the so-called “swimmer’s air” (otitas externa). This bacterium can also cause infection of the cornea in people who wear contact lenses. Besides, it is one of the main causes of urinary tract infections, surgical infections and burn wound infections. P. aeruginosa is an opportunistic pathogen. This means that it is able of making use of the problems of the host’s immune system to penetrate into the organism and develop infections.
P. aeruginosa is the species most commonly associated with human diseases, ear and skin infections acquired in contaminated pools, hot tubs and whirlpools. It causes serious nosocomial (hospital-acquired) infections in cystic fibrosis and burn patients, and immunocompromised individuals [13]. P. aeruginosa is a common environmental organism and it can be found in faeces, soil, water and sewage. It can multiply in water environment and also on organic surfaces that are in contact with water [1,2]. The cells of such ubiquitous bacterial species as P. aeruginosa when responding to favourable nutrient conditions and being stuck to the present surfaces at the time of binary fission and exopolymer production form strong biofilms [14,15]. P. aeruginosa biofilms cause persistent infections in the individuals with health problems. For example, most people with the genetic cystic fibrosis disease suffer from chronic P. aeruginosa biofilm infections of the lungs [9]. P. aeruginosa biofilms are developed communities with individual bacterial cells embedded in an extracellular polysaccharide matrix and are inherently resistant to antimicrobial treatment [15,16].
4. Conclusions
The investigations carried out show that the quality of centralised urban drinking water is good; however, some cases of increased microbial contamination were determined. Lithuanian users are supplied with drinking water from deep aquifers in a centralised way. The environment exerts no considerable impact on water from deep aquifers. Unfortunately, groundwater is not safe for consumption.
The majority of violations of the requirements set forth in the Lithuanian Hygiene Standard HN 24:2003 were discovered in individual water supply systems, especially in dug wells. Contamination with coliform bacteria was discovered in 72.9% of all the wells under investigation. Contamination of water with Escherichia coli 54.8% and contamination with intestinal enterococci 56.2% were found in all samples of well water studied.
The reason for micro-biological contamination of wells is that polluted surface water can get into the well water. It is the owners of a homestead themselves that most often contaminate drinking water. Traditionally a well is dug in the centre of the homestead, close to dwelling houses and outside buildings, kitchen gardens. Groundwater can be contaminated if nearby soils is fertilized or if pollutants get directly into a well.
Violations of the requirements laid down in the Lithuanian Hygiene Standard HN 24:2003 were discovered in centralised urban drinking water where contamination with coliform bacteria was detected in 11.5% of the samples. All the samples were contaminated with intestinal rods (Escherichia coli) 0.2% and intestinal enterococci 0.1%.
The Lithuanian Hygiene Standard 24:2003 Pseudomonas aeruginosa contamination is controlled only in pre-packed drinking water (they cannot be found in 250 ml of water) [12]. Since P. aeruginosa do not require any nutrients, they can survive and reproduce in water prepacked drinking water containing very small amounts of organic substances). P. aeruginosa can be discovered in drinking water, especially in pre-packed water, they can be related to complaints about the taste, odour and turbidity. The bacteria release poisonous endotoxin and enzymes therefore Pseudomonas is constantly monitored in bottled drinking water. Publicly supplied drinking water is neither regulated nor controlled with respect to P. aeruginosa.
Acknowledgements
We express our deep gratitude to the staff of the National Public Health Surveillance Laboratory (NPHSL), Kaunas Department, Microbiology subunit, for assistance provided in our investigations.
REFERENCES
- World Health Organization, “Guidelines for DrinkingWater Quality,” Incorporating First Addendum, Vol. 1, Recommendations, 3rd Edition, 2006, pp. 121-144, 221- 247.
- World Health Organization, “Guidelines for DrinkingWater Quality,” 4th Edition, 2011.
- V. Kiguoliene, “Ką Reikia Žinoti Apie Geiamąjį Vandenį,” 2013.
- L. Dijkshoorn, A. Nemec and H. Seifert, “An Increasing Threat in Hospitals: Multidrug-Resistant Acinetobacter baumannii,” Nature Reviews Microbiology, Vol. 5, No. 12, 2007, pp. 939-951. http://dx.doi.org/10.1038/nrmicro1789
- J. J. LiPuma, B. J. Currie, G. D. Lum and P. A. R. Vandamme, “Burkholderia, Stenotrophomonas, Ralstonia, Cupriavidus, Pandoraea, Brevundimonas, Comamonas, Delftia, and Acidovorax,” In: P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry and M. A. Pfaller, Eds., Manual of Clinical Microbiology, 9th Edition, ASM Press, Washington DC, 2007, pp. 749-769.
- S. C. Su, M. Vaneechoutte, L. Dijkshoorn, Y. F. Wei, Y. L. Chen and T. C. Chang, “Identification of Non-Fermenting Gram-Negative Bacteria of Clinical Importance by an Oligonucleotide Array,” Journal of Medical Microbiology, Vol. 58, No. 5, 2009, pp. 596-605. http://dx.doi.org/10.1099/jmm.0.004606-0
- P. R. Murray, E. J. Boran, M. A. Pfallek, F. C. Tenover and R. H. Yolken, “Manual of Clinical Microbiology,” 6th Edition, ASM Press, Washington DC, 2005, pp. 509- 530.
- M. David and D. V. M. Moore, “Reference Paper: Pseudomonas and the Laboratory Animal,” 1997.
- E. Banin, M. L. Vasil and E. P. Greenberg, “Iron and Pseudomonas aeruginosa biofilm formation,” Proceedings of the National Academy of Sciences of the United states of America, Vol. 102, No. 31, 2005, pp. 11076- 11081. http://dx.doi.org/10.1073/pnas.0504266102
- LST EN ISO 9308-1:2001, “Water Quality—Detection and Enumeration of Escherichia coli and Coliform Bacteria—Part 1: Membrane Filtration Method,” ISO 9308- 1:2000.
- LST EN ISO 16266:2008, “Water Quality—Detection and Enumeration of Pseudomonas aeruginosa—Method by Membrane Filtration,” ISO 16266:2006.
- Lithuanian Hygiene Norm HN 24:2003, Drinking Water Safety and Quality Requirements.
- V. T. C. Penna, S. A. M. Martins and P. G. Mazzola, “Identification of Bacteria in Drinking and Purified Water during the Monitoring of a Typical Purification System,” BMC Public Health, Vol. 2, 2002, p. 13. http://dx.doi.org/10.1186/1471-2458-2-13
- K. D. Mena and C. P. Gerba, “Risk Assessment of Pseudomonas aeruginosa in Water,” Reviews of Environmental Contamination and Toxicology, Vol. 201, 2009, pp. 71-115 http://dx.doi.org/10.1007/978-1-4419-0032-6_3
- J. W. Costerton, Z. Lewandowski, D. E. Caldwell, D. R. Korber and H. M. Lappin-Scott, “Microbial Biofilms,” Annual Review of Microbiology, Vol. 49, 1995, pp. 711- 745. http://dx.doi.org/10.1146/annurev.mi.49.100195.003431
- J. W. Costerton, Philip S. Stewart and E. P. Greenberg, “Bacterial Biofilms: A Common Cause of Persistent Infections,” Science, New Series, Vol. 284, No. 5418, 1999, pp. 1318-1322.

