Advances in Bioscience and Biotechnology
Vol.5 No.3(2014), Article ID:42623,8 pages DOI:10.4236/abb.2014.53026
Identification of an extracellular infection-induced glyceraldehyde-3-phosphate dehydrogenase of the phytopathogenic proteobacterium Pseudomonas syringae pv tomato DC3000
![]()
1Laboratory of Physiology & Genetics Molecular, Faculty of Sciences, Ain Chock, Casablanca, Morocco
2Institute for Plant Biochemistry and Photosynthesis (IBVF), CSIC-Universidad de Sevilla, Seville, Spain
Email: *ab.soukri@gmail.com
Copyright © 2014 Bouchra Elkhalfi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Bouchra Elkhalfi et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 8 November 2013; revised 12 January 2014; accepted 24 January 2014
Keywords: GAPDH; Secretome; MALDI-TOF; Gene Expression; Infective State; Bacterial Speak; RNA
ABSTRACT
According to molecular biology, genomic and proteomic data, the phytopathogenic gamma-proteobacterium Pseudomonas syringae pv. tomato DC3000 produces a number of proteins that may promote infection and draw nutrients from plants. Remarkably, P. syringae DC3000 strain possesses three paralogous gap genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzymes with different predicted molecular sizes and metabolic functions. As GAPDH was shown to be a virulence factor in other microbial pathogens, in the current study, we analyzed the expression levels of each paralogous gap gene by realtime PCR to understand the actual impact of their protein products on P. syringae virulence. We found that all of them were strongly induced during the infection process. Nevertheless, proteomic analysis of culture supernatants revealed that only Class I GAPDH1 encoded by the gap1 gene was identified as an extracellular protein in infective cells. These results strongly suggest that this GAPDH should play a role in the infective process, including its well-know enzymatic function in the glycolytic metabolic pathway. GAPDH; Secretome; MALDI-TOF; Gene Expression; Infective State; Bacterial Speak; RNA
1. INTRODUCTION
The gammaproteobacterium Pseudomonas syringae pv. tomato DC3000 is an aerobic, rod-shaped, polar, motile and a Gram negative bacteria [1-3], which showed a fluorescent pigmentation in King B medium and oxidatively metabolizes glucose. This pathogenic is also characterized by Levan-positive colonies on nutrient agar medium supplemented with sucrose, exhibits positive hypersensitive reaction in the presence of tobacco leaves; it is oxidase, arginine-dehydrolase negative and does not produce pectolytic enzymes (were potato rot-negative) [4,5].
Disseminated primarily by water, P. syringae pv. tomato DC3000 has the potential to move through plug greenhouses very quickly; thus, infested seedlings could become an important inoculum source for field epiphytotics [6,7]. Consequently, this bacterium can survive up to 20 years in the crevices and cavities of the coat of tomato seeds causing bacterial speck, a disease characterized by small brown-black leaf spots sometimes surrounded by chlorotic margins, and dark superficial specks on green fruits specks on ripe fruit may become sunken, and are surrounded by a zone of delayed ripening [2,8,9]. Stunting and yield loss, particularly if young plants are infected, considerably reduced the market value of speckled fruit [9].
This pathogenicity of P. syringae DC3000 strain has been the subject of several studies, and it has emerged as an important model organism in molecular plant pathology because of its genetic tractability [10-12], and its genome has been fully sequenced [9]. Remarkably, the pathogenicity of DC3000 resembles that of most animal and other plant pathogens in the Gammaproteobacteria, which relys on a Type III secretion system (TTSS) to release virulence effector proteins into host cells [13,14].
Other studies have shown that DC3000 strain encodes a wide range of proteins that should be involved in virulence [12]. One of these proteins is glyceraldehyde-3- phosphate dehydrogenase (GAPDH), a highly conserved enzyme during evolution with a key role in the glycolytic and gluconeogenic pathways. GAPDH catalyzes the redox-linked reversible phosphorylation of glyceraldehyde-3-phosphate to 1,3-diphosphoglycerate, in the presence of NAD(P)+ and inorganic phosphate [15]. Most glycolytic Class I GAPDHs so far studied are homotetrameric proteins with 34 - 38 kDa subunits, and the native oligomer has a molecular weight of 140 - 150 kDa [16,17]. In addition to its classic metabolic roles, GAPDH has been claimed to be involved in a number of diverse cellular processes unrelated to glycolysis (“moonlighting protein”), and many of these alternative roles are dependent on the ability of GAPDH to modify its subcellular localization. GAPDH was used as a model standard protein, or control, in gene regulation and catalytic mechanism related studies, because of its high degree of structural conservation across species [18]. Furthermore, with the advent of the molecular biotechnology, GAPDH recently emerged as a useful model for innovative methods to advance our understanding of the mechanisms through which cells organize and express their genetic information, despite its general consideration as a housekeeping enzyme [19]. Regarding its possible specific role in pathogenic organisms, the presence of a surface-associated GAPDH has been reported in all streptococcal strains tested so far [20,21], and in other diverse microorganisms like Candida albicans, Schistosoma bovis, Mycoplasma genitalium, and Staphylococcus spp. [22]. GAPDH was shown to be secreted by several pathogenic bacteria, like Streptococcus pyogenes and a number of enteropathogenic Escherichia coli strains [20,21,23]. It was claimed that secreted GAPDH has a role in signal transduction to their host cells, rendering them probably more susceptible to bacterial infection [21,24]. Three gap genes encoding distinct GAPDH proteins are present in the genome of P. syringae pv. tomato DC3000 [25]. Whether any of these bacterial GAPDHs are extracellular proteins involved in the plant infection process remains to be established. In any case, their possible secretion and association to the cell membrane might have biotechnological implications.
