American Journal of Plant Sciences
Vol.4 No.2A(2013), Article ID:28463,10 pages DOI:10.4236/ajps.2013.42A054
Functional Analysis of a Wilt Fungus Inducible PR10-1 Gene from Cotton
![]()
1Plant Biotechnology Center, Fudan-SJTU-Nottingham Plant Biotechnology R&D Center, College of Agriculture and Life Science, Shanghai Jiao Tong University, Shanghai, China; 2Plant Genomics Research Center, Mianyang Normal University, Mianyang, China.
Email: *kjzuo@sjtu.edu.cn
Received December 13th, 2012; revised January 15th, 2013; accepted January 22nd, 2013
Keywords: Pathogenesis-Related Protein; Gossypium barbadense; Ribonuclease Activity; Incompatible Response
ABSTRACT
Early stage expression of PR10 combined with phytoalexins contributed to Verticillium wilt resistance in cotton. In order to analysis the activities of PR10 proteins during pathogens’ infection, we cloned a Verticillium-induced PR10 (GbPR10-1) gene from cotton (Gossypium barbadense) and compared its expression patterns and domains with other PR10 proteins. Bioinformatics indicated that GbPR10-1 showed the lowest similarity with other 12 different PR10 genes in cotton (Upland and sea-island cotton). Expression profiles showed that GbPR10-1 gene instantly up-regulated after infection by V. dahliae in the sea-island cotton plants. GbPR10-1 was also induced by environmental stimulus including heat, submergence and salt, and ethylene but not by ABA and salicylic acid. The GbPR10-1 protein expressed in E. coli BL21 demonstrated that it had a low ribonuclease-like activity in vitro, and could inhibit V. dahliae hyphae growth but not its spores. Comparison analysis of GbPR10-1 (from resistant species) and GhPR10-1 (from susceptible species) responding to V. dahliae infection, only GbPR10-1 gene was strongly induced in the sea-island cotton plants (incompatible response), indicating that PR10-1 genes was linked to resistance signal. In summary, the earlier activation of GbPR10-1 gene, as the index of resistance response, would be aid to block the V. dahliae attack in cotton.
1. Introduction
Verticillium dahliae is a kind of destructive soil-borne fungus, which brings about severe loss in cotton, tomato, potato and other crops [1,2]. When V. dahliae pathogens penetrated directly into cotton epidermal cells, parenchyma cells of the vascular tissues of resistant variety were reinforced by the callose, cellulose and polysaccharides [1-5]. The high concentration of terpenoids and phenolic substances were accumulated in these cells, which then coated pathogens within the vessels to stop fungal ingression [6-10]. Recent documents also showed that these phytoalexins were probably strengthened by early stage expression of PR10 in the non-host reaction [7,11,12]. The RNase activity of PR10 protein was deduced to degrade pathogens RNA when fungus entered into cells, and inhibited hyphae growth within intercellular spaces following stomatal entry in sorghum [13,14].
PRs were one of major products which represented the change of those genes’ regulations occurred in the hypersensitive reaction [15,16]. PR10 is a small family protein, which a few PR10 members were proven to be differentially expressed upon infection and related to hypersensitive reaction [17-20]. In our previous study, we found that only one PR10 transcript was quickly induced upon pathogens’ infection [21]. In cotton there are more than 8 different PR10 members [22,23]. So the key problem is to know which PR10 member in cotton was activated to express more quickly under fungal attack. In order to investigate the roles of PR10 family during the defense process, we cloned the PR10-1 gene from G. barbadense and analyzed its expression patterns under different stresses. We reported the sequence, and characterized the function of GbPR10-1 gene expressed in E. coli. We also discussed their roles of PR10 proteins through comparing their homologues’ expression in different Gossypium species (resistant and susceptible) infected with V. dahliae.
2. Materials and Methods
2.1. Plant Materials and Different Stresses Treatments
Gossypium barbadense L. variety 7124 (resistant to Verticillium dahliae Kleb.) and G. hirsutum L. variety Ejing-1 (susceptible) were used in this study. Their seeds were first sterilized with 75% ethanol, and then rinsed with distilled water. After sterilization, the seeds were put in the pasteurized sands for germination and growth in the 15 cm × 10 cm × 10 cm plastic pots. The seedlings had grown at 25˚C - 28˚C and watered with Hoagland solution for 10 - 15 days. When plants had 2 - 3 leaves, the seedlings were subjected into various stresses.
A kind of defoliating V. dahliae isolate from infected cotton plants was used as the pathogen in this experiment. A single spore from the potato-dextrose agar (PDA) plate was inoculated into the Czapek Broth (NaNO3, 0.3% w/v; MgSO4, 0.1% w/v; KH2PO4, 0.1% w/v; FeSO4, 0.0002% w/v; KCl, 0.1% w/v; Sucrose, 3% w/v; pH 6.0) to incubate for 5 - 8 days until the concentration of spores reached about 1.0 × 107 spores/ml. The suspension liquid was adjusted to 1 × 105 spores/ml with sterile distilled water to be used in inoculation experiment afterwards referring to the report [2]. The seedlings were infected with V. dahliae by root-dip inoculation into the suspension solution of fungal conidia for 5 min, and returned to their original pots for interval harvesting infected roots.
