Journal of Environmental Protection
Vol. 3 No. 1 (2012) , Article ID: 16859 , 6 pages DOI:10.4236/jep.2012.31007
Enrichment and Isolation of CO2-Fixing Bacteria with Electrochemical Reducing Power as a Sole Energy Source
![]()
1Department of Biological Engineering, Seokyeong University, Seoul, South Korea; 2Department of Radiation Biology, Environmental Radiation Research Group, Korea Atomic Energy Research Institute, Daejeon, South Korea.
Email: *baakdoo@skuniv.ac.kr
Received October 3rd, 2011; revised November 8th, 2011; accepted December 18th, 2011
Keywords: Electrochemical Reducing Power; CO2; Autotroph; Alcaligenes sp.; Pyrosequencing
ABSTRACT
Enrichment of bacteria capable of growing with electrochemical reducing power and CO2 was accomplished using a plate-type electrochemical bioreactor (PEB). A bacterial source obtained from wastewater treatment reactant and forest soil was cultivated on carbonate-based mineral agar medium prepared in the PEB (PEB-carbonate agar). According to the pyrosequencing analyses, the abundance of Betaproteobacteria and Gammaproteobacteria at the phylum level, and Achromobacter, Alcaligenes, and Pseudomonas at the genus level were selectively increased after the electrochemical enrichment culture. Finally, one genus of bacterium that was autotrophically grown on the PEB-carbonate agar was identified as Alcaligenes. This bacterium may be useful to fix atmospheric CO2 with electrochemical energy obtained from the solar cell.
1. Introduction
Atmospheric carbon dioxide has been increased and was reached approximately to 392 mg/L by volume as of 2011 [1]. The balance between the CO2 generated by heterotrophs and that fixed by autotrophs is unlikely to be profoundly altered in natural ecosystem; however, the concentration of CO2 in the atmosphere has been increased continuously via fossil fuel combustion and increase of desertification. Very little of solar energy reached to Earth surface is converted to chemical energy (organic compounds) by phototrophs but most of that is disappeared from Earth as radiant heat. The solar light is most plentiful and clean energy that can be converted to electric energy without increase of atmospheric CO2 balance. Electric energy can be converted to biochemical reducing power through electrochemical redox reaction of neutral red (NR) [2,3]. The atmospheric CO2 generated from fossil fuel combustion may be fixed by the CO2-fixing bacteria capable of growing with the electro-chemical reducing power. The bacterial cells cultivated using the electrochemical reducing power and atmospheric carbon dioxide may be not useful for industrial, nutritional, and pharmacological purpose but may be effective for conversion of carbon dioxide to the chemically stable biopolymers without combustion of fossil fuel and occupation of ecological habitats.
Chemoautotrophic bacteria regenerate NAD(P)H in coupling with the oxidation of , SO, or H2 [4-9]. NAD(P)H functions as a driving force for the assimilation of CO2 into biomolecules in both autotrophic and heterotrophic (mixotrophic) organisms [10-13]. The regeneration of NADH can also be catalyzed in coupling with the oxidation of the electrochemically reduced NR without resorting to enzymatic catalysis [2,14]. The electrochemically reduced NR activated the ethanol production of Zymomonas mobilis, the denitrification metabolism of Ochrobactrum sp., and the metabolic ammonium oxidation of Nitrosomonas sp. [3,15,16]. All of these reactions may be activated in proportion to high NADH/NAD+ balance. The NADH/NAD+ balance in the bacterial cytoplasm can’t be measured biochemically, but may be estimated based on the types and production of metabolites. The electrochemically reduced NR catalyzed the regeneration of NADH in vitro test [17] and induced the increase of metabolites generated in coupling with NADH oxidation in the bioreactor [18]. The objective of this study is to isolate the bacteria capable of growing autotrophically with electrochemical reducing power and CO2, as well as to compare the variations in the bacterial community before and after the electrochemical enrichment culture. The NR was employed as an electron mediator to induce transfer of electrochemical reducing power from electrode to bacterial cells. Pyrosequencing technique was employed to monitor the enriched bacteria and to minutely analyze the bacterial community variations before and after the electrochemical enrichment culture.
