Advances in Bioscience and Biotechnology
Vol.4 No.1(2013), Article ID:26938,11 pages DOI:10.4236/abb.2013.41007
HSF1-mediated oxidative stress response to menadione in Saccharomyces cerevisiae KNU5377Y3 by using proteomic approach
![]()
1Advanced Bio-Resource Research Center, Department of Biology, Kyungpook National University, Daegu, South Korea
2Department of Microbiology, Kyungpook National University, Daegu, South Korea
Email: 92kis@hanmail.net
Received 20 October 2012; revised 23 November 2012; accepted 3 January 2013
Keywords: Saccharomyces cerevisiae KNU5377Y3; HSF1; Gene Expression; Menadione; Redox Homeostasis
ABSTRACT
The hat shock transcription factor HSF1 in the yeast Saccharomyces cerevisiae regulates a wide range of genes and functions in diverse cellular reactions. To investigate the physiological response of HSF1 in the presence of menadione (MD) in S. cerevisiae KNU 5377Y3, wild-type (k3wt) and isogenic hsf1 mutant (k3h1) cells were introduced. HSF1 was induced when k3wt cells were exposed to the superoxide-generating agent MD and k3h1 cells were hypersensitive to MD. Under MD stress, k3h1 cells down-regulated the expression of metabolic enzymes (Hxk, Fba1, Pgk1, Eno2, and Adh1), antioxidant enzymes (Trx2 and porin), and molecular chaperones and their cofactors (Hsp104, Ssb1, Hsp60, Hsp42, Hsp26, Hsp12, Cpr1, and Sti1). In addition, k3h1 cells increased cellular hydroperoxide levels and protein carbonylation under MD stress as compared to k3wt cells. However, there was a moderate difference in the wild-type (b3wt) and mutant (b3h1) cells derived from S. cerevisiae S288C under the same conditions. Thus, these results show that HSF1 is an important component of the stress response system, acting as an activator of cell rescue genes in S. cerevisiae KNU5377Y3, and its expression protects the cells from MD-induced oxidative damage by maintaining redox homeostasis and proteostasis in the presence of MD.
1. INTRODUCTION
Oxidative stress caused via endogenousand/or exogenous environmental changes can damage a wide range of cellular components such as carbohydrates, lipids, nucleic acids, and proteins. To protect or adapt against oxidative damage, cells have evolved a variety of cellular defense ystems, including the regulated expression of specific transcription factors. There are a number of transcriptional control systems that regulate the stress response in Saccharomyces cerevisiae, some of which are specific to one type of stress, or even a subset of stress responses, and some of which are more general. The main transcription factors involved in regulating the oxidative stress in yeast are Yap1, Skn7, and Msn2/4 [1-4]. Yap1 and Skn7 activate the expression of proteins that scavenge and destroy reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), superoxide anion (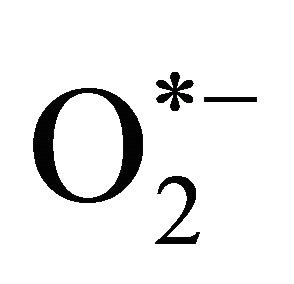 ), and hydroxyl radical (HO*). Msn2/4 controls the expression of a small number of antioxidants as well as heat shock proteins (Hsp), molecular chaperones, metabolic process enzymes, and members of the ubiquitinproteasome degradation systems [5]. Some minor stressrelated transcription factors in S. cerevisiae include Yap2p (Cad2p), which is turned on in response to toxic compounds; Yap4p (Cin5p/Hal6p) and Yap6p (Hal7p), which are turned on in response to osmotic stress; and Yap8p/Acr1p, which is turned on in response to arsenic stress [2].
), and hydroxyl radical (HO*). Msn2/4 controls the expression of a small number of antioxidants as well as heat shock proteins (Hsp), molecular chaperones, metabolic process enzymes, and members of the ubiquitinproteasome degradation systems [5]. Some minor stressrelated transcription factors in S. cerevisiae include Yap2p (Cad2p), which is turned on in response to toxic compounds; Yap4p (Cin5p/Hal6p) and Yap6p (Hal7p), which are turned on in response to osmotic stress; and Yap8p/Acr1p, which is turned on in response to arsenic stress [2].
HSF1 is a transcriptional activator that mediates eukaryotic gene expression in response to heat shock, pathogen infection, and inflammation, pharmacological agents, and other environmental stresses [6], thereby playing a central role in the regulation of cellular homeostasis [7]. Although genes encoding heat shock proteins and molecular chaperones are the best characterized targets of HSF1, recent genome-wide localization of S. cerevisiae HSF1 has revealed novel HSF1 targets involved in a wide range of cellular functions [3,7]. HSF1 activates multiple signaling pathways in response to oxidative stress; the degree to which a given pathway is activated is highly dependent on the nature and duration of the stress, as well as the cell type [3]. More recent work has extended these findings to suggest that HSF1 is actually linked to oxidative stress tolerance. When cells are exposed to the ROS generator menadione (2-methyl-1,4-naphtoquinone; MD), HSF1 increases the mRNA expression of CUP1, a gene encoding metallothionein [8] and ERO1, a gene encoding thiol oxidase, which is required for oxidative protein folding in the endoplasmic reticulum [5,9]. In mice, HSF1 knock-out caused an alteration of redox homeostasis and increased oxidative damage [1].
In a previous study, we identified that HSF1 of the S. cerevisiae strain KNU5377Y is induced in the presence of MD and activates a stress defense mechanism distinct from the mechanism induced when reference strain BY4741 (derived from S288C) was exposed to oxidants [10]. Among these oxidants, MD is a quinine compound that undergoes redox cycling. One-electron transfer, mainly from the mitochondrial respiratory chain, forms semiquinone radicals that can rapidly reduce O2, thereby generating superoxide radical intracellularly and regenerating the quinine [4,10]. The current study was carried out to obtain further information on the relationship between HSF1 expression and stress response at the translational level in S. cerevisiae KNU5377Y3. Our results demonstrate that HSF1 expression in KNU 5377 Y3 leads to the activation of cell rescue genes via a mechanism different from that observed in BY4743 when cells were challenged with MD. Thus, the results obtained suggest another HSF-mediated stress responsive mechanism that is regulated by an oxidative-stress regulon, unlike previously studied systems in BY4743.