For the salt treatment, the plants were stressed by the Hoagland solution with addition NaCl of 100 mM. High temperature stress was imposed by transferring plants into 45˚C artificial atmospheric bank. For aerobic stresses, the whole plants were submerged into Hogaland solution. Plant growth regulator ABA was first dissolved into 50% ethanol and then diluted into the solution to a final concentration of 10 μM to spray cotton leaves and to submerge their roots. For ethylene treatment, plants were placed in a 10-litre jar with 200 mg/L 40% Ethephon spray.
The roots of the plants were harvested in 0, 1, 3, 7, 12, 24, 48, 72 hours after different stress treatments. All the roots were used to extract total RNA immediately.
2.2. Total RNA and DNA Extraction
Total RNA was isolated from above roots following the method described by the brochure of Trizol kit (GIBCOBRL, CA, USA). PolyA+ RNA was purified by oligo- (dT)n cellulose affinity chromatogy (Invitrogen, CA, USA). Double stranded cDNAs were then prepared from polyA+ RNA according to the manufacturers’ recommendations (Takara, Liaolin, China). Genomic DNA was isolated with CTAB method according to the method of Sambrook et al. [24].
2.3. Southern Blotting and RT-PCR Analysis
G. babardense genomic DNA (10 μg) was digested with EcoRI, BamHI and PstI endonucleases. Fragments were separated with 0.8% agarose gel and blotted onto HybondN+ membrane (Amersham, England, UK). The probe was prepared with a segment of GbPR10-1 coding segment (92 - 570 nt). The Gene Images random primer labeling module and Gene Images CDP-Star detection module were used for probe labeling, hybridization, and detection procedures (GIBCO-BRL, CA, USA).
Total RNA (1 μg) after digesting genomic DNA with DNaseI was added into a tube containing first-strand cDNA synthesis and PCR reaction buffer mixture according to the brochure of Takara RT-PCR kit (Takara, Dalian, China). The primers used in the RT-PCR were PR10-1(5’-C103GA GTT ATG AGT TTG AGG TAA-3’) and PR10-2 (5’-G239AC TAG CAT CAC CTT CGA GCT-3’), which could amplify 140 bp fragment of different PR10-1 genes in both sea-island and upland cotton varieties. After the first cDNA strand was synthsized, the PCR reactions went on quickly without pulling out tubes or adding any components. The PCR reaction program was 94˚C, 30 seconds; 58˚C, 30 seconds; 72˚C, 30 seconds; 25 cycles. PCR products (10 μl) were then electrophorsized in 1.2% agarose gel with 10 μg/ml ethidium bromide. The quantity of products was analyzed with gene analysis software package in the White/Ultraviolet Trans-illuminator (UVP®, CA, USA). The results were then used for determining the expression levels of PR10 gene in different intervals.
The template is used to format your paper and style the text. All margins, column widths, line spaces, and text fonts are prescribed; please do not alter them. You may note peculiarities. For example, the head margin in this template measures proportionately more than is customary. This measurement and others are deliberate, using specifications that anticipate your paper as one part of the entire proceedings, and not as an independent document. Please do not revise any of the current designations.
2.4. Bioinformatics Analysis of PR10 Protein Family in Cotton
The plasmid DNA of the positive clones were isolated and sequenced by Sangon Company in China. Sequence similarity was analyzed with BLAST based on GenBank (http://www.ncbi.nlm.nih.gov). The amino acid sequence encoded by GbPR10-1 and its homological proteins were aligned using the ClustalX programs.
PR10 family proteins were screened using tblastX on GenBank (http://www.ncbi.nlm.nih.gov). When the similarity between two sequences is more than 99.5%, these two sequences were considered as the same sequence. All of nucleotide sequences (EST and CDS from G. babardense and G. hirsutum) which probably encode PR10 proteins were then assigned into PR10-1, PR10-2 etc. These PR10 protein sequence were then used in bioinformatics analysis afterwards. Phylogenetic tree was constructed by using the neighbor-joining method [25,26].
2.5. Expression, Purification of GbPR10-1 Protein and Its Ribonuclease Activity Analysis
The ORF of GbPR10-1 gene was amplified from pMD18- T vector (Takara, Japan) plasmid which contained the full-length cDNA of GbPR10-1 gene. The PCR products digested by NotI/SalI were sub-cloned into the plasmid pET32-a. The plasmid of a positive clone was then transformed into the E. coli strain BL21 cell to get the recombinant expression vector. Isopropyl-1-thio-b-D-gactopyranoside (IPTG, 4 mM in the end) was added to the LB solution with Ampecillin (50 mg/L) when OD600 of the cultures reached 0.6. The cells were further cultured for 3 h and harvested by centrifugation at 4000 rpm 4˚C for 10 min.
The pellets were resolved in 50 mM Tris-HCl (pH 7.5) buffer containing 0.5 M NaCl, and 1 mM PMSF and lysised by sonication. Following centrifugation at 15,000 rpm 4˚C for 30 min, the supernatant was loaded onto SDS-PAGE gel to identify whether GbPR10-1 protein expressed highly or not in vitro. The supernatant of highly expressed clone was then loaded into a His-tag Sepharose-Fast-Flow gel column (Amersham-Pharmacia Biotech, USA) and washed with a gradient of 100 mM to 1M solution to collect recombinant protein.
RNase activity of the purified negative and recombinant GbPR10-1 protein was carried out at 37˚C. The reaction mixture contained 6 μg cotton total RNA and 4 μg proteins in 10 mM imidazole, 5 mM NaCl, and 10 mM Tris-HCl (pH 7.5) buffer. After 15 and 30 minutes incubation, the proteins were removed from the reaction mixtures by extraction with phenol-chloroform. The control mixture described above without GbPR10-1 protein was also incubated for 15 and 30 minutes at room temperature. The experimental results were observed on 1.2% agarose gel to determine the enzyme activity of protein.