, SO, or H2 [4-9]. NAD(P)H functions as a driving force for the assimilation of CO2 into biomolecules in both autotrophic and heterotrophic (mixotrophic) organisms [10-13]. The regeneration of NADH can also be catalyzed in coupling with the oxidation of the electrochemically reduced NR without resorting to enzymatic catalysis [2,14]. The electrochemically reduced NR activated the ethanol production of Zymomonas mobilis, the denitrification metabolism of Ochrobactrum sp., and the metabolic ammonium oxidation of Nitrosomonas sp. [3,15,16]. All of these reactions may be activated in proportion to high NADH/NAD+ balance. The NADH/NAD+ balance in the bacterial cytoplasm can’t be measured biochemically, but may be estimated based on the types and production of metabolites. The electrochemically reduced NR catalyzed the regeneration of NADH in vitro test [17] and induced the increase of metabolites generated in coupling with NADH oxidation in the bioreactor [18]. The objective of this study is to isolate the bacteria capable of growing autotrophically with electrochemical reducing power and CO2, as well as to compare the variations in the bacterial community before and after the electrochemical enrichment culture. The NR was employed as an electron mediator to induce transfer of electrochemical reducing power from electrode to bacterial cells. Pyrosequencing technique was employed to monitor the enriched bacteria and to minutely analyze the bacterial community variations before and after the electrochemical enrichment culture.
2. Materials and Methods
2.1. Plate Type Electrochemical Bioreactor
The PEB was prepared to charge electrochemical reduceing power to agar medium by modification of glass petri dish (diameter 120 mm, height 12 mm), as is shown in Figure 1. A sintered glass filter (pore 1 m - 1.6 m, diameter 30 mm, thickness 2 mm, Duran) modified with cellulose acetate film (thickness 35 m, Electron Microscopy Science) was placed between anode and cathode compartment to limit water transfer while permitting proton transfer. DC-2 V of electricity was charged to the titanium plate cathode (0.89 mm thickness, 99.7% purity, VWR) equipped in the PEB bottom to induce the electrochemical reduction reaction of neutral red (NR) contained in a carbonate-based mineral agar medium (carbonate agar), which was composed of 4.2 g/L of NaHCO3, 2 g/L of NH4Cl, 1 g/L of K2HPO4, 1 g/L of KH2PO4, 0.00058 g/L (200 M) of NR, 20 g/L of agar, and 2ml/L of trace mineral stock solution that was prepared by previously described method [15].
2.2. Source of Bacterial Community
50 g of wet humus soil that was obtained from Bukhan mountain national park (Seoul, Korea) was suspended in 50 ml of aerobic wastewater treatment reactant that was obtained from a terminal disposal plant of sewage (Seoul, Korea), and filtered with Whatman No. 1 filter paper. Precipitant obtained from the filtrate by centrifugation for 30 min at 4˚C and 5000 ×g (relative centrifugal force) was suspended in 5 ml of phosphate-buffered saline, which was employed as a bacterial source for the electrochemical enrichment culture. 300 mL of the bacterial source was inoculated into 5 of PEB-carbonate agars by spreading method and then cultivated for 10 days in a CO2 incubator (Figure 1) at 25˚C. 99% of CO2 was initially charged into the CO2-incubator (20 L) and CO2-reservoir (10 L) and periodically exchanged with fresh CO2 at 24 h intervals.