2. MATERIALS AND METHODS
2.1. Yeast Growth Conditions
Yeast cells were grown aerobically in a YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) at 30˚C overnight, transferred to fresh YPD medium, and further cultured by shaking at 160 rpm [11]. Once a mid-log phase (OD600 = 2.0) was reached, cells were challenged directly with 0.4 mM MD (water-insoluble form; 2- methyl-1,4 naptoquinone; Sigma) for 1 h at 30˚C. Cells were harvested by centrifugation and the pellets were washed twice with a cold phosphate buffered saline (PBS), washed once with chilled distilled water, and then used for the subsequent experiments.
2.2. Stress Sensitivity to Menadione
For cell viability, mid-log phase cells were exposed to 0.0, 0.2, 0.4, 0.6, and 0.8 mM MD for 1 h, and then were diluted, spread on YPD agar plates, and incubated for 2 - 3 days. Cell viability was calculated by measuring colony-forming units (CFU), and the number of colonies grown in YPD liquid medium without MD treatment was set to 100%. For growth rate analysis, cells (1 × 106 cells per ml) were inoculated in YPD media and simultaneously treated with 45 μΜ MD. Optical density was measured at 600 nm at 2 h intervals for the indicated time. For the spotting assay, mid-log phase cells (OD600 = 2.0) were challenged with 0.4 mM MD at 30˚C for 1 h in YPD liquid media, and then diluted to 10−5. An aliquot (5 μl) of each diluted sample was spotted onto YPD agar medium to monitor sensitivity. Wild-type (k3wt) and its isogenic Hsf1 mutant hsf1Δ (k3h1) cells, wild-type (b3wt) and hsf1Δ mutant (b3h1) cells from S. cerevisiae BY 4743, and wild-type (WT) and porin mutant (por1Δ) cells from S. cerevisiae BY4741 were used for stress sensitivity to MD.
2.3. Sequence Analysis
The mid-log phase cells grown for 6 h at 30˚C in a YPD liquid medium were harvested by centrifugation. Total RNA was extracted using the SV Total RNA Isolation system (Promega) according to the manufacturer’s instructions. cDNA synthesis from total RNA was performed using an AccuPower RT Premix (Bioneer). The reaction mixture contained 0.1 μg of template RNA and 200 pmol of oligo dT18 primer in a total volume of 20 μl [12]. The reaction was conducted according to the manufacturer’s instructions. The HSF1 coding region was amplified from the cDNA by PCR using Taq and Pfu polymerases (Roche) with the following primers: HSF1- A (5’-TGCAGTTCATGCATATTAAGTGAGT-3’) and HSF1-D (5’-AGTCAATATAAGTACGCCAACTTGC-3’). PCR reaction conditions were as follows: initial denaturation at 94˚C for 3 min, followed by 30 cycles of 1 min at 94˚C, 1 min at 54˚C, 4 min at 72˚C, and a final extension of 10 min at 72˚C. The PCR product was extracted from 1.0% agarose gel using Wizard® SV Gel and PCR Clean-Up System (Promega) and inserted into the TOPO TA cloning vector (Invitrogen) according to manufacturer’s protocol. The cloned plasmid was sequenced using the M13 primer set. The sequenced nucleotide was translated using European Molecular Biology Laboratory (EMBL)-European Bioinformatics Institute (EBI) Transeq (http://www.ebi.ac.uk/Tools/st/emboss_transeq/) software. Alignment of the predicted amino acid sequence was carried out using DAN Data Bank of Japan (DDBJ) ClustalW (http://clustalw.ddbj.nig.ac.jp/) software.
2.4. Western Blot Analysis
Cell lyates were prepared by lysing cells in a lysis buffer [50 mM Tris-HCl, pH 7.4, 5% glycerol, 2% sodium dodecyl sulfate (SDS), 1.5% β-mercaptoethanol, 1 mM PMSF, and EDTA-free protease inhibitor cocktail (Roche)] and crude proteins were extracted using glass beads [13]. SDS-polyacrylamide gel electrophoresis (SDSPAGE) was performed using the Laemmli method [14]. Denatured proteins (25 μg) were loaded into 10%, 12%, or 15% polyarylamide gels and then electro-phoretically transferred to a PVDF membrane (Bio-Rad). The PVDF membranes were incubated for 1.5 h at room temperature in a blocking buffer containing TBST (0.05% Tween-20, 10 mM Tris-HCl, pH 7.6, 150 mM NaCl), 5% non-fat skim milk, and 0.02% sodium azide. The blotted membranes were incubated overnight at 4˚C with the following primary antibodies: anti-Hsp104, and antiHsp60, (Stressgen), anti-glucose-6-phosphate dehydrogenase, and anti-Zpr1 (Sigma), anti-hexokinase (Abcam), anti-porin (Invitrogen), anti-tubulin (Millipore), and antiHsp82, anti-Ssa1, anti-Ssb1, anti-Hsp42, anti-Hsp26, anti-Hsp12, anti-Sti1, anti-Grp, and anti-Cpr1 [10] antibodies, After washing 4 times for 40 min with TBST, the membranes were incubated for 1.5 h at room temperature with antirabbit IgG (H + L) (Promega) or antimouse IgG (Millipore) secondary antibodies conjugated to HRP, washed 4 times for 40 min with TBST, and developed using enhanced chemiluminescence western blot detection kit (GE Healthcare).
2.5. Two-Dimensional Gel Electrophoresis and Protein Identification
The mid-log phase yeast cells were exposed to 0.4 mM MD for 1 h at 30˚C with shaking.