In this study we analyzed the expression level of each paralogous gap gene of P. syringae pv. tomato DC3000 both in the infective and resting states by one-step quantitative RT-PCR, looking for possible differences in mRNA expression between these two physiological status. Besides, the protein secretome of infective cells was analyzed by MALDI-TOF mass spectrometry in order to identify which of the three GAPDHs of this phytopathogenic bacterium exhibits an extracellular localization. In this way, the overexpressed Class I GAPDH1 was identified in the secretome of infective cells, suggesting a possible role of this protein in the virulence of P. syringae pv. tomato DC3000.
2. MATERIALS AND METHODS
2.1. Organisms and Growth Conditions
Pseudomonas syringae pv tomato DC3000 cells used in this study were grown in LB medium with glucose (3 g/l) at 28˚C in the presence of rifampicin 50 µg/ml, in the darkness with agitation. For infection studies in solid and liquid medium we adjusted the bacterial suspensions to 106 CFUs per milliliter with sterile distilled water.
2.2. Plant Material
Round Tomato (Solanum lycopersicum L.) leaflets from two-months old plants were detached with their petioles, from a biological culture of tomato plants grown under natural outdoors conditions in region of Sidi-Rehal (Casablanca, Morocco).
2.3. Plant sterilization Method
Leaflets were treated as follows: soaking in 10% (v/v) commercial bleach-sterilized distilled water for 2 min followed by soaking in 70% (v/v) ethanol-sterilized distilled water for 1 min; then rinsed tree times in sterilized distilled water and placed in sterile Petri dishes until de time of infection.
2.4. Bacterial Infection Method
Detached leaflets were inoculated by injection with a 106 of bacterial suspension (from either solid or liquid LB medium) and maintained at 28˚C under a 16 h photoperiod, with high relative humidity. All infection experiments were repeated at least three times. No-infected detached leaflets inoculated with sterile distilled water and maintained in LB medium were used in all experiments as controls.
2.5. Total RNA Isolation
Total RNAs was successfully extracted from bacterial suspensions with Aurum™ Total RNA Mini Kit (BioRad). The final product yielded 260/280 nm ratios in the range 1.8 - 2.05 and its purity was confirmed by agarose gel electrophoresis. The nucleic acid concentration was determined using a Nanodrop spectrophotometer (Thermo Scientific).
2.6. Real-Time PCR
Real-time quantitative PCR based on the use of the Power SYBR Green PCR Master Mix (Applied Biosystems) was performed in an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Reaction mixtures (12.5 µl) contained 6.5 µl of 2x SYBR Green RT PCR reaction mixture, 0.25 µl of iScript reverse transcriptase for one-step RT-PCR, 300 nM of each primer and 10 ng of isolated RNA solution.
2.7. Primers Used
Specific primers for each paralogous gap gene of P. syringae DC3000 strain were designed using Primer Premier 5.0 program (Biosoft International).
gap1, PSPTO_1287 (gi|28868497)
Fw: GAAGAAGTCAACGCGCTGATG Rv: CCCAGGACCTTGGAGTGCT
gap2, PSPTO_2102 (gi|28869306)
Fw: GCACTCGGACCTGCACAAG Rv: CACCACTTCCTGCGAGTTCAC
gap3, PSPTO_0386 (gi|28867616)
Fw: TTTCTGGTCCAAGGCTGGTG Rv: AAGCCCCACTCGTTATCGAAC
2.8. Concentration of Extracellular Proteins
A 10-ml sample from overnight cultures of P. syringae pv tomato DC3000 strain grown in LB medium with glucose (3 g/l) in presence of tomato plant leaflets, was centrifuged for 15 min at 5000 g at 4˚C. The cell-free supernatant was mixed with ice-cold 100% (w/v) trichloroacetic acid up to a final concentration of 7% (w/v) trichloroacetic acid. The protein precipitation was accomplished by incubation of the mixture on ice for 1 h, and then washed with cold acetone. The precipitated protein was collected by centrifugation and eventually resuspended in 50 µl of SDS-PAGE sample buffer containing: 1% (p/v) SDS, 10% (v/v) glycerol, 1% (v/v) 2-mercaptoethanol, 0.01% (w/v) bromophenol blue and Tris-HCl (60 mM, pH 6.8). A saturated Tris solution was added to neutralize the sample.
2.9. Protein Gel Electrophoresis (1D SDS-PAGE)
Prior to SDS-PAGE analysis, protein samples were incubated at 100˚C for 3 min. Then, the protein samples were loaded onto a 12% polyacrylamide gel and separated on a Mini Protean II cell (Bio-Rad, Hercules, CA), according to the method of Laemmli [26]. SDS-PAGE was performed at 120 V for 1 h, using a Tris-glycine buffer (25 mM Tris; 192 mM Glycine; 0.1% SDS; pH 8.3). The apparent molecular masses of the proteins were estimated using the Precision Plus Protein Standard (BioRad) and the analysis software Quantity One (Bio-Rad). Western blots after SDS-PAGE and transfer to nitrocellulose membranes were carried out as previously described [27] using anti-GAPDH rabbit antibodies [25].