3. Results
3.1. GbPR10-1 Gene Cloning and Block Analysis
Our previous report showed that an EST (Gb125, accession number CB066575) highly similar to other PR10 gene was strongly induced after V. dahliae inoculation [21]. This EST was than used as the probe to screen a sea-island variety 7124 root cDNA library. Seven clones were isolated in which two clones were full-length cDNA after two-round screening. The cDNA clone (Figure 1) contained an insert of 745 nt (nucleotide) with a complete coding frame of 477 bp from 91 nt to 568 nt. The start codon ATG site and a stop codon TAA site were recognized after comparison with other PR10 genes and confirmed by other homologous sequences in GenBank. Within 3’-untranslated region one polyadenylation

Figure 1. The full-length cDNA sequence and deduced amino acid sequences of sea-island cotton GbPR10-1 protein. Nucleotide positions are given on the left side of the sequence in the 5’ to 3’ orientation. The start codon ATG was underlined and the stop codon TAG was underlined italically. The deduced amino acid sequence is shown beneath the nucleotide sequence and the amino acids are numbered on the right side of the sequence. The motif GDASPGSIVK was underlined boldly and italically. The polyadenylation signal AATAAA is double-underlined. The cDNA clone has been deposited to GenBank (Acc. No. AY241395).
signal AATAAA was found at 68 bp upstream from the poly-(A)n tail. Blast search showed that this gene was 98%, 86%, and 76% identical to GaPR10 (Accession NO. AF416652.1), GhPR10 (Accession NO. AF305065.1) and Betv1-sc3 (Accession NO. AF2122374C) at nucleotide level respectively, in which Betv1-sc3 was a typical PR10 protein. According to rule of gene nomenclature, this gene was then called as GbPR10-1 (Figure 2).
The deduced GbPR10-1 protein was 159 aa in length with a molecular weight of 17.9 kDa, which is a highly hydrophilic acid protein with predicted pI 4.95. The comparison of amino acid sequences revealed that GbPR10-1 shared 98%, 79%, 51% and 52% homology to G. arboum, G. hirsutum, Corylus avellana and Betula pendula PR10 proteins respectively. The conserved sequence K-A-X-EX-Y-L was also found in C-terminal from 145 aa to 151 aa (Figure 1). Several amino acids influencing PR10 protein RNase activity were detected in Gly51, Lys55, and Glu96, but the conserved RNase binding site P-loop structure G-X-G-G-X-G in Betv1 protein was not found in GbPR10-1 protein [27]. This motif is known as “P-loop” (phosphate-binding loop) and is frequently found with variations in protein kinases as well as in nucleotidebinding proteins [28]. Therefore, it indicated that RNase activity of GbPR10-1 protein was influenced by the change of this motif.
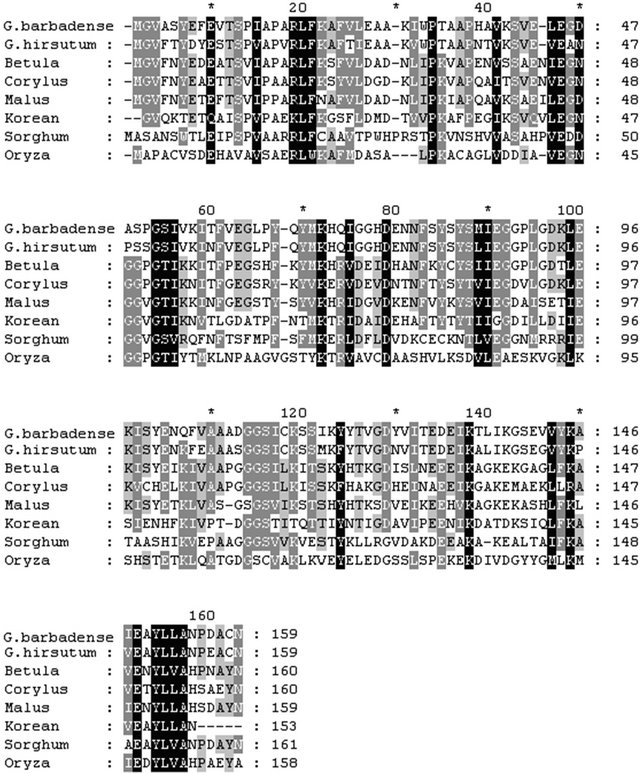
Figure 2. Alignment of amino acid sequences of PR10 protein from different plant species. The predicted amino acid sequence of GbPR10-1 protein was aligned with PR10 polypeptide sequences from G. arboum, G. hirsutum, Corylus avellana and Betula pendula using the Clustal multiple alignment program. Gaps to optimize alignments are designated by dashes (-). Asterisks (*) indicate consensus amino acid identity among all organisms. Dots (. or :) indicate positions of conservative amino acid replacement. Black blocks showed that these amino acids were extremely conserved in all the protein. Gray blocks showed that all these protein have these amino acids. The boxed numbers at the bottom refer to the position in the amino acid sequence.
Genomic walking showed that GbPR10-1 gene has no intron in the coding sequence. Southern analysis (Figure 3) indicated that there are two copies in the EcoRI lane, and two copies in the BamHI digestion lanes. Endonucleases restriction analysis showed that only one BamHI digestion site in the probe region (from nucleotide 300 to nucleotide 700), indicating there is one copy GbPR10-1 gene in the G. babardense genome.