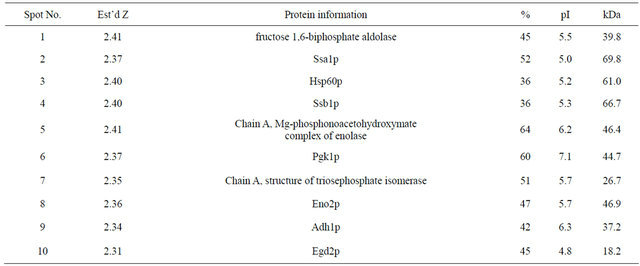
After washing twice with cold PBS, harvested cells were agitated 5 times for 5 min using a MicroMixer in a lysis buffer [80 mM Tris-HCl, pH 8.0, 2% SDS, 1.5% β-mercaptoethanol , 5% glycerol, 1 mM PMSF, and EDTA-free protease inhibitor cocktail (Roche)] and an equal amount of glass beads (400 - 600 micron; Sigma). Following centrifugation at 13,000 rpm for 20 min at 4˚C, the cleared supernatants were incubated with DNase/ RNase/Mg mix on ice for 15 min before boiling for 5 min and cooling on ice for 5 min. Proteins in the supernatants were precipitated by trichloroacetic acid (final 10%) for 1 h on ice, centrifuged at 15,000 rpm for 20 min at 4˚C, and then washed 5 times with HPLCgrade ethanol containing 0.07% β-mercaptoethanol. The washed pellets were dried using a Speed Vac and were resuspended in sample buffer [9.5 M urea, 4% CHAPS, 0.1 M dethiothreitol (DTT), 40 mM Tris and 0.2% Bio-Lyte (3 - 10; Bio-Rad)] for 1 h and centrifuged at 15,000 rpm for 30 min. The cleared supernatants were transferred to a new tube, and the protein concentration was measured by a modified Bradford method [15] using a protein assay reagent (Bio-Rad). First-dimensional isoelectric focusing (IEF) was carried out at 20˚C on commercial immobilized pH gradient (IPG) strips (17 cm, pH 4 - 7; Bio-Rad), with a maximum current limitation of 50 μA/strip using Protean IEF Cell (Bio-Rad). The protein sample (1.0 mg) was loaded onto the bottom of an IPG strip. Running conditions were conducted according to the manufacturer’s protocol. After equilibration following IEF, the strips were loaded on a 12% SDSPAGE gel. Electrophoresis was performed on 1.0-mm thick gels at a constant current of 20 mA per gel at 18˚C. After electrophoresis, the gels were fixed, stained with Coomassie Brilliant Blue R-250 (Sigma) for 3 h, and then destained. Spots upregulated in wild type KNU 5377Y3 (k3wt) cells were excised for matrix associated laser-desorption ionization time-of-flight mass-spectrometry (MALDI-TOF MS) analysis. Protein identification was carried out using ProFound (http://prowl.Rockefeller.edu/profound_bin/WebProFound) software. The occurrence of a degraded protein species was assessed on the basis of the simultaneous occurrence of the following events: the definition of protein identification (Pro-Found’s Est’d Z score > 1.6) and an evident discrepancy, with respect to the expected Mr values of the intact protein (ΔMr > 30%).
2.6. Redox State and Protein Oxidation
The intracellular hydroperoxide level was spectrophotometrically determined by ferrous ion oxidation in the presence of a ferric ion indicator, xylenol orange (FOX) as described previously [10]. To measure the ROS levels, cells in the exponential growth phase were incubated for 20 min at 30˚C with 100 μM dichlorodihydrofluorescein diacetate (DCFHDA) and 100 μM rhodamine 123 (DHR 123), exposed to 0.4 mM or 20 mM H2O2 for 1 h with shaking, washed twice with PBS buffer, and then resuspended in the same buffer. Cells loaded with the fluorescent probes were imaged by fluorescence microscopy (excitation, 488 nm; emission, 525 nm). Carbonyl content was also measured by the spectrometric method reported previously [16].
2.7. Statistical Analysis
All biochemical experiments were carried out in at least 3 independent repetitions. Results are expressed as mean standard deviation (SD). The results of the spotting assay, growth kinetics, and redox state are representative of at least 2 independent experiments carried out under identical conditions.
3. RESULTS
3.1. Identification and Sequence Analysis of S. cerevisiae KNU5377Y3 HSF1
The complete sequence of HSF1 from diploid S. cerevisiae KNU5377Y3 was analyzed. The complete HSF1 ORF encodes a protein consisting of 833-amino-acids, with a predicted molecular weight of approximately 93 kDa. The deduced amino acid sequence was 98.2% identical to that of the diploid S. cerevisiae S288C. Fifteen amino acids from KNU5377Y3 HSF1 were substituted as compared those of S288C HSF1: S72P, S123T, K125E, D139N, W254R, N283S, S305I, Q377K, G433A, E579D, K614E, W630L, N799K, K802E, and A831V (Figure 1). These results suggest that the genetic variation of the KNU5377Y3 HSF1 gene could explain why previous reports showed a different stress response in yeast cells exposed to unfavorable conditions.
3.2. HSF1 Upregulation Attenuates Stress Sensitivity and Cells Expressing Its Isogenic Mutant Are Hypersensitive to Menadione in Diploid S. cerevisiae KNU5377Y3
To examine whether HSF1 is activated under MDinduced oxidative stress in wild-type cells of S. cerevisiae KNU5377Y3 (k3wt), semi-quantitative RT-PCR and western blot were conducted. As shown in Figure 2(a), Hsf1 expression in k3wt cells was highly induced at the translational level when yeast cells were exposed to 0.4 mM MD for 1 h. However, there was a distinct difference in Hsf1 expression in wild-type cells of S.
cerevisiae BY4743 (b3wt) in the absence and presence of MD. To investigate if the Hsf1 activation in k3wt cells is involved in the stress response to MD, KNU5377Y3 cells expressing the isogenic Hsf1 mutant hsf1Δ (k3h1) were introduced. In addition, wild-type (b3wt) and hsf1Δ mutant (b3h1) cells from S. cerevisiae BY4743 were used as positive control cells. Each genotype used is described in Table 1. Under MD stress, the k3h1 cells were dose-dependently hypersensitive as compared to the k3wt cells. Although this notable distinction was not observed in both the b3wt and b3h1 cells, there were minor differences in both cells (Figure 2(b)). This difference in stress response to MD was strongly supported by growth kinetics and spotting assay. In the presence of 45 μM MD, the k3wt cells showed a faster recovery than the k3h1 cells over the indicated time. However, these significant differences were not found in the b3wt and b3h1 standard cells under the same conditions (Figure 2(c)). These observations were confirmed by the agar plate assay. The k3wt cells recovered rapidly in the presence of 0.4 mM MD as compared to the k3h1 cells, while cell survival was not affected in the b3wt and b3h1 cells under the same MD stress (Figure 2(d)). In contrast to most findings reported to date, our results indicate that HSF1 in S. cerevisiae KNU5377Y3 is induced by MD stress and is a critical component of the stress response.