2.3. Identification of Enriched Bacterium
30 of the bacterial colonies grown on the PEB-carbonate agars after enrichment culture were randomly picked and transferred to fresh PEB-carbonate agars, then cultivated under a 99% CO2 atmosphere for 10 days. The chromosomal DNA was directly extracted from the bac-
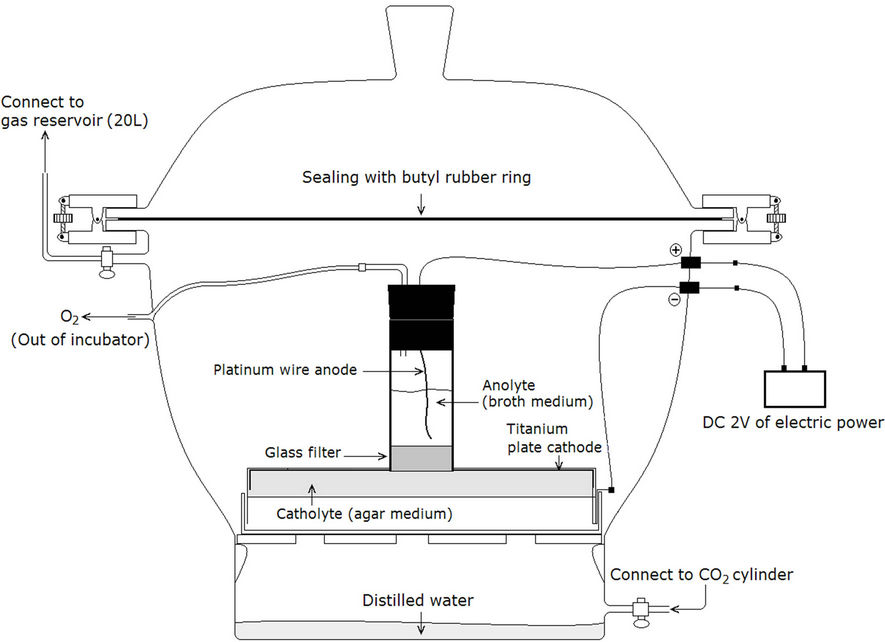
Figure 1. Schematic structure of a plate-type electrochemical bioreactor (inside part) for the electrochemical culture of bacteria and a CO2 incubator for maintenance of CO2 atmosphere around the plate-type electrochemical bioreactor.
teria grown autotrophically on the fresh PEB-carbonate agar. 16S-rDNA was amplified via the PCR technique that was previously employed [19]. The amplified 16S-r DNA was sequenced by a professional sequencing system (Macrogen, Seoul, Korea). The species-specific identity of the amplified 16S-rDNA was determined using the GenBank database system.
2.4. Analysis of Bacterial Community
All of the bacterial colonies grown on 5 of the PEBcarbonate agars were collected using autoclaved phosphate-buffered saline. Bacterial pellets were obtained from the bacterial source before the enrichment culture and bacterial suspension collected from the PEB-carbonate agar after the enrichment culture by centrifugation for 30 min at 5000 ×g and 4˚C. DNAs were extracted from the pellets using a DNA isolation kit (Ultra Clean Water DNA Isolation Kit, Mo Bio Laboratories, USA), which were used as the template for pyrosequencing. The pyrosequencing was conducted using a 454 Life Sciences GS FLX series genome sequencer (Roche, Manheim, Germany) at the Macrogen Bioinformatics Research Institute (http://macrogen.co.kr/eng, Seoul, Korea) via a turnkey-based project. A GS FLX fusion primer designed for the 454 pyrosequencing was composed of primer A (5’-CGTATCGCCTCCCTCGCGCCA-3’), primer B (5’-CTATGCGCCTTGCCAGCCCGCTC-AG-3’), linker (5’-TCAG-3’), bar codes (5’-ATCAGACACG-3’; 5’-ACG CTCGACA-3’), and template-specific sequence (27F and 518R). The bacterium-specific forward primer 27F (5’-primer A-linker-barcode- GAGTTTGATCMTGGCTCAG-3’) and reverse primer 518R (5’-primer B-linker-bar-code- WTTACCGCGGCTGCTGG-3’) were employed to amplify the variable region of the 16S-rDNA. Only those sequences that were >300 bp in length with a quality score of >20 with no ambiguous characteristics were included in the analysis [20]. The classifiable sequences obtained by the pyrosequencing were identified using the Ribosomal Database Project.
3. Results and Discussion
Only 2 of the 30 bacterial colonies that were transferred to the fresh PEB-carbonate agar grew autotrophically with the electrochemical reducing power and CO2. Both of them were identified as genus Alcaligenes based on 16S-rDNA sequence homology. The isolates were registered in the GenBank database system, from which the accession numbers (HQ738483 and HQ738484) were obtained. The bacteria grown with the electrochemical reducing power and CO2 on the fresh PEB-carbonate agar must be an autotroph because the fresh PEB-carbonate agar is impossible to be contaminated with the organic compounds originated from the bacterial source. Some of the genus Alcaligenes was reported to autotrophically grow [4,6,21,22] and metabolically oxidize H2 to regenerate the biochemical reducing power during autotrophic growth under an H2-CO2 atmosphere [23]. Genus Alcaligenes grown on the fresh PEB-carbonate agar is supposed to grow autotrophically with the electrochemical reducing power instead of H2 under CO2 atmosphere.
The community diversity and richness after the enrichment culture were reduced greatly in comparison with those before the enrichment culture based on the number of pyrotags, operational taxonomic units (OTUs), and the results of Chao’s study [24,25], as is shown in Table 1. The 16S rRNA pyrotags obtained by the pyrosequencing were a little lower before than after the enrichment culture; however, the OTUs’ diversity and Chao’s richness were decreased greatly after the enrichment culture. Based on the OTUs and Chao’s results, most of bacteria originated from the humus soil and the wastewater treatment reactant may be died out but some of bacteria are presumed to grow or survive during enrichment culture with the electrochemical reducing power under CO2 atmosphere.
Practically, the number of bacterial phyla analyzed at a sequence similarity level above 95% was 19 before the enrichment culture but was decreased to be 2 after the enrichment culture. Betaproteobacteria and Gammaproteobacteria were enriched selectively but other bacterial phyla were not detected after the enrichment culture, as is shown in Table 2. However, the bacteria with physiological functions related to the metabolism for CO2-fixing can’t be verified based on the phyla analysis.
Evaluation at the genus level revealed that members of the bacterial genera included in the bacterial source were greatly simplified after the enrichment culture, as is shown in Table 3. Fifty four of bacterial genera before the enrichment culture but 4 of them after the enrichment culture were identified with 16S rDNA pyrotags analyzed at a sequence similarity level above 99%. The genus Bacillus (38.35%) was the highest predominant bacterial group before the enrichment culture but was not detected after the enrichment culture. The abundances of genera Achromobacter, Alcaligenes, and Pseudomonas were profoundly increased after the enrichment culture; however, the abundance of genus Stenotrophomonas was reduced after the enrichment culture. The genus Achromobacter, a facultative chemoautotroph [26-28], is expected to grow mixotrophically with organic compounds,

Table 1. Bacterial diversity analyzed at 3% of genetic distance level and 95% sequence similarity level before and after enrichment culture.

Table 2. Phyla diversity of the bacterial community analyzed at higher than 95% sequence similarity level before and after enrichment culture.
electrochemical reducing power, and CO2 based on the results that it was predominant on the PEB-carbonate agar for the enrichment culture but was not grown after transferred to the fresh PEB-carbonate agar. The genera Pseudomonas and Stenotrophomonas, which are typical heterotrophs, are assumed to grow limitedly or maintain themselves on the PEB-carbonate agar with the organic compounds originated from the bacterial source.
Variations of diversity analyzed at the phylum and genus level, along with Chao’s richness estimates (Table 1), indicated that a substantial fraction of the heterotrophic bacteria included in the bacterial source is possible to be died and consumed as nutrients for the survived heterotrophic (mixotrophic) bacteria during enriched; however, the bacteria capable of assimilating CO2 with the electrochemical reducing power may be enriched predominantly and some heterotrophic bacteria may survive by exploiting the low concentration of organic compounds originated from the bacterial source composed of wastewater, humus soil, and microbial biomass.