3.2. GbPR10-1 Genes Show the Lowest Similarity with Other Putative PR10 Proteins in Cotton
Recent data showed that different PR10 protein member is involved in controlling the growth or pathogens stimuli. In order to analysis relationship among different members in cotton, we screened GeneBank using tblastx and found that there are 9 different members in upland cotton. Phylogenetic tree of these putative PR-10 proteins from G. barbadense and G. hirsutum showed that all of these PR10 proteins could be classified into three subfamilies (Figure 4). Subfamily 2 was composed of PR10-2, PR10- 3, PR10-5 and PR10-6 proteins. Subfamily 3 was composed of PR10-4, PR10-7, PR10-8 and PR10-9 proteins. PR10-1, which showed the lowest similarity with other PR10 proteins, was assigned into subfamily 1. Expression pattern of subfamily 3 showed that these genes were highly expressed in developing cotton fiber (data not shown). The Expression patterns of PR10 genes in subfamily 2 were not typical. Subfamily 1 is composed of only one PR10 protein (Figures 4 and 5), indicating that the function of GbPR10-1 gene needs to be analyzed in detail.
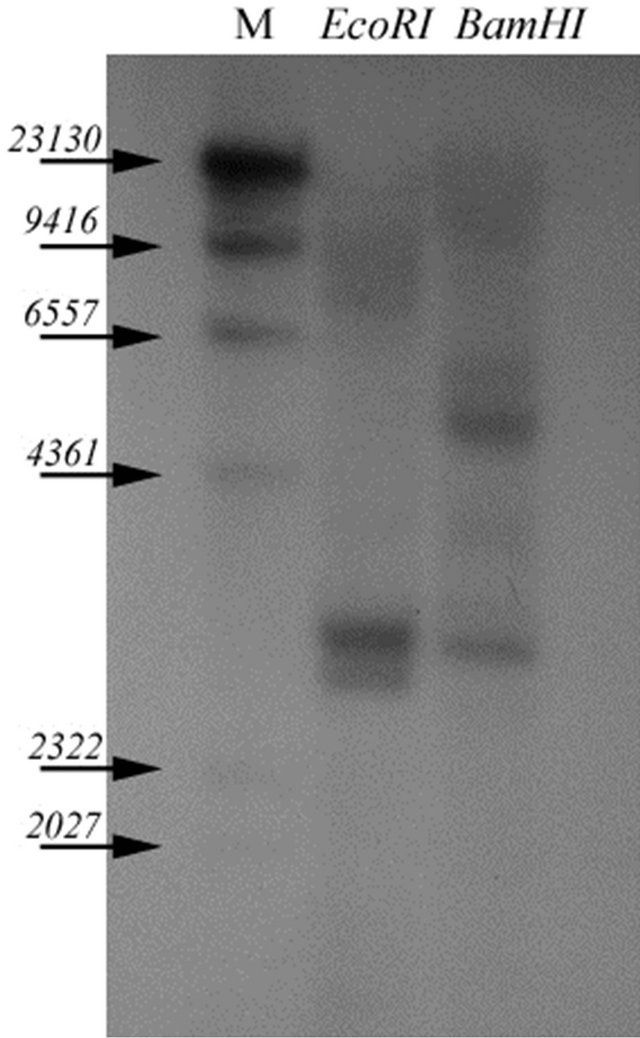
Figure 3. Genomic DNA southern blotting analysis of GbPR10- 1 gene in G. barbadense. Each lane contained 10 µg of genomic DNA digested with EcoRI and BamHI respectively. The blot was probed with full-length of GbPR10-1 gene cDNA. Molecular marker was shown on the first lane, their molecular weight are 23,130, 9416, 6557, 4361, 2322, 2027 bp respectively.
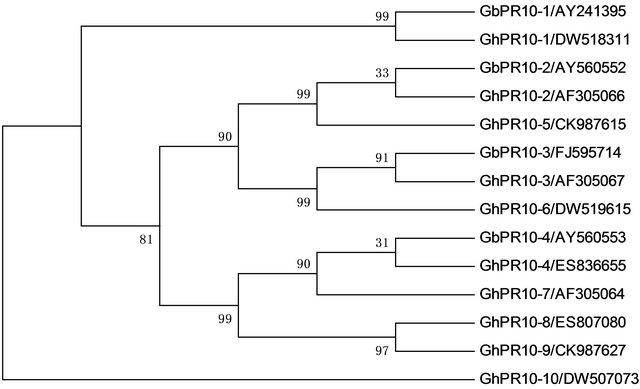
Figure 4. Phylogenetic tree of PR-10 genes from Gossypium barbadense and Gossypium hirsutum. The phylogenetic tree is constructed by Maximum Parsimony method, and the numbers at each node represent the bootstrap values (with 1000 replicates). (Accession number was shown after genes’ name).