2.10. MALDI-TOF Mass Spectrometry Identification of Proteins
1) Sample preparation. Protein bands of interest were automatically excised from 1D SDS-PAGE gels after electrophoresis and Coomassie Brilliant Blue R-250 staining. The gel pieces were washed with distilled water (50 µl) during 5 min, then destained with 50 µl acetonitrile twice and vacuum dried during 15 min. The gel pieces were then chemically reduced with 10 mM dithiothreitol in 50 mM ammonium bicarbonate (Ambic) for 30 min at 56˚C, incubated with acetonitrile during 5 min, and then alkylated with 55 mM iodoacetamide in 25 mM Ambic at room temperature for 20 min in the dark. After that, gel pieces were washed with 25 mM Ambic twice, followed by acetonitrile, and dried under vacuum. Gel pieces were dried by shrinking in acetonitrile followed by vacuum drying. Then, all gel pieces were incubated with 12.5 ng/µl of sequencing-grade modified trypsin (Promega) in 50 mM Ambic for 30 min at 4˚C. After digestion, the supernatants were recovered. Peptides were then incubated overnight with 10 µl of 50 mM Ambic at 37˚C, and eventually extracted from the gel pieces twice with a 0.5% (v/v) trifluoroacetic acid at room temperature during 15 min.
2) MALDI-TOF mass spectrometry. analyses were performed on an Autoflex mass spectrometer (Bruker Daltonics) in the Servicio de Proteómica of IBVF (CSICUS). All mass spectra were calibrated externally with a peptide mass standard kit (Bruker) and internally with trypsin autolysis peaks.
3) Database search. The peptide mass fingerprinting and peptide fragment ion data obtained from MALDITOF analyses were used to search for protein candidates in the NCBI database and genomes databases by using Mascot (Matrix Science) software programs.
3. RESULTS AND DISCUSSION
Plant pathogenic bacteria possess a large number of genes that allow them to grow and cause disease on plants. Analyses of the molecular basis of pathogenesis in P. syringae. pv. tomato DC3000 reveal a complex and intimate interaction between bacteria and plant cells that depends on the coordinated expression of multiple pathogenicity and virulence factors. These include cytotoxins, polysaccharides, extracellular proteins, and the translocation of many of these proteins into plant cells by Type III (Hrp) secretion system [13,14,28].
Glyceraldehyde-3-phosphate dehydrogenase was described as one extracellular protein in a number of pathogenic microorganisms, both prokaryotes and eukaryotes [22]. Remarkably, genome sequence analysis revealed the occurrence in the phytopathogenic proteobacterium P. syringae. pv. tomato DC3000 of three paralogous gap genes located in distant regions of the 6.5-Mb chromosome with different genomic organization: namely at ca. 1.41 (gap1), 2.28 (gap2) and 0.43 (gap3) Mb from the replication origin. Moreover, these paralogous genes are predicted to encode three GAPDH enzymatic proteins with distinct molecular and catalytic features, namely two Class I enzymes having different molecular mass subunits and one class III D-erythrose-4-phosphate dehydrogenase/GAPDH bifunctional enzyme [25].
In the present study total RNA was extracted as described in the Material and Methods section from both normal (bacteria in nutritive medium) and infective (bacteria in contact with tomato plant) P. syringae. pv. tomato DC3000 cells. RNA yields being in the range 70 - 100 μg per bacterial cell pellet. The determined A260/A280 ratios ranged between 2.0 and 2.2 indicating high RNA purity and absence of proteins and polysaccharide contaminations. The integrity of extracted RNA preparations was further analyzed by migration in agarose gel electrophoresis (data not shown). Then the expression level of the three paralogous gap genes was subsequently determined by one-step real time-PCR. The obtained results clearly show that in the infective state there is a clear up-expression of transcripts of the three gap genes in comparison with the normal state of the bacterium (Figure 1). Moreover, we noticed a slight expression difference between the three gaps genes in this physiological state (data not shown).