Figure 1. HSF1 sequence alignment of predicted amino acids between S. cerevisiae KNU5377 (KNU5377) and S. cerevisiae S288C (S288C). Red letters indicate no match. “*” indicates that the residues in that column are identical in all sequences in the alignment, “:” indicates the presence of conserved substitutions, and “.” indicates the presence of semiconserved substitutions.
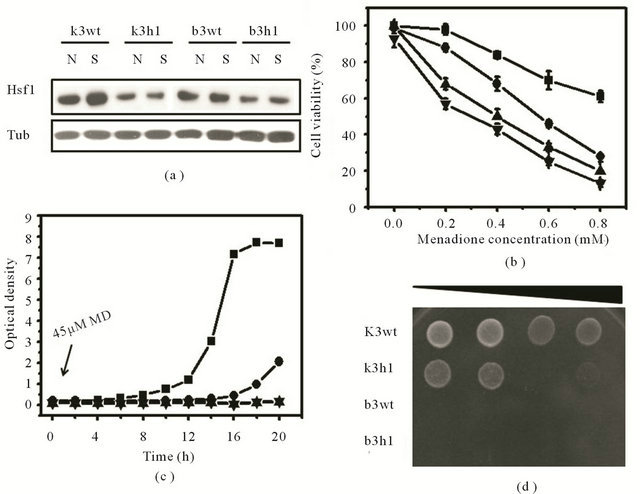
Figure 2. Expression changes of HSF1, growth kinetics, and cell survival in response to MD stress. (a) Translation analysis of Hsf1 in the wild-type and the hsf1∆ mutant cells from S. cerevisiae KNU5377Y3 and S. cerevisiae BY4743 in the absence (N) and presence (S) of 0.4 mM MD for 1 h by western blot. Tubulin (Tub) protein was used as a loading control. For cell survival assay, cell viability at different concentrations of MD (b), growth kinetics in the presence of 45 μM MD (c), and spotting assay using serial dilution (10−2 to 10−5) after 0.4 mM MD for 1 h (d) were done as described in Materials and Methods. Square, k3wt cells; circle, k3h1 cells; upward triangle, b3wt cells; downward triangle, b3h1 cells.
Table 1. S. cerevisiae strains used in this study.
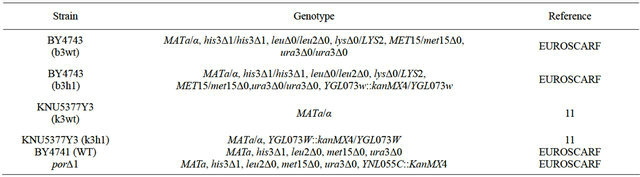
*EUROSCARF: European Saccharomyces cerevisiae archives for functional analysis.
3.3. HSF1 Activates Cell Rescue Systems in Stress Response to Menadione in S. cerevisiae KNU5377Y3
HSF1 has been largely known as a transcription factor and several HSF-mediated stress responsive studies at the transcriptional level have been reported to date [4-7]. However, a survey of HSF1-controlled stress responses at the translational levels has been not conducted even though there can be a distinct difference between transcripts and proteins. Based on these facts, we further analyzed the effects of HSF1 during the stress response by using a proteomic approach. To achieve this, MS analysis following 2-D gel electrophoresis and western blot were performed. In the k3wt cells, MD treatment led to the accumulation of various proteins, including fructose-1,6-biphosphate aldolase (Fba1), heat shock protein 70 family (Hsp70; Ssa1), Hsp60p, enolase II (Eno2), 3- phosphoglycerate kinase (Pgk1), alpha subunit of the heteromeric nascent polypeptide-associated complex (Egd2), and alcohol dehydrogenase isoform 1 (Adh1), while the k3h1 cells down-regulated these proteins under the same conditions (Figure 3). To widely confirm HSF1-regulated target proteins, western blot was carried out. The k3wt cells had elevated expression of metabolic enzymes, including hexokinase (Hxk), and antioxidant enzymes including thioredoxin isoform 2 (Trx2) and porin (Por) under MD stress, while k3h1 cells down-regulated these proteins in the absence and presence of MD. Hexokinase and Porin expression in the b3wt and b3h1 cells increased in a HSF1-independent manner. However, Trx2 expression in b3h1 cells was repressed in the presence and absence of MD compared to that of the b3wt cells, even though the expression mildly increased under MD stress (Figure 4(a)). In particular, repression of porin protein increased stress sensitivity (Figure 4(b), upper panel) and cytosolic and mitochondrial ROS levels (Figure 4(b), lower panel) when yeast cells were treated with 0.2 mM MD for 1 h. In addition, the k3wt cells up-regulated molecular chaperones and their associated cofactors, including Hsp104, Hsp70 (Ssb1), Hsp60, Hsp 42, Hsp26, Cpr1, and Sti1 in the presence of MD, whereas k3h1 cells decreased expression of these proteins under the same stress condition. However, Hsp82, Grp, and Zpr1 expression in the k3h1 cells increased under MD stress. Although Hsp12 expression in the k3wt cells did not change, the expression in the k3h1 cells was reduced with and without MD treatment. The expression of most proteins, except Ssb1, increased in both the b3wt and b3h1 cells in the absence or presence of MD. The expression intensity of b3h1 cells was higher than that of b3wt cells under MD treatment. Ssb1 expression was reduced in the b3h1 cells under MD stress (Figure 4(c)). Therefore, our findings indicate that HSF1 in S. cerevisiae KNU5377Y3 cells regulates cell rescue systems through a mechanism different from the known regulation mechanism.