3.3. GbPR10-1 Gene Responds to Verticillium Attack and Abiotic Stresses
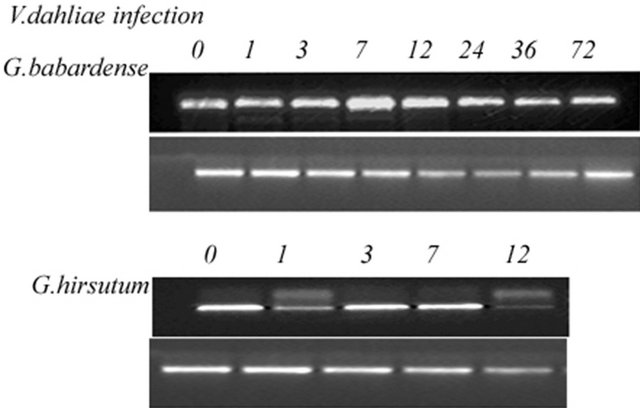
To examine the biotic and abiotic stresses on GbPR10-1 gene expression in the cotton plant, we analyzed the expression change using RT-PCR strategy with total RNA from the roots of plants subjected to different stress conditions. Plants grown in pots until two weeks were treated with salt, high temperature, ABA, anaerobic and pathogens attack. In all experiments, two primers PR10-1 and PR10-2 were used to amplify the coding region of PR10 and they displayed different patterns of expression under different conditions (Figure 6).
The expression pattern showed that GbPR10-1 gene transcripts did not decline after the fungus treatment; it subsequently increased and reached the maximum levels at 7 hours then decreased gradually to the normal level at 72 h. Differently, the expression of GhPR10 firstly declined after pathogens inoculation, return to normal condition in the 48 h after inoculation. For G. hirsutum variety Ejing-1, it was too sensitive to endure long-time pathogens attack and plants were wilt in 12 h post-inoculation and died after 72 hours treatment, so the total RNA were not extracted till 12 h after the pathogen treatment. However, the GhPR10 gene expression change was still clearly detected. The highest expression level of GhPR10 transcripts after inoculation was not up-regulated, and its expression levels under different intervals were much lower than that of GbPR10-1 in 7124.
Meanwhile, 1 µg of total RNA from G. barbadense with 100 mM NaCl was also adopted to analyze GbPR10- 1 gene expression. Compared to untreated plants, the expression levels of the GbPR10-1 had increased about three-fold and reached maximum levels till 24 h and this expression level did not decrease in 72 h after inoculation.

Figure 5. Multiple alignments of PR-10 proteins from Gossypium barbadense and Gossypium hirsutum of based on amino acid sequences. Black and grey blocks showed that these amino acids were different from other protein. The red boxes showed that all these protein share same or similar amino acids.

(A) (B)
Figure 6. Expression profiles of PR10 gene under abiotic and biotic stresses in cotton. Total RNA (1 µg/lane) was isolated at 0, 1 h, 3 h, 7 h, 12 h, 24 h, 48 h, 72 h after exposure to different stresses respectively, and subjected to one-step RT-PCR amplification (upper panel). The entire experiments were repeated twice using total RNA isolated from the roots of cotton as templates. Ubiquitin cDNA was used to normalize the amount of templates added in PCR reactions (lower panel). (A) GbPR10-1 gene expression profile under biotic stress including heat, salt, submergenece, and ABA; (B) Comparison of PR10 gene expression in sea-island and upland cotton under V. dahliae attack.
Upon exposure of plants to ABA solution, the transcript level of GbPR10-1 remained relatively constant in root of cotton. That means that GbPR10-1 gene does not respond to ABA induction.
The seedling was too young to endure long-time heat treatment and plants were wilted after 24 h high temperature and then died, so the total RNA were extracted till 24 h after the heat treatment. During the high-temperature stress (45˚C), the GbPR10-1 gene transcripts of G. barbadense were not declined in the first 7 h and subsequently increased and reached the highest level at 24 h.
3.4. GbPR10-1 Protein Has Rnase Activity and Restricts Verticillium Hyphal Growth in Vitro
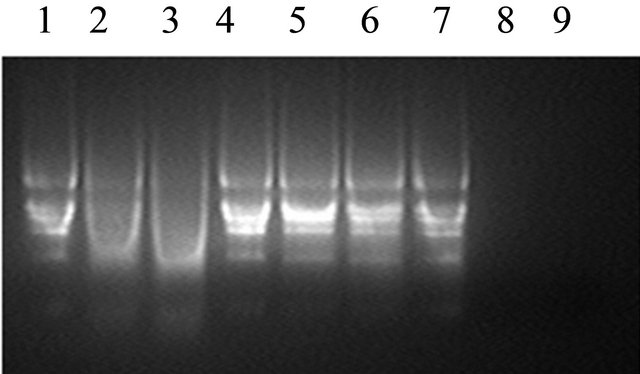
GbPR10-1 fusion protein was expressed in E. coli after induction with IPTG for 4 hr. The result showed that recombinant GbPR10-1 is about 34 kDa (Figure 7) in the lane 3 which was consist with the putative size of the fusion protein. This fusion protein isolated by an affinity column was incubated with the V. dehaliae RNA, and it could cleave the total RNA into fragments slowly (Figure 8(a)). After incubation with V. dahliae on the plate for 4 days, GbPR10-1 show very low inhibition activity on hyphal growth of V. dahliae but not for its spore germination (Figure 8(b)).