These results are in agreement with previous reports claiming that the GAPDH is among the proteins involved in virulence of many pathogenic bacteria [29], and with
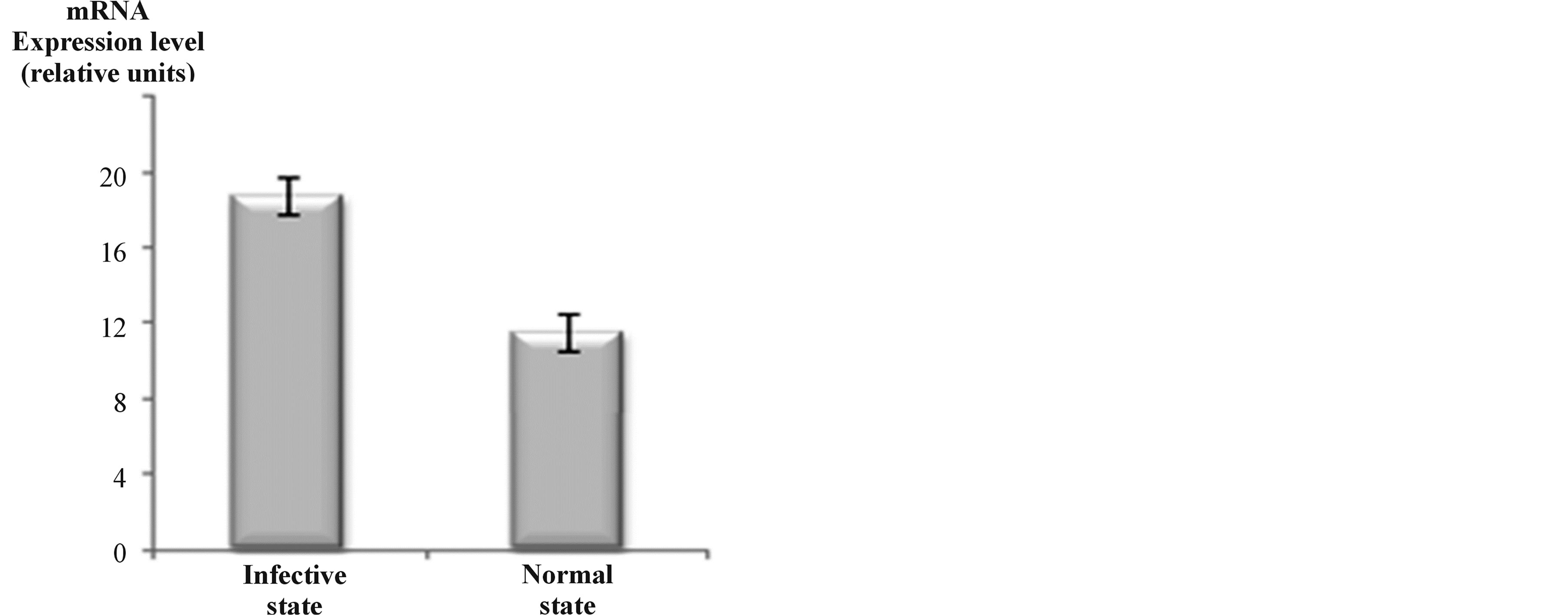
Figure 1. P. syringae pv. tomato DC3000 infective cells have higher expression levels of all the three gap genes when compared with normal cells, as estimated by real-time quantitative PCR analysis. Bars represent mean values ± SD of three independent experiments. Significant statistical differences in Student’s test (p ≤ 0.05).
others suggesting that a secreted GAPDH has a role in signal transduction to the host cells, rendering them more susceptible to bacterial infection [21,24].
However, three GAPDH-encoding genes are present in the genome of P. syringae pv. tomato DC3000 [25], it remains to be established whether any of these GAPDHs are actually extracellular proteins involved in the plant infection process. As described, the pathogenicity of DC3000 resembles those of most animal and plant microbial pathogens of the Gammaproteobacteria, which in the infective state secrete a number of proteins across their cell envelopes [30-32]. Plant-pathogen interactions involve complex processes including translocation of proteins across two cell membranes, the cytoplasmic and the outer membrane. For that, analyzing the P. syringae pv. tomato DC3000 secretome in the infective state is therefore of particular importance.
In order to ascertain the possible occurrence of an extracellular GAPDH during the infection process and identify which of the three GAPDHs is secreted, the extracellular proteins (secretome) released by P. syringae DC3000 cells in the infective state were analyzed by a proteomic approach in concentrates of culture supernatants. After protein resolution of concentrates by SDSPAGE, automatic excision of selected protein bands from Coomasie blue-stained gels was performed, followed by MALDI-TOF peptide mass fingerprinting as described in Materials and Methods.
A number of protein bands with apparent molecular masses in the range 20 - 200 kDa were resolved in SDSPAGE gels (Figure 2). In this figure we noticed the presence of a major band with the expected molecular mass for the GAPDH subunit (36.3 kDa). To verify this finding, this band with others in the range 20 - 50 kDa (see asterisks Figure 2) were selected for MALDI-TOF analysis.
The result of this analyses confirm that the suspected band (36.3 kDa) contain the gap1 gene product GAPDH1 as a main component (see Figure 2 and Table 1). We have previously cloned the gap1 gene and expressed its protein product in E. coli BL21, being a member of the GAPDH family classified into the ubiquitous Class I enzymes that utilize NAD + (EC 1.2.1.12) [25]. This finding has allowed us to know which of the three GAPDHs from Pseudomonas syringae pv tomato DC3000 is extracellular.