3.4. HSF1 Deficiency Causes Enhancement of Cellular ROS Levels and Protein Oxidation in S. cerevisiae KNU5377Y3
To investigate whether HSF1 affects cellular redox state in the presence of MD, cellular hydroperoxide levels and protein carbonyl contents were measured under MD stress. The level of intracellular hydroperoxides was evaluated by FOX assay and fluorescence microscopy with a cytosolic oxidant-sensitive probe DCFHDA. An intensity increase in DCF fluorescence was observed in both k3wt and k3h1 cells when they were exposed to MD, and this increase was more pronounced in the k3h1 cells. A similar result was detected in both the b3wt and b3h1 cells. Overall, the DCF fluorescence intensity increased from k3h1 to b3h1 to b3wt to k3wt cells in the presence of MD. In addition, we observed a moderate release of probe dye outside the k3h1 and b3h1 cells (Figure 5(a)). To confirm these findings, hydroperoxide levels using FOX reagent were also measured. Cellular hydroperoxide level in the k3h1 cells was 2-fold higher than that in the k3wt cells when yeast cells were exposed to MD, whereas a moderate increase was observed in the b3h1 cells under the same stress condition as compared to the b3wt cells. However, the hydroperoxide levels in both the b3wt and b3h1 cells were lower than those of the k3h1 cells under MD stress. There was a small increase in the ROS levels in the k3h1 cells under normal conditions as compared to the k3wt cells and the b3wt cells (Figure 5(b)). To determine whether increased ROS levels affect protein oxidation under MD stress, protein carbonyl contents were measured spectrophotometrically using dinitrophenylhydrazine (DNPH) in the absence and presence of MD. A significant increase in the carbonyl contents was observed in the k3h1 cells, with average levels approximately 1.5-fold higher than that in the k3wt cells during MD stress. An increase of the carbonyl contents in the k3h1 cells was detected under MD-free conditions. On the other hand, the carbonyl content of the b3h1 cells was slightly increased in the presence of MD as compared to that in the b3wt cells (Figure 5(c)). These results suggest that hsf1∆ deficiency in S. cerevisiae KNU5377Y3 increases protein oxidation following imbalance of the redox state.
4. DISCUSSION
HSF1 of diploid S. cerevisiae KNU5377Y3 displayed genetic variation of 15 amino acids compared to that of the diploid S. cerevisiae S288C (Figure 1). HSF1 expression in wild-type S. cerevisiae KNU5377Y3 (k3wt) cells was induced under MD stress and the isogenic hsf1∆ mutant (k3h1) cells were hypersensitive to MD. However, there was no phenotypic difference in the wild-type (b3wt) and hsf1∆ mutant (b3h1) cells of S. cerevisiae BY4743 derived S. cerevisiae S288C in the presence of MD (Figure 2). According to previous studies, most of HSF1 activation in yeast is induced by diamide, transition to the stationary phase, depletion of nitrogen or carbon sources, heat shock [17], and ROS generated by H2O2 treatment [4]. In Drosophila, a single HSF molecule responds directly to heat and to oxidative stress (H2O2) [18]. HSF1 of mammalian cells is essential for cell protection from stresses induced by heat shock and oxidative stress [19], or protection against acute cell injury, pharmaceutical agents, infection and inflammation [20]. In addition, HSF is essential for extended
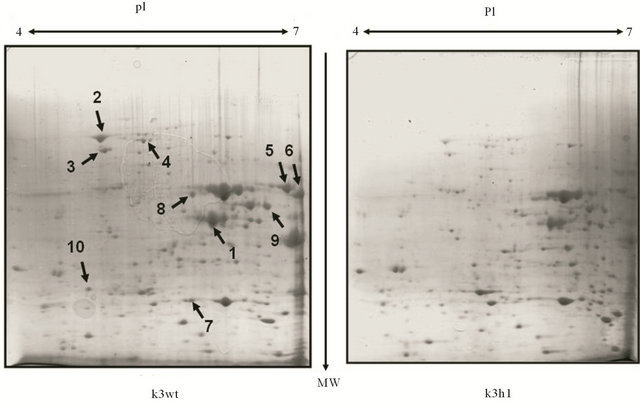
Figure 3. Comparative proteome analysis between k3wt cells and k3h1 cells after 0.4 mM MD treatment for 1 h. Protein (1 mg) was loaded on the IPG strip gel. After isoelectric focusing, equilibration, and SDS-PAGE (12%), the gels were stained with CBB R-250 and destained. Spots of interest were cut for MALDI-TOF MS analysis. Spots upregulated only in k3wt cells were arrowed and numbered. pI, isoelectric point; MW, molecular weight.
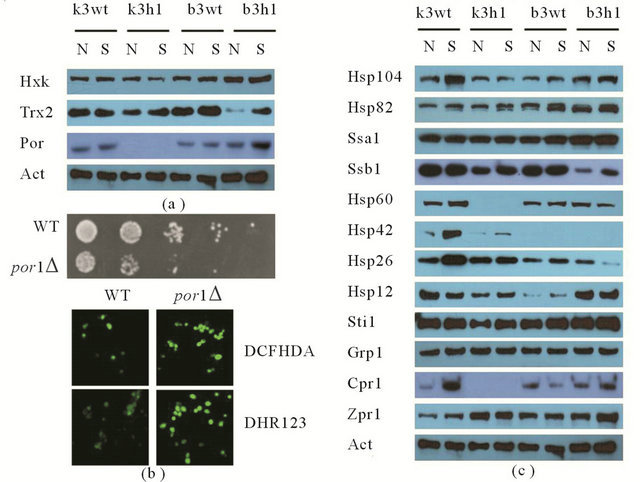
Figure 4. Expression regulation by HSF1 using western blot. (a) Expression changes of metabolic and antioxidant enzymes in the absence and presence of 0.4 mM MD; (b) Stress sensitivity (upper panel) and redox state (lower panel) of wild-type (WT) cells and por1∆ mutant (por1∆) cells of S. cerevisiae BY4741 under 0.2 mM MD stress; (c) Expression analysis of molecular chaperones and their cofactors under 0.4 mM MD stress. Actin (Act) protein was used as a loading control for western blot. N, MD-untreated cells; S, MDtreated cells.
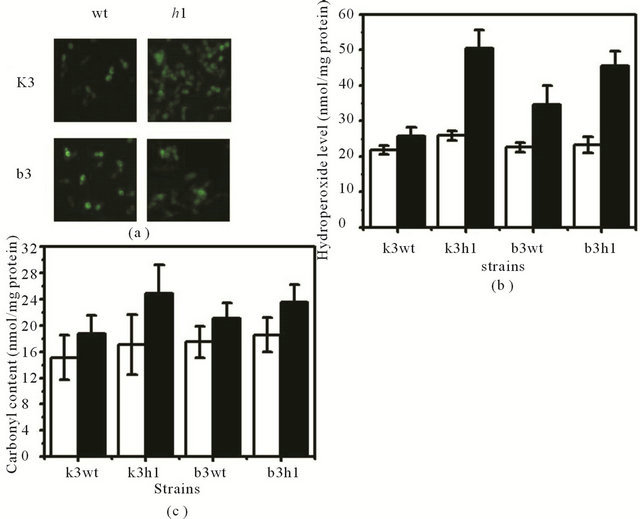
Figure 5. Measurement of cellular ROS level and carbonyl content under MD-induced oxidative stress. (a) Measurement of in vivo molecular oxidation. DCF fluorescence was measured in yeast cells exposed to MD for 1 h. Fluorescence images were obtained by fluorescence microscopy; (b) Cellular ROS level was measured by the spectrophotometric method using the FOX reagent; (c) Carbonyl contents were measured using the spectrophotometric assay. The levels are represented as nmol/mg protein. White bar, MD-untreated cells; black bar, MDtreated cells.
longevity in Caenorhabditis elegans [21], oogenesis, early larval development, survival of acute stress response in Drosophila melanogaster [22], and extraembryonic development and stress resistance in mice [17, 23]. Escherichia coli HSF (sigma32) provides stress tolerance to both highand low-temperatures [24]. HSF in plants is required for thermotolerance [25] and stress tolerance to high-light stress and heat-shock, and H2O2 [26] in Arabidopsis and soybeans [27]. Taken together, unlike HSF1 of S. cerevisiae BY4743, our results indicate that HSF1 of S. cerevisiae KNU5377Y3 is induced by MD stress and is a key factor of intrinsic tolerance under MD-induced oxidative stress.
Under MD stress, k3wt cells up-regulated metabolic enzymes, including hexokinase (Hxk), fructose-1,6-biphosphate aldolase (Fba1), enolase II (Eno2), 3-phosphoglycerate kinase (Pgk1), and alcohol dehydrogenase isoform 1 (Adh1), and antioxidant enzymes, including thioredoxin 2 (Trx2) and porin (Por) in the presence of MD, while the k3h1 cells down-regulated these proteins (Table 2, Figures 3 and 4(a)). However, the expression of most proteins analyzed in the b3h1 cells was higher, except Trx2, as compared to the b3wt cells. Trx2 expression in the k3wt cells was HSF1-dependent. Hxk1 and Trx2 are involved in the stress response to menadione [28,29]. Deletion or downregulation of these antioxidant agents lead to a number of oxygen-dependent phenotypes, including oxygen sensitivity, slow growth, hypersensitivity to superoxide generating agents such as MD or paraquat, which are believed to accelerate aging, and auxotrophy for methionine and lysine [30]. Furthermore, the mitochondrial function requires maintaining redox homeostasis, which is mediated primarily by the voltagedependent anion channel (VDAC; porin pore). The VDAC releases superoxide anions from the mitochondria to the cytosol [31], and its deletion increases stress sensitivity, following excess ROS accumulation in the cytosol and mitochondria (Figure 4(b)). In the k3h1 cells, the elevated ROS makes it difficult to modify neutralizing ROS or to repair oxidative damage (Figure 5), which is associated with sensitivity to MD-induced oxidative stress. Thus, our findings suggest that HSF1 in S. cerevisiae KNU5377Y3 cells regulates the expression of metabolic and antioxidant enzymes in response to MD stress.
The k3wt cells over-expressed molecular chaperones
Table 2. Identified proteins by MALDI-TOF MS analysis after 2-D PAGE.
and their cofactors, including the alpha subunit of the heteromeric nascent polypeptide-associated complex (Egd2), Hsp104, Hsp70 (Ssb1), Hsp60, Hsp42, Hsp26, Cpr1, and Sti1, when the cells were challenged to MD stress, whereas the k3h1 cells down-regulated these proteins under the same conditions. However, Hsp82, Grp, and Zpr1 expression in the k3h1 cells increased greatly under MD stress as compared to the k3wt cells. In contrast, these results, except Ssb1, were not observed in the b3wt and b3h1 cells.