4. Discussion
Verticillium wilt resistance was associated with the HR reaction. Following the inoculation of fungus, a series of chemical compounds such as phytolexins, ethylene, and ROS was implicated in this course of HR [11,12]. In our previous study [21], we had isolated and characterized hundreds of differentially expressed transcripts responding to the pathogens attacking. Most of them were related to oxidative burst, phytolexins synthesis and transcription regulation, in which only one kind of PR gene family (GbPR10-1) was highly expressed. In this study we reported the cloning of a novel PR10 gene, GbPR10-1,
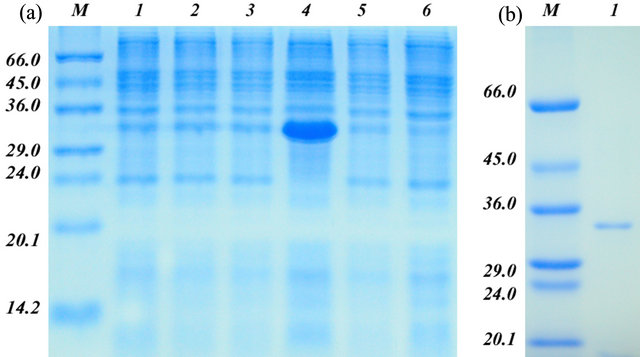
Figure 7. The recombinant expression of GbPR10-1 protein and purification. (a) The recombinant expression of GbPR10- 1 protein without purification. M: protein marker, their molecular weight were shown on the left (kDa); 1: Noninduced E. coli BL21 cell; 2: Induced E. coli BL21 protein; 3: Non-induced E. coli BL21 carrying the GbPR10-1 insert in PET-32a; 4: Induced E. coli BL21 carrying the GbPR10-1 insert in PET-32a; 5: Non-induced E. coli BL21 carrying PET-32a plasmid without GbPR10-1 insertion; 6: Induced E. coli BL21 carrying PET-32a plasmid without GbPR10-1 insertion; (b) The recombinant expression of GbPR10-1 protein after purification. M: Protein marker, their molecular weights were shown on the left (kDa); 1: Purified recombinant GbPR10-1 protein.
 (a)
(a) (b)
(b)
Figure 8. (a) Ribonuclease analysis of GbPR10-1 protein. Lane 1: control RNA; Lane 2: cotton root total RNA digested by recombinant fusion protein GbPR10-1 for 15 min; Lane 3: cotton root total RNA digested by recombinant fusion protein GbPR10-1 for 30 min; Lane 4: control RNA; Lane 5: cotton root total RNA incubation with protein purification solution for 15 min; Lane 6: cotton root total RNA incubation with protein purification solution for 30 min; Lane 7: control RNA; Lane 8: cotton root total RNA digested by 10 U RNase for 15 min; Lane 9: cotton root total RNA digested by 10 U RNase for 30 min; (b) Inhibition activity analysis of GbPR10-1 recombinant protein. 1, 2: 2 μg and 4 μg GbPR10-1 purification recombinant protein; 3, 4: 10 and 20 μL protein purification solution.
from G. barbadense by screening its infected root cDNA library. Characterizations of GbPR10-1 gene including sequence analysis, molecular evolution analysis, expression profiles and its function in vitro were also investigated.
Evolutionary analysis of its putative amino acid sequences revealed that the cotton GbPR10-1 and potato PR10 protein shared the same taxonomic group, and was closely categorized (Figure 2). The significant difference between GbPR10-1 protein and other PR10 proteins was that GbPR10-1 protein has a G-D-A-S-P-G-S-I-V-K motif instead of an RNase activity G-X-G-G-X-G motif. This motif change may contribute to low degradation RNA activity of recombinant GbPR10-1 in vitro. Anyway, this result was not exactly consisted with the analysis data of recombinant PR10 protein from Asian cotton in vitro [29]. Sea-island cotton was the offspring from G. abroeum crossed with G. raimondii based on the evolutionary phylogeny. Several amino acid variations were found between GbPR10-1 and GaPR10, and these amino acid substitutions may contribute to their difference of biological function.
Expression profile investigation showed that PR10 transcripts under biotic and abiotic stresses were different. High concentration of NaCl could cause rapid and transient increase of GbPR10-1. GbPR10-1 gene could also respond to high temperature stress, but there was not evident increase of its transcripts under submergence. GbPR10- 1 expression profiles were different under phytohormones including MJ, JA, ethylene and ABA treatments. MJ and JA could shortly up-regulate GbPR10-1 expression (data not shown), but GbPR10-1 gene did not respond to ABA and ethylene treatment. The transient enrichment of PR10 gene only induced by jasmonic acid instead of ethylene and salicylic acid was observed in the Asian cotton [30]. Combined with genome-wide microarray analysis in Arabidopsis, PR10 was activated by those abiotic and biotic stresses, which may share common activating signals or they have the interactions in their signal transduction pathways [31].
In order to gain insight into the expression of PR10 genes under pathogens’ attack, we examined PR10 gene expression in both compatible and incompatible hostpathogens interaction. Because there are 9 isoforms (or even more) in cotton, we designed specific primers to analysis PR10-1 gene expression after aligning different PR10 genes from two cotton species. In the case of incompatible interaction (sea-island cotton-V. dahliae), the accumulation of the PR10 transcripts was instantly upregulated after inoculation with pathogens, and it reached the highest level at 7 h post-inoculation. In the compatible interaction (upland cotton-V. dahliae), the expression of PR10 gene was inhibited after pathogens inoculation, then return to normal expression level at 7 h post-inoculation. GhPR10-1 gene was not induced by the inoculation of fungus ingression. In the former study, GhPR10-1 genes (AA659993, AA659999) expressed transiently and their expression levels were very limitedly up-regulated in the roots upon infection with defoliating and un-defoliating V. dahliae strains. In rice, the JIOsPR10 transcript was also found to be transiently induced 24 h after inoculation with the fungus M. grisea in case of an incompatible interaction, while increase in mRNA level was only recorded at 120 h in compatible situation [18]. This result demonstrated a differential pattern of PR10 gene induction when considering a compatible and incompatible interaction between the pathogens and cotton roots.