Bioinformatic searches in NCBI database allow us to identify a number of extracellular proteins in the secretome of this phytopathogenic bacterium. As shown in Table 1, the Class I GAPDH1 was identified among other secreted proteins of P. syringae DC3000 cells. A similar scenario has been reported for pathogenic E. coli strains, namely the GAPDH A is secreted to the periplasmic compartment and the extracellular medium [20-
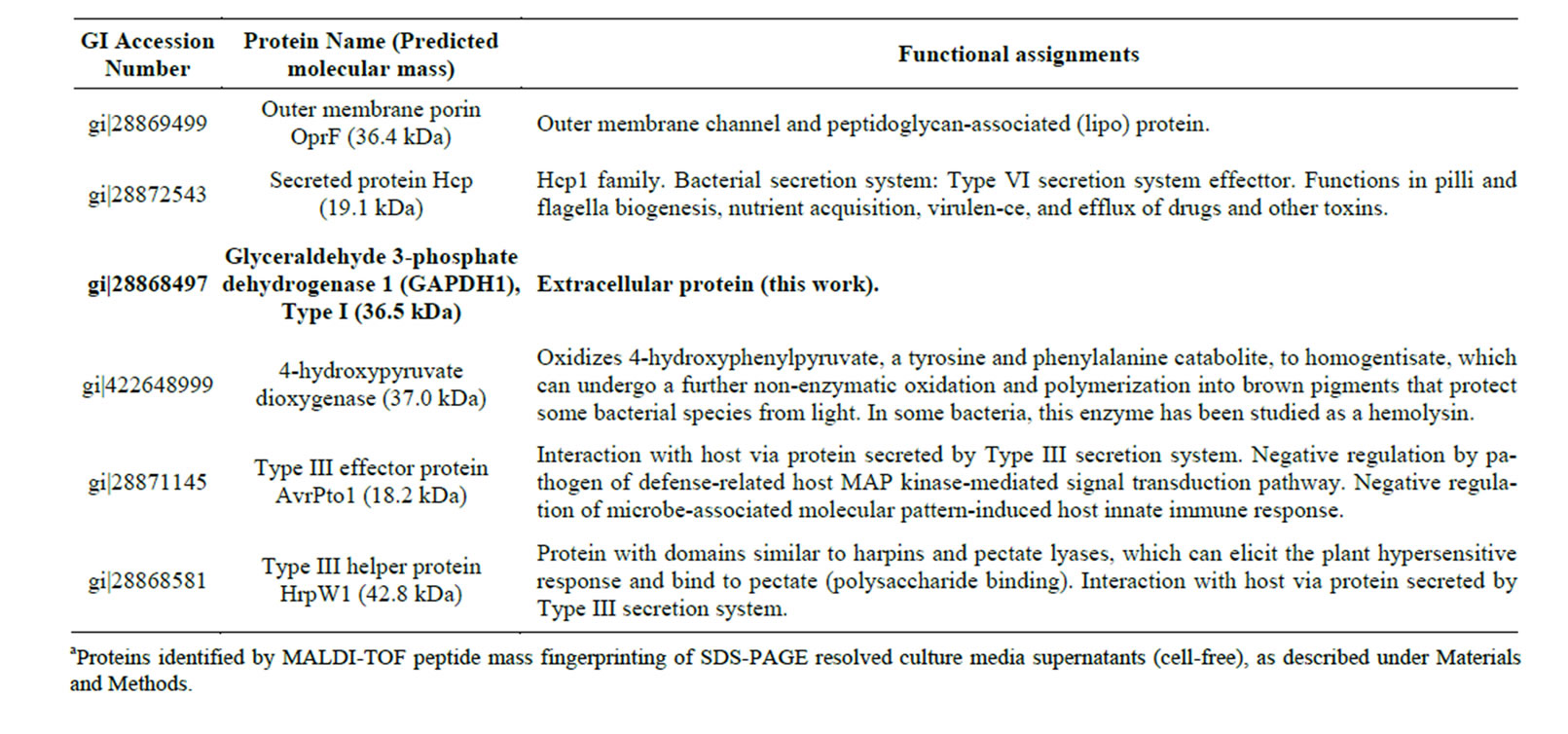
Table 1. Some extracellular proteins produced by Pseudomonas syringae pv tomato DC3000 cells in the infective statea.
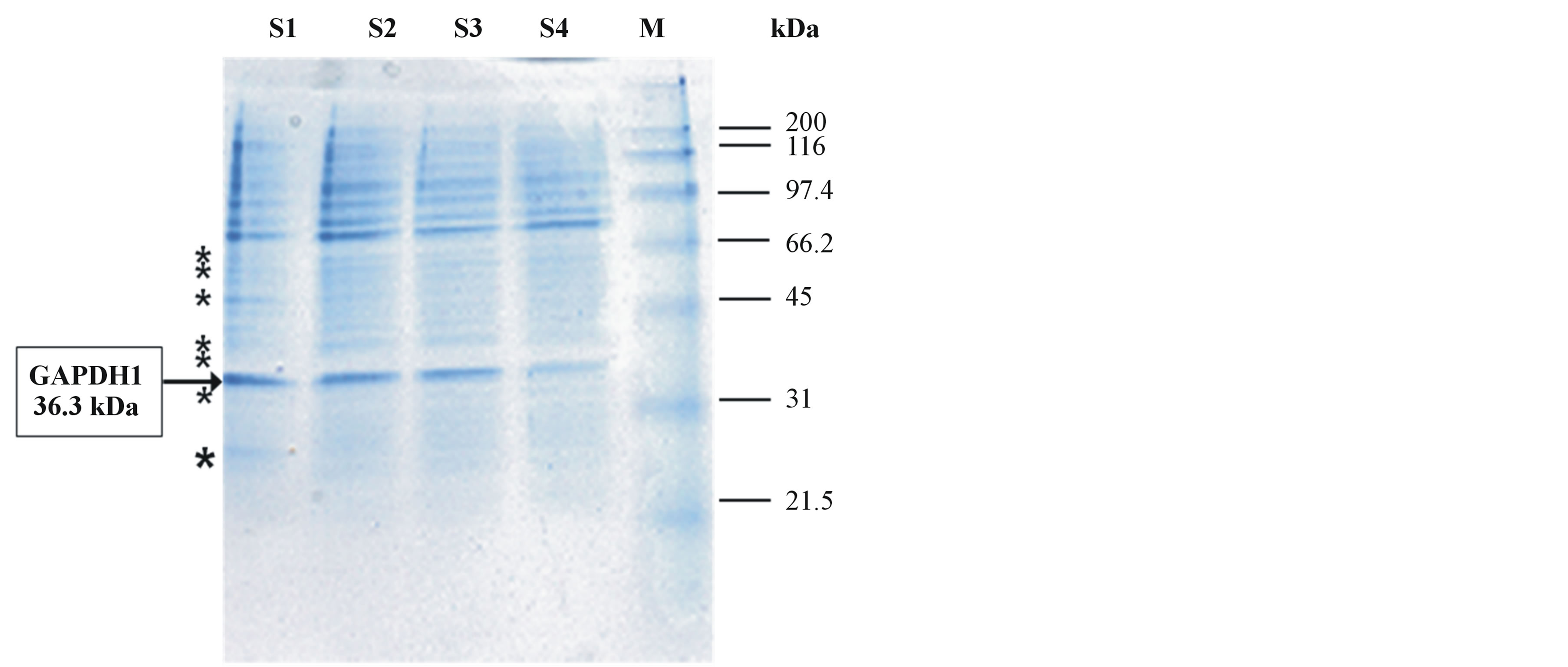
Figure 2. Coomassie Brilliant Blue-stained SDS-PAGE electrophoretogram showing the protein pattern of the secretome of P. syringae pv tomato DC3000 cells in the infective state. Protein concentrates (ca. 50 µg protein per lane) from culture supernatants of four independent experiments (S1-S4) were analyzed (S1-4 samples of Ps syringae celles in the infective state). The molecular masses of protein markers (M) are shown on the right side. The arrow on the left side indicates a major protein band with the expected molecular mass of the GAPDH subunit (36.3 kDa), of which the gap1 gene product GAPDH1 was identified as a main component by MALDI-TOF analysis. Asterisks indicate the approximate positions of other selected protein bands identified by MALDI-TOF analyses (see Table 1).
22]. Moreover, a post-translational modification of the secreted GAPDH has been described in the parasitic protist Entamoeba histolytica, while an extracellular form of GAPDH [E.C. 1.2.1.12] was reported as the major secreted protein produced by the endocarditis isolate S. gordonii FSS2 [33].
As is shown in Table 1, the proteomic analysis of P. syringae pv tomato DC3000 culture concentrates allowed us to identify other extracellular proteins already described to be involved in virulence of other pathogenic bacteria, the secretion of which having been previously demonstrated. As the secreted protein Hcp, which play an important role in bacterial fitness, allowing microbial pathogens to survive under conditions where it has to compete with other