4. Conclusion
Genus Alcaligenes was not grown autotrophically without chemical electron donor that is hydrogen but grew with the electrochemical reducing power under CO2 atmosphere. The plate type electrochemical bioreactor designed to enrich and isolate the autotrophic bacteria with electrochemical reducing power and CO2 was first devel-
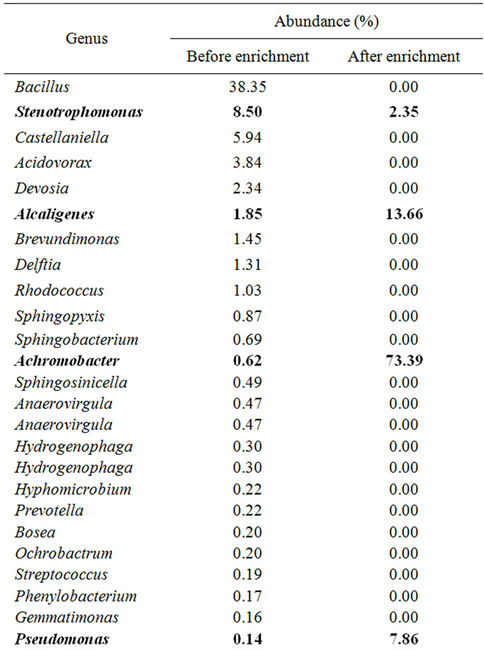
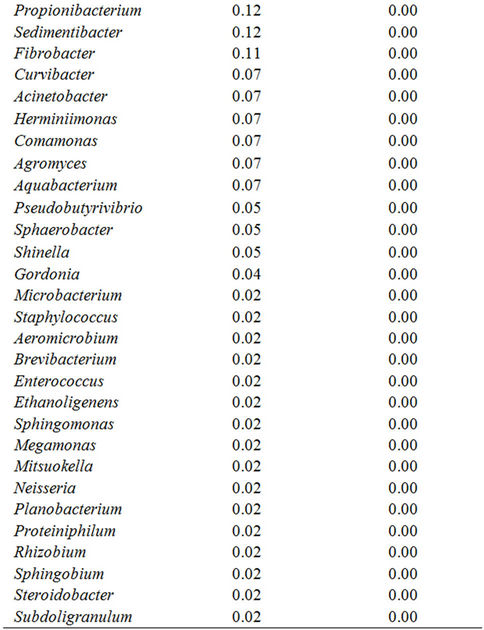
Table 3. Genera diversity of the bacterial community analyzed at higher than 99% sequence similarity level before and after enrichment culture.
oped in this preliminary research. Thus, we suggest that bacteria grown with the electrochemical reducing power as a sole energy source and CO2 as a sole carbon source may be classified as “electroautotroph”. The electroautotroph may be useful to fix atmospheric CO2 with the electrochemical reducing power but without combustion of fossil fuel. Practically, liquid medium may be substituted for agar plate medium and a cylinder type electrochemical bioreactor [29] may be substituted for the plate type electrochemical bioreactor to apply the electroautotroph to a system for bioelectrochemical fixation of atmospheric CO2.
5. Acknowledgements
This work was supported by the New & Renewable Energy of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) grant funded by the Korea governmental Ministry of Knowledge Economy (2010T1001100334).
REFERENCES
- P. Tans, “Trends in Atmospheric Carbon Dioxide,” 2011. http://en.wikipedia.org/wiki/Carbon_dioxide_in_Earth’s_atmosphere
- D. H. Park and J. G. Zeikus, “Utilization of Electrically Reduced Neutral Red by Actinobacillus succinogenes: Physiological Function of Neutral Red in MembraneDriven Fumarate Reduction and Energy Conservation,” Journal of Bacteriology, Vol. 181, No. 7, 1999, pp. 2403- 2410.
- B. Y. Jeon, S. Y. Kim, Y. K. Park and D. H. Park, “Enrichment of Hydrogenotrophic Methanogens in Coupling with Methane Production Using Electrochemical Bioreactor,” Journal of Microbiology and Biotechnology, Vol. 19, No. 12, 2009, pp. 1665-1671.
- C. M. Doyle and D. J. Arp, “Regulation of H2 Oxidizing Activity and Hydrogenase Protein Levels by H2, O2, and Carbon Substrates in Alcaligenes latus,” Journal of Bacteriology, Vol. 169, No. 10, 1987, pp. 4463-4468.
- U. Jahn, H. Huber, W. Eisenreich, M. Hügler and G. Fuchs, “Insight into the Autotrophic CO2 Fixation Pathway of the Archaeon Ignicoccus hospitalis: Comprehensive Analysis of the Central Carbon Metabolism,” Journal of Bacteriology, Vol. 189, No. 11, 2007, pp. 4108- 4119. doi:10.1128/JB.00047-07
- L. Leadbeater and B. Bowien, “Control of Autotrophic Carbon Assimilation in Alcaligenes eutrophus by Inactivation and Reactivation of Phosphoribulokinase,” Journal of Bacteriology, Vol. 157, No. 1, 1984, pp. 95-99.
- N. Ohmura, K. Sasaki, N. Matsumoto and H. Saiki, “Anaerobic Respiration Using Fe, S, and H2 in the Chemoautotrophic Bacterium Acidithiobacillus ferroxidans,” Journal of Bacteriology, Vol. 184, No. 8, 2002, pp. 2081- 2087. doi:10.1128/JB.184.8.2081-2087.2002
- W. H. Ramos-Vera, I. A. Berg and G. Fuchs, “Autotrophic Carbon Dioxide Assimilation in Thermoproteales Revisited,” Journal of Bacteriology, Vol. 191, No. 13, 2009, pp. 4286-4297. doi:10.1128/JB.00145-09
- S. Schouten, M. Strous, M. M. M. Kuypers, W. I. C. Rijpstra, M. Bass, C. J. Schubert, M. S. M. Jetten and J. S. S. Damsté, “Stable carbon Isotopic Fractionations Associated with Inorganic Carbon Fixation by Anaerobic Ammonium-Oxidizing Bacteria,” Applied and Environmental Microbiology, Vol. 70, No. 6, 2004, pp. 3785- 3788. doi:10.1128/AEM.70.6.3785-3788.2004
- R. J. Cogdell, N. W. Isaacs, T. D. Howard, K. McLuskey, N. J. Fraser and S. M. Prince, “How Photosynthetic Bacteria Harvest Solar Energy (Mini Review),” Journal of Bacteriology, Vol. 181, No. 13, 1999, pp. 3869-3879.
- F. J. Small and S. A. Ensign, “Carbon Dioxide Fixation in the Metabolism of Propylene and Propylene Oxide by Xanthobacter Strain Py2,” Journal of Bacteriology, Vol. 177, No. 21, 1995, pp. 6170-6175.
- F. R. Tabita, “Molecular and Cellular Regulation of Autotrophic Carbon Dioxide Fixation in Microorganisms,” Microbiological Reviews. Vol. 52, No. 2, 1988, pp. 155- 189.
- R. K. Thauer, K. Jungermann and K. Decker, “Energy Conservation in Chemotrophic Anaerobic Bacteria,” Bacteriological Reviews. Vol. 41, No. 1, 1977, pp. 100-180.
- D. H. Park, M. Laivenieks, M. V. Guettler, M. K. Jain and J. G. Zeikus, “Microbial Utilization of Electrically Reduced Neutral Red as the Sole Electron Donor for Growth and Metabolite Production,” Applied and Environmental Microbiology, Vol. 65, No.7, 1999, pp. 2912- 2917.
- B. Y. Jeon, H. N. Seo, S. W. Kang and D. H. Park, “Effect of Electrochemical Redox Reaction on Biochemical Ammonium Oxidation and Chemical Nitrite Oxidation,” Journal of Microbiology and Biotechnology, Vol. 20, No. 3, 2010, pp. 485-493.
- W. J. Lee and D. H. Park, “Electrochemical Activation of Nitrate Reduction to Nitrogen by Ochrobactrum sp. G3-1 Using a Noncompartmented Electrochemical Bioreactor,” Journal of Microbiology and Biotechnology, Vol. 19, No. 8, 2009, pp. 836-844.