When the fungus ingress the cotton plant, the pathogens always first colonize on the root surface [4,5], then directly penetrate into the root or the stem through wounded areas. Early studies showed that the PR protein and phytoalexins were detected much more early in roots of resistant lines than susceptible lines. These phytoalexins can concurrently accumulate for several days in the vascular tissues in the resistant infected plants and they were toxic to wilt pathogens and contributed to occlude hyphae expanding [6,30]. In this study, we analyzed the GbPR10-1 protein antifungal activity in vitro, when the extraction was inoculated on the plate, which showed the growth of fungus was destroyed by the recombinant PR10 protein. After 4 days co-culture, the effect of PR10 suppress the extending of pathogens could be detected. Based on above analysis, we probably concluded that earlier activation of PR10 genes in sea-island cotton is important for blocking the penetration of the fungus. The earlier activation of PR10 genes was regulated by MJ or SJ signal molecule, which then activates resistance response to reduce symptoms (leaf wilt and vascular discoloration) caused by V. dahliae [14,32,33]. Anyway, the exact genetic analysis of this gene should be combined with unraveling the roles of PR-activated proteins such as transcription factors.
5. Acknowledgements
This work was funded by NSFC (31071458), China Transgenic Program (2011AA100605) and 973 project in China.
REFERENCES
- A. A. Bell, “Mechanisms of Disease Resistance in Gossypium Species and Variation in Verticillium dahliae,” In: G. A. Constable and N. W. Forrester, Eds., Challenging the Future, Proceedings of the World Cotton Research Conference, 1-5 February 1995, Commonwealth Scientific and Industrial Research Organization, Melbourne, pp. 225-235.
- C. S. Kenneth and E. G. William, “A Root-Injection Method to Assess Verticillium Wilt Resistance of Peppermint (Menth x piperita L.) and Its Use in Identifying Resistant Somaclones of cv. Black Mitcham,” Euphytica, Vol. 106, No. 3, 1999, pp. 223-230.
- F. Daayf, M. Nicole, B. Boher, A. Pando and J. P. Geiger, “Early Vascular Defense Reactions of Cotton Roots Infected with a Defoliating Mutant Strain of Verticillium dahliae,” European Journal of Plant Pathology, Vol. 103, No. 2, 1997, pp. 125-136. doi:10.1023/A:1008620410471
- N. A. Garas and A. C. J. Waiss, “Differential AccumuLation and Distribution of Antifungal Sesquiterpenoids in Cotton Stems Inoculated with Verticillium dahliae,” Phytopathol, Vol. 76, 1996, pp. 1011-1017. doi:10.1094/Phyto-76-1011
- N. A. Garas, S. Wilhem and J. E. Sagen, “Relationship of Cultivar Resistance to Distribution of Verticillium dahliae in Inoculated Cotton Plants and to Growth of Single Condia on Excised Stem Segments,” Phytopathology, Vol. 76, No. 10, 1986, pp. 1005-1010. doi:10.1094/Phyto-76-1005
- M. E. Mace, R. D. Stipanoyic and A. A. Bell, “Toxicity and Role of Terpenoid Phytoalexins in Verticillium wilt Re-Sistance in Cotton,” Physiological Plant Pathology, Vol. 26, No. 2, 1985, pp. 209-218. doi:10.1016/0048-4059(85)90021-9
- M. J. R. Mould and M. C. Heath, “Ultra-Structural Evidence of Differential Changes in Transcription, Translation, and Cortical Microtubules during in Planta Penetration of Cells Resistant or Susceptible to Rust Infection,” Physiological and Molecular Plant Pathology, Vol. 55, No. 4, 1999, pp. 225-236. doi:10.1006/pmpp.1999.0224
- W. C. Muller and A. T. Morgham, “Ultra-Structure of the Vascular Responses of Cotton to Verticillium dahlia,” Canadian Journal of Botany, Vol. 71, No. 1, 1993, pp. 32- 36.
- I. A. Dubery and V. Slater, “Induced Defence Responses in Cotton Leaf Disks by Elicitors from Verticillium dahliae,” Phytochem, Vol. 44, No. 8, 1997, pp. 1429-1434. doi:10.1016/S0031-9422(96)00635-8
- R. Rakwal, S. Katsuhiko, K. A. Ganesh and Y. Masami, “Protein Phosphatase Inhibitors Activate Defense Responses in Rice (Oryza sativa) Leaves,” Physiologia Plantarum, Vol. 111, No. 2, 2001, pp. 151-157. doi:10.1034/j.1399-3054.2001.1110204.x
- S. R. McLusky, M. H. Bennett, M. H. Beale, M. J. Lewis, P. Gaskin and J. W. Mansfield, “Cell Wall Alterations and Localized Accumulation of Feruloyl-3’-methoxytyamine in Onion Epidermis at Sites of Attempted Penetration by Botrytis allii Are Associated with Actin Polarision, Peroxidase Activity and Suppression of Flavonoid Biosynthesis,” The Plant Journal, Vol. 17, No. 5, 1999, pp. 523-534. doi:10.1046/j.1365-313X.1999.00403.x
- R. Meyer and I. A. Dubery, “High-Affinity Binding of a Protein-Lipopolysaccharide Phytotoxin from Verticillium dahliae to Cotton Membranes,” FEBS Letters, Vol. 335, No. 2, 1993, pp. 203-206. doi:10.1016/0014-5793(93)80730-I
- S.-C. C. Lo, J. D. Hipskind and R. L. Nicholson, “cDNA Cloning of a Sorghum Pathogenesis-Related Protein (PR10) and Differential Expression of Defense-Related Genes Following Inoculation with Cochliobolus heterostropohus or Colletotrichum sublineolum,” Molecular Plant-Microbe Interactions, Vol. 12, No. 6, 1999, pp. 479-489. doi:10.1094/MPMI.1999.12.6.479
- L. F. Li, “Multilayer-Coated Diffraction Gratings: Differential Method of Chandezon et al. Revisited: Errata,” Journal of the Optical Society of American A, Vol. 13, No. 3, 1996, p. 534. doi:10.1364/JOSAA.13.000543
- D. J. Bowels, “Defense-Related Proteins in Higher Plants,” Annual Review of Biochemistry, Vol. 59, 1990, pp. 87-89.