microorganisms for resources [34]. And the helper protein Hrp, a member of a protein family of secreted proteins encoded by the hypersensitive response and pathogenicity genes (hrp) that control the ability of major groups of plant pathogenic bacteria to elicit the hypersensitive response (HR) in resistant plants and to cause disease in susceptible plants [14,35,36]. A number of Hrp proteins share significant similarities with components of Type III secretion apparatus and flagellar assembly apparatus in animal pathogenic bacteria [13,14,30,36-39]. In fact, Type III effector protein AvrPto1, a secreted bacterial protein involved in negative regulation of defense-related host signal transduction pathways, was also identified in the P. syringae pv tomato DC3000 secretome (see Table 1).
Overall, our proteomic data should be considered a non-exhaustive representative sample of the P. syringae pv tomato DC3000 secretome. The concurrent release of Class I GAPDH1 with a number of virulence-involved secreted proteins strongly suggests that this dehydrogenase should be a bona fide extracellular protein that may have an active role in P. syringae pv tomato DC3000 pathogenicity.
4. CONCLUSION
In this study, we determined the expression levels of the three gap genes from the phytopathogenic bacterium P. syringae pv tomato DC3000 under infective and normal physiological conditions by one-step Real-Time PCR. Then, the secretome of infective cells was analyzed by MALDI-TOF peptide mass fingerprinting. It was found that there is a clear up-expression of the transcripts of the three gap genes in the infective state, in comparison with the normal state. However, only the Class I GAPDH1 isoform was found as an extracellular protein in infective cells, concurrently with a number of secreted proteins that are involved in the virulence of P. syringae pv. tomato DC3000.
ACKNOWLEDGEMENTS
Support of the Collaborative grant A1/043076/11 of the Spanish AECID (MAEC), and Moroccan CNRST (URAC42) is acknowledged. Authors thanks to Dr. Rocio Rodriguez Sanchez (Servicio de Proteómica del IBVF, CSIC-Universidad de Sevilla) for her skillful technical assistance.