Most proteins expressed HSF1-independently under MD stress (Figure 4(c)). It has been known that the bulk of Hsps and some glycolytic enzymes such as phosphoglycerate kinase and enolase are induced by HSF1 during temperature shift [32,33]. Recently, it has been reported that HSF1 in yeast is able to activate the transcription of target genes (CUP1, BTN2, SIS1, HSP10, SGT2, and SSA3) in response to superoxide anion-generating agents menadione and potassium superoxide (KO2) and the thiol-oxidizing compound diamide, as well as in response to heat shock [5]. In this study, we identified several new HSF1 target genes, including Egd2, Hsp104, Ssb1, Hsp60, Hsp42, Hsp26, Hsp12, Cpr1, and Sti1 under MD stress in S. cerevisiae KNU 5377Y3. Molecular chaperones are known to be involved in many cellular processes and pathways such as protein translocation across membranes, ribosomal RNA processing, and ER-associated protein degradation [34]. Chaperones that participate broadly in protein refolding, such as Hsp70 family (Ssa and Ssb), Hsp90, Hsp104, and sHsps (Hsp40, Hsp26, and Hsp12), promote the folding process through cycles of substrate binding and by cofactor proteins [35]. Hsp104 and Hsp70 act together with Hsp26 in protein refolding after stress-induced unfolding [36]. In addition, Hsp90 depends on its association with a variety of cochaperones and cofactors. The co-chaperones include Hsp70 and Hsp40 (Hsp42) [37], and cyclophilins Cpr6 and Cpr7 [38]. The formation of a complex between Hsp70 and Hsp90 is mediated through the association of both chaperones with an adaptor protein termed as an activator of the Ssa protein Sti1 [39] and an essential ER chaperone Grp (Grp94) that functions as an interaction domain of Hsp90 [10,40]. An imbalance of chaperone machinery systems in the k3h1 cells creates a difficulty in protein (re)folding under unstressed and stressed conditions when they are accompanied by elevated protein oxidation (Figure 5(c)). Protein carbonylation is an irreversible and irreparable modification. The hsf1 mutant yeast cells defective in transcriptional activation were sensitive to MD and diamide [5]. In mice, hsf1 knock-out alters cardiac redox homeostasis and increases oxidative damage via elevated superoxide anion production [1]. HSF-knockout cells attenuate cellular processes such as normal growth, activation of HSP genes in response to stress, embryonic development, inflammatory responses, fertility, and resistance to stress-induced apoptosis [1,20,32]. In addition, HSF-knockout in flies and worms affects normal growth and development, aging, and stress resistance [10,32]. To sum up our results, HSF1 in S. cerevisiae KNU5377Y3 cells regulates gene expression of various molecular chaperones which promote protein folding/ refolding under MD stress by minimizing MD-induced oxidative damage, leading to a better stress response.
In conclusion, our results suggest that the discovery of HSF-mediated target genes at the translational level could be an effective approach because protein expression in the k3h1 cells is down-regulated under normal conditions. The identification of novel HSF1 target genes during MD stress reveals a more diverse function for HSF1 than previously reported, suggesting that HSF1 affects the gene expression of various cell rescue systems, including molecular chaperones, antioxidant enzymes, and carbohydrate metabolism, playing an important role in oxidative stress and development by MD.
5. ACKNOWLEDGEMENTS
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ0080602012), Rural Development Administration, and funded by the Global Frontier Program (2011-0031341) of the Ministry of Education, Science and Technology (MEST), Republic of Korea.
REFERENCES
- Yan, L.J., Christians, E.S., Liu, L., Xiao, X., Sohal, R.S. and Benjamin, I.J. (2002) Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO Journal, 21, 5164-5172. doi:10.1093/emboj/cdf528
- Rodrigues-Pousada, C.A., Nevitt, T., Menezes, R., Azevedo, D., Pereira, J. and Amaral, C. (2004) Yeast activator proteins and stress response: An overview. FEBS Letters, 567, 80-85. doi:10.1016/j.febslet.2004.03.119
- Finkel, T. and Holbrook, N.J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature, 408, 239-247. doi:10.1038/35041687
- Lee, S., Carlson, T., Christian, N., Lea, K., Kedzie, J., Reilly, J.P. and Bonner, J.J. (2000) The yeast heat shock transcription factor changes conformation in response to superoxide and temperature. Molecular Biology of the Cell, 11, 1753-1764.
- Yamamoto, A., Ueda, J., Yamamoto, N., Hashikawa, N. and Sakurai, H. (2007) Role of heat shock transcription factor in Saccharomyces cerevisiae oxidative stress response. Eukaryotic Cells, 6, 1373-1379. doi:10.1128/EC.00098-07
- Wu, C. (1995) Heat shock transcription factors: Structure and regulation. Annual Review of Cell and Developmental Biology, 11, 441-469. doi:10.1146/annurev.cb.11.110195.002301
- Hahn, J.S., Neef, D.W. and Thiele, D.J. (2006) A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Molecular Microbiology, 60, 240-251. doi:10.1111/j.1365-2958.2006.05097.x
- Liu, X.D. and Thiele, D.J. (1996) Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes and Development, 10, 592-603. doi:10.1101/gad.10.5.592
- Takemori, Y., Sakaguchi, A., Matsuda, S., Mizukami, Y. and Sakurai, H. (2006) Stress-induced transcription of the endoplasmic reticulum oxidoreductin gene ERO1 in the yeast Saccharomyces cerevisiae. Molecular Genetics and Genomics, 275, 89-96. doi:10.1007/s00438-005-0065-9
- Kim, I.S., Jin, I. and Yoon, H.S. (2011) Decarbonylated cyclophilin A Cpr1 protein protects Saccharomyces cerevisiae KNU5377Y when exposed to stress induced by menadione. Cell Stress and Chaperones, 16, 1-14. doi:10.1007/s12192-010-0215-9
- Kim, I.S., Yun, H.S., Choi, H.J., Sohn, H.Y., Yu, C.B., Kim, J.K. and Jin, I.N. (2006) Construction of hsf1 knockout-mutant of a thermotolerant yeast strain Saccharomyces cerevisiae KNU5377. Korean Journal of Life Science, 16, 454-458.
- Kim, I.S., Moon, H.Y., Yun, H.S. and Jin, I. (2006) Heat shock causes oxidative stress and induces a variety of cell rescue proteins in Saccharomyces cerevisiae KNU5377. Journal of Microbiology, 44, 492-501.
- Kim. I., Yun, H. and Jin, I. (2007) Comparative proteomic analyses of the yeast Saccharomyces cerevisiae KNU5377 strain against menadione-induced oxidative stress. Journal of Microbiology and Biotechnology, 17, 207-217.