- H. S. Kang, B. K. Na and D. H. Park, “Oxidation of Butane to Butanol Coupled to Electrochemical Redox Reaction of NAD+/NADH,” Biotechnology Letters, Vol. 29, No. 8, 2007, pp. 1277-1280. doi:10.1007/s10529-007-9385-7
- B. Y. Jeon and D. H. Park, “Improvement of Ethanol Production by Electrochemical Redox Combination of Zymomonas mobilis and Saccharomyces cerevisieae,” Journal of Microbiology and Biotechnology, Vol. 20, No. 1, 2010, pp. 94-100.
- B. Y. Jeon, T. S. Hwang and D. H. Park, “Electrochemical and Biochemical Analysis of Ethanol Fermentation of Zymomonas mobilis KCCM11336,” Journal of Microbiology and Biotechnology, Vol. 19, No. 7, 2009, pp. 666- 674.
- S. M. Huse, J. A. Huber, H. G. Morrison, M. L. Sogin and M. D. Welch, “Accuracy and Quality of Massively Parallel DNA Pyrosequencing,” Genome Biology, Vol. 8, No. 7, 2007, p. R143. doi:10.1186/gb-2007-8-7-r143
- A. Freter and B. Bowien, “Identification of a Novel Gene, aut, Involved in Autotrophic Growth of Alcaligenes eutrophus,” Journal of Bacteriology, Vol. 176, No. 17, 1994, pp. 5401-5408.
- I. Reutz, P. Schobert and B. Bowien, “Effect of Phosphor-Glycerate Deficiency on Heterotrophic and Autotrophic Carbon Metabolism of Alcaligenes eutrophus,” Journal of Bacteriology, Vol. 151, No. 1, 1982, pp. 8-14.
- C. Hogrefe, D. Römermann and B. Friedrich, “Alcaligenes eutrophus Hydrogenase Gene (Hox),” Journal of Bacteriology, Vol. 158, No. 1, 1984, pp. 43-48.
- P. Y. Hong, C. Hwang, F. Ling, G. L. Andersen, M. W. LeChevallier and W. T. Liu, “Pyrosequencing Analysis of Bacterial Biofilm Communities in Water Meters of a Drinking Water Distribution System,” Applied and Environmental Microbiology, Vol. 76, No. 16, 2010, pp. 5631- 5635. doi:10.1128/AEM.00281-10
- C. Simon, A. Wiezer, A. W. Strittmatter and R. Daniel, “Phylogenetic Diversity and Metabolic Potential Revealed in a Glacier Ice Metagenome,” Applied and Environmental Microbiology, Vol. 75, No. 23, 2009, pp. 7519-7526. doi:10.1128/AEM.00946-09
- I. R. Hamilton, R. J. Burris, P. W. Wilson and C. H. Wang, “Pyruvate Metabolism, Carbon Dioxide Assimilation, and Nitrogen Fixation by an Achromobacter Specie,” Journal of Bacteriology, Vol. 89, No. 3, 1965, pp. 647-653.
- O. Meyer, K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck and M. Karaut, “Utilization of Carbon Monoxide by Aerobes: Recent Advances,” FEMS Microbiology Reviews, Vol. 87, No. 3-4, 1990, pp. 253-260. doi:10.1111/j.1574-6968.1990.tb04921.x
- A. K. Romanova, A. V. Nozhevnikova, J. G. Leonthev and S. A. Alekseeva, “Pathways of Assimilation of Carbon Oxides in Carboxydobacteria Seliberia carboxydohydrogena and Achromobacter carboxydus,” Microbiology, Vol. 46, No. 5, 1977, pp. 885-889.
- B. Y. Jeon, I. L. Jung and D. H. Park, “Enrichment of CO2-Fixing Bacteria in Cylinder-Type Electrochemical Bioreactor with Built-In Anode Compartment,” Journal of Microbiology and Biotechnology, Vol. 21, No. 6, 2011, pp. 590-598.
NOTES
*Corresponding author.