REFERENCES
- Bashan, Y., Okon, Y. and Henis, Y. (1985) Detection of cutinases and pectic enzymes during infection of tomato by Pseudomonas syringae pv. tomato. Phytopathology, 75, 940. http://dx.doi.org/10.1094/Phyto-75-940
- Bashan, Y. and De-Bashan, L.E. (2002) Protection of tomato seedlings against infection by Pseudomonas syringae pv. tomato by using the plant growth-promoting bacterium Azospirillum brasilense. Applied and Environmental Microbiology, 68, 2637-2643. http://dx.doi.org/10.1128/AEM.68.6.2637-2643.2002
- Badel, J.L., Nomura, K., Bandyopadhyay, S., Shimizu, R., Collmer, A. and He, S.Y. (2003) Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp Type III secretion system in a chaperone-dependent manner. Molecular Microbiology, 49, 1239-1251. http://dx.doi.org/10.1046/j.1365-2958.2003.03647.x
- Preston, G.M. (2001) Pseudomonas syringae pv. tomato: The right pathogen, of the right plant, at the right time. Molecular Plant Pathology, 1, 263-275. http://dx.doi.org/10.1046/j.1364-3703.2000.00036.x
- Elkhalfi, B., Essari, A., Serrano, A. and Soukri. A. (2013) Antibacterial activity of plant methanolic extracts on a field isolate of Pseudomonas syringae pv tomato from the casablanca region (Morocco). Advances in Bioscience and Biotechnology, 4, 1-9.
- Rico, A. and Preston, G.M. (2008) Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Molecular Plant-Microbe Interactions, 21, 269-282. http://dx.doi.org/10.1094/MPMI-21-2-0269
- Schneider, R.W. and Grogan, R.G. (1977) Tomato leaf trichomes, a habitat for resident populations of Pseudomonas tomato. Phytopathology, 67, 898-902. http://dx.doi.org/10.1094/Phyto-67-898
- Bashan, Y. and de-Bashan, L.E. (2002) Reduction of bacterial speck (Pseudomonas syringae pv. tomato) of tomato by combined treatments of plant growth-promoting bacterium, Azospirillum brasilense, streptomycin sulfate, and chemo-thermal seed treatment. European Journal of Plant Pathology, 108, 821-829. http://dx.doi.org/10.1023/A:1021274419518
- Buell, C.R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I.T., Gwinn, M.L., et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences, 100, 10181-10186. http://dx.doi.org/10.1073/pnas.1731982100
- Feil, H., Feil, W.S., Chain, P., Larimer, F., DiBartolo, G., Copeland, A., et al. (2005) Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America, 102, 11064-11069. http://dx.doi.org/10.1073/pnas.0504930102
- Fouts, D.E., Abramovitch, R.B., Alfano, J.R., Baldo, A.M., Buell, C.R., Cartinhour, S., et al. (2002) Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proceedings of the National Academy of Sciences, 99, 2275-2280. http://dx.doi.org/10.1073/pnas.032514099
- Buell, C.R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I.T., Gwinn, M.L., et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences, 100, 10181- 10186. http://dx.doi.org/10.1073/pnas.1731982100
- Hueck, C.J. (1998) Type III Protein Secretion Systems in Bacterial Pathogens of Animals and Plants. Microbiology and Molecular Biology Reviews, 62, 379-433.
- Zhao, Y., Thilmony, R., Bender, C.L., Schaller, A., He, S.Y. and Howe, G.A. (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. The Plant Journal, 36, 485-499. http://dx.doi.org/10.1046/j.1365-313X.2003.01895.x
- Soukri, A., Hafid, N., Valverde, F., Elkebbaj, M.S. and Serrano, A. (1292) Evidence for a posttranslational covalent modification of liver glyceraldehyde-3-phosphate dehydrogenase in hibernating jerboa ( Jaculus orientalis). Biochimica et Biophysica Acta (BBA): Protein Structure, 1996, 177-187. http://dx.doi.org/10.1016/0167-4838(95)00200-6
- Soukri, A., Valverde, F., Hafid, N., Elkebbaj, M.S. and Serrano, A. (1995) Characterization of muscle glyceraldehyde-3-phosphate dehydrogenase isoforms from euthermic and induced hibernating Jaculus orientalis. Biochimica et Biophysica Acta (BBA): General Subjects, 1243, 161-168. http://dx.doi.org/10.1016/0304-4165(94)00137-M
- Petersen, J., Brinkmann, H. and Cerff, R. (2003) Origin, Evolution, and Metabolic Role of a Novel Glycolytic GAPDH Enzyme Recruited by Land Plant Plastids. Journal of Molecular Evolution, 57, 16-26. http://dx.doi.org/10.1007/s00239-002-2441-y
- Zheng, L., Roeder, R.G. and Luo, Y.S. (2003) Phase Activation of the Histone H2B Promoter by OCA-S, a Coactivator Complex that Contains GAPDH as a Key Component. Cell, 114, 255-266. http://dx.doi.org/10.1016/S0092-8674(03)00552-X
- Dheda, K., Huggett, J.F., Bustin, S.A., Johnson, M.A., Rook, G. and Zumla, A. (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques, 37, 112-119.