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. doi:10.1038/227680a0
- Ramagli, L. and Rodriguez, L. (1985) Quantitation of microgram amounts of protein in two-dimensional polyacrtlamide gel electrophoresis sample buffer. Electrophoresis, 6, 559-563. doi:10.1002/elps.1150061109
- Kim, I.S., Yun, H.S., Park, I.S., Sohn, H.Y., Iwahashi, H. and Jin, I.N. (2006) A knockout strain of CPR1 induced during fermentation of Saccharomyces cerevisiae KNU5377 is susceptible to various types of stress. Journal of Bioscience and Bioengineering, 102, 288-296. doi:10.1263/jbb.102.288
- Hahn, J.S. and Thiele, D.J. (2004) Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. The Journal of Biological Chemistry, 279, 5169-5176. doi:10.1074/jbc.M311005200
- Zhong, M., Orosz, A. and Wu, C. (1998) Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Molecualr Cell, 2, 101-108. doi:10.1016/S1097-2765(00)80118-5
- Ahn, S.G. and Thiele, D.J. (2003) Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes and Development, 17, 516-528. doi:10.1101/gad.1044503
- Christians, E.S., Yan, L.J. and Benjamin, I.J. (2002) Heat shock factor 1 and heat shock proteins: Critical partners in protection against acute cell injury. Critical Care Medicine, 30, S43-S50. doi:10.1097/00003246-200201001-00006
- Garigan, D., Hsu, A.L., Fraser, A.G., Kamath, R.S., Ahringer, J. and Kenyon, C. (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heatshock factor and bacterial proliferation. Genetics, 161, 1101-1112.
- Sorensen, J.G., Kristensen, T.N., Kristensen, K.V. and Loeschcke, V. (2007) Sex specific effects of heat induced hormesis in Hsf-deficient Drosophila melanogaster. Experimental Gerontology, 42, 1123-1129. doi:10.1016/j.exger.2007.09.001
- Xiao, X., Zuo, X., Davis, A.A., McMillan, D.R., Curry, B.B., Richardson, J.A. and Benjamin, I.J. (1999) HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO Journal, 18, 5943-5952. doi:10.1093/emboj/18.21.5943
- Kourennaia, O.V. and Dehaseth, P.L. (2007) Substitution of a highly conserved histidine in the Escherichia coli heat shock transcription factor, sigma32, affects promoter utilization in vitro and leads to overexpression of the biofilm-associated flu protein in vivo. Journal of Bacteriology, 189, 8430-8436. doi:10.1128/JB.01197-07
- Charng, Y.Y., Liu, H.C., Liu, N.Y., Chi, W.T., Wang, C.N., Chang, S.H. and Wang, T.T. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiology, 143, 251-262. doi:10.1104/pp.106.091322
- Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K. and Shigeoka, S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant Journal, 48, 535-547. doi:10.1111/j.1365-313X.2006.02889.x
- Zhu, B., Ye, C., Lu, H., Chen, X., Chai, G., Chen, J. and Wang, C. (2006) Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). Journal of Plant Research, 119, 247-256. doi:10.1007/s10265-006-0267-1
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D. and Brown, P.O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Molecular Biology of the Cell, 11, 4241-4257.
- Tucker, C.L. and Fields, S. (2004) Quantitative genomewide analysis of yeast deletion strain sensitivities to oxidative and chemical stress. Comparative and Functional Genomics, 5, 216-224. doi:10.1002/cfg.391
- Tan, S.X., Teo, M., Lam, Y.T., Dawes, I.W. and Perrone, G.G. (2009) Cu, Zn superoxide dismutase and NADP(H) homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Molecular Biology of the Cell, 20, 1493-1508. doi:10.1091/mbc.E08-07-0697
- Han, D., Antunes, F., Canali, R., Rettori, D. and Cadenas, E. (2003) Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. The Journal of Biological Chemistry, 278, 5557- 5563. doi:10.1074/jbc.M210269200
- Hahn, J.S., Hu, Z., Thiele, D.J. and Iyer, V.R. (2004) Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Molecular and Cellular Biology, 24, 5249-5256. doi:10.1128/MCB.24.12.5249-5256.2004
- Yamamoto, A., Mizukami, Y. and Sakurai, H. (2005) Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. The Journal of Biological Chemistry, 280, 11911-11919. doi:10.1074/jbc.M411256200
- Gong, Y., Kakihara, Y., Krogan, N., Greenblatt, J., Emili, A., Zhang, Z. and Houry, W.A. (2009) An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: Implications to protein folding pathways in the cell. Molecular Systems Biology, 5, 275. doi:10.1038/msb.2009.26
- Hartl, F.U. and Hayer-Hartl, M. (2002) Molecular chaperones in the cytosol: From nascent chain to folded protein. Science, 295, 1852-1858. doi:10.1126/science.1068408
- Haslbeck, M., Miess, A., Stromer, T., Walter, S. and Buchner, J. (2005) Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. The Journal of Biological Chemistry, 280, 23861-23868. doi:10.1074/jbc.M502697200
- Zhao, R., Davey, M., Hsu, Y.C., Kaplanek, P., Tong, A., Parsons, A.B., Krogan, N., Cagney, G., Mai, D., Greenblatt, J., Boone, C., Emili, A. and Houry W.A. (2005) Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell, 120, 715-727. doi:10.1016/j.cell.2004.12.024
- Mayr, C., Richter, K., Lilie, H. and Buchner, J. (2000) Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. The Journal of Biological Chemistry, 275, 34140-34146. doi:10.1074/jbc.M005251200
- Wegele, H., Haslbeck, M., Reinstein, J. and Buchner, J. (2003) Sti1 is a novel activator of the Ssa proteins. The Journal of Biological Chemistry, 278, 25970-25976. doi:10.1074/jbc.M301548200
- Chu, F., Maynard, J.C., Chiosis, G., Nicchitta, C.V. and Burlingame, A.L. (2006) Identification of novel quaternary domain interactions in the Hsp90 chaperone, GRP94. Protein Science, 15, 1260-1269. doi:10.1110/ps.052065106
NOTES
*Both corresponding authors contributed equally to this work.