- L. C. Van Loon, W. S. Pierpoint, T. Boller and V. Conejero, “Recommendation for Naming Plant PathogenesisRelated Proteins,” Plant Molecular Biology Reporter, Vol. 12, No. 3, 1994, pp. 245-264. doi:10.1007/BF02668748
- M. H. Walter, J. W. Liu, J. Wunn and D. Hess, “Bean Ribonuclease-Like Pathogenesis-Related Protein Genes (Ypr10) Display Complex Patterns of Developmental, Dark-Induced and Exogenous-Stimulus Dependent Expression,” European Journal of Biochemistry, Vol. 239, No. 2, 1996, pp. 281-293. doi:10.1111/j.1432-1033.1996.0281u.x
- N. S. Jwa, G. K. Agrawal, R. Rakwal, C. H. Park and V. S. Agrawal, “Molecular Cloning and Characterization of a Novel Jasmonate Inducible Pathogenesis-Related Class 10 Protein Gene, JISsPR10, from Rice (Oryza sativa L.) Seedling Leaves,” Biochemical and Biophysical Research Communications, Vol. 76, 1996, pp. 1011-1017.
- M. Hashimoto, L. Kisseleva, S. Sawa, T. Furukawa, S. Komatsu and T. Koshiba, “A Novel Rice PR10 Protein, RSOsPR10, Specifically Induced in Roots by Biotic and Abiotic Stresses, Possibly via the Jasmonic Acid Signaling Pathway,” Plant Cell Physiology, Vol. 45, No. 5, 2004, pp. 550-559. doi:10.1093/pcp/pch063
- F. Smit and I. A. Dubery, “Cell Wall Reinforcement in Cotton Hypocotyls in Response to Verticillium dahlia Elicitor,” Phytochemistry, Vol. 44, No. 5, 1997, pp. 811- 815. doi:10.1016/S0031-9422(96)00595-X
- K. J. Zuo, J. Wang, W. Wu, Y. Chai, X. Sun and K. X. Tang, “Identification and Characterization of Differentially Expressed ESTs of Gossypium barbadense Infected by Verticillium dahliae with Suppression Subtractive Hybridization,” Molecular Biology, Vol. 39, No, 2, 2005, pp. 191-191. doi:10.1007/s11008-005-0028-6
- K. H. Melissa, J. L. Karin and R. L. Bruce, “Identification of Disease Response Genes Expressed in Gossypium hirsutum,” Plant Molecular Biology, Vol. 40, No. 2, 1999, pp. 286-296.
- H. G. McFadden, R. Chapple, R. de Feyter and E. Dennis, “Expression of Pathogenesis-Related Genes in Cotton Stems in Response to Infection by Verticillium dahlia,” Physiological and Molecular Plant Pathology, Vol. 58, No. 3, 2001, pp. 119-131.
- J. Sambrook, E. F. Fritsch and T. Maniatis, “Molecular Cloning, a Laboratory Manual,” 2nd Edition, Cold Spring Harbor laboratory Press, New York, 1989.
- Y. Tateno, N. Takezaki and M. Nei, “Relative Efficiencies of the Maximum-Likelihood, Neighbor-Joining, and Maximum-Parsimony Methods When Substitution Rate Varies with Site,” Molecular Biology and Evolution, Vol. 11, No. 2, 1994, pp. 261-277.
- K. Tamura, J. Dudley, M. Nei and S. Kumar, “MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0,” Molecular Biology and Evolution, Vol. 24, No. 8, 2007, pp. 1596-1599. doi:10.1093/molbev/msm092
- M. Gajhede, P. Osmark, F. E. Poulsen, H. Ipsen, J. N. Larsen, R. J. J. van Neerven, C. Schou, H. Lǿwenstein and M. D. Spangfor, “X-Ray and NMR Structure of Betv1, the Origin of Birch Pollen Allergy,” Nature Structural & Molecular Biology, Vol. 3, No. 12, 1996, pp. 1040- 1045. doi:10.1038/nsb1296-1040
- M. Saraste, P. R. Shibbald and A. Wittinghofer, “The PLoop—A Common Motif in ATPand GTP-Binding Proteins,” Trends in Biochemical Sciences, Vol. 15, No. 11, 1990, pp. 430-434. doi:10.1016/0968-0004(90)90281-F
- X. J. Zhou, S. Lu, Y. H. Xu, J. W