- Aguilera, L., Ferreira, E., Giménez, R., Fernández, F.J., Taulés, M., Aguilar, J., et al. (2012) Secretion of the housekeeping protein glyceraldehyde-3-phosphate dehydrogenase by the LEE-encoded Type III secretion system in enteropathogenic Escherichia coli. The International Journal of Biochemistry & Cell Biology, 44, 955-962. http://dx.doi.org/10.1016/j.biocel.2012.03.002
- Kenny, B. and Finlay, B.B. (1995) Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proceedings of the National Academy of Sciences, 92, 7991-7995. http://dx.doi.org/10.1073/pnas.92.17.7991
- Alvarez, A.H., Martinez-Cadena, G., Silva, M.E., Saavedra, E. and Avila, E.E. (2007) Entamoeba histolytica: ADP-ribosylation of secreted glyceraldehyde-3-phosphate dehydrogenase. Experimental Parasitology, 117, 349-356. http://dx.doi.org/10.1016/j.exppara.2007.04.016
- Eichenbaum, Z., Green, B.D. and Scott, J.R. (1996) Iron starvation causes release from the group A streptococcus of the ADP-ribosylating protein called plasmin receptor or surface glyceraldehyde-3-phosphate-dehydrogenase. Infection and Immunity, 64, 1956-1960.
- Pancholi, V. and Fischetti, V.A. (1997) Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: Signal transduction between streptococci and pharyngeal cells. The Journal of Experimental Medicine, 186, 1633-1643. http://dx.doi.org/10.1084/jem.186.10.1633
- Elkhalfi, B., Araya-Garay, J.M., Rodríguez-Castro, J., Rey-Méndez, M., Soukri, A. and Serrano Delgado, A. (2013) Cloning and heterologous overexpression of three gap genes encoding different glyceraldehyde-3-phosphate dehydrogenases from the plant pathogenic bacterium Pseudomonas syringae pv. tomato strain DC3000. Protein Expression and Purification, 89, 146-155. http://dx.doi.org/10.1016/j.pep.2013.02.005
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. http://dx.doi.org/10.1038/227680a0
- Iddar, A., Serrano, A. and Soukri, A. (2002) A phosphate-stimulated NAD (P)+-dependent glyceraldehyde- 3-phosphate dehydrogenase in Bacillus cereus. FEMS Microbiology Letters, 211, 29-35. http://dx.doi.org/10.1111/j.1574-6968.2002.tb11199.x
- Yuan, J. and He, S.Y. (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. Journal of Bacteriology, 178, 6399-6402.
- Nelson, D., Goldstein, J.M., Boatright, K., Harty, D.W.S., Cook, S.L., Hickman, P.J., et al. (2001) pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: Purification, characterization, and cloning of the gene encoding this enzyme. Journal of Dental Research, 80, 371-377. http://dx.doi.org/10.1177/00220345010800011301
- Tommassen, J., Filloux, A., Bally, M., Murgier, M. and Lazdunski, A. (1992) Protein secretion in Pseudomonas aeruginosa. FEMS Microbiology Letters, 103, 73-90. http://dx.doi.org/10.1111/j.1574-6968.1992.tb05824.x
- Pugsley, A.P. (1993) The complete general secretory pathway in gram-negative bacteria. Microbiological Reviews, 57, 50-108.
- Nelson, D., Goldstein, J.M., Boatright, K., Harty, D.W.S., Cook, S.L., Hickman, P.J., Potempa, J., Travis, J. and Mayo, J.A. (2001) pH-regulated secretion of a glyceroldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: Purification, characterization, and cloning of the gene encoding this enzyme. Journal of Dental Research, 80, 371-377. http://dx.doi.org/10.1177/00220345010800011301
- Haapalainen, M., Mosorin, H., Dorati, F., Wu, R.F., Roine, E., Taira, S., Nissinena, R., Mattinena, L., Jacksonc, R., Pirhonena, M. and Lin, N.C. (2012) Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. Journal of Bacteriology, 194, 4810- 4822. http://dx.doi.org/10.1128/JB.00611-12
- Charkowski, A.O., Alfano, J.R., Preston, G., Yuan, J., He, S.Y. and Collmer, A. (1998) The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. Journal of Bacteriology, 180, 5211-5217.
- Peñaloza-Vázquez, A., Preston, G.M., Collmer, A. and Bender, C.L. (2000) Regulatory interactions between the Hrp Type III protein secretion system and coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000. Microbiology, 146, 2447-2456.
- Tampakaki, A.P., Fadouloglou, V.E., Gazi, A.D., Panopoulos, N.J. and Kokkinidis, M. (2004) Conserved features of Type III secretion. Cellular Microbiology, 6, 805-616. http://dx.doi.org/10.1111/j.1462-5822.2004.00432.x
- Roine, E., Wei, W., Yuan, J., Nurmiaho-Lassila, E.L., Kalkkinen, N., Romantschuk, M. and He, S.Y. (1997) Hrp pilus: An hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America, 94, 3459-3464. http://dx.doi.org/10.1073/pnas.94.7.3459
- Kvitko, B.H., Ramos, A.R., Morello, J.E., Oh, H.S. and Collmer, A. (2007) Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of Type III secretion system effectors. Journal of Bacteriology, 189, 8059-8072. http://dx.doi.org/10.1128/JB.01146-07
ABBREVIATION LIST
GAPDH: glyceraldehyde-3- phosphate dehydrogenase;
TTSS: type III secretion system;
NAD: Nicotinamide adenine dinucleotide;
LB: Luria Broth;
TCA: trichloroacetic acid;
Hrp: hypersensitive response and pathogenicity;
NOTES
*Corresponding author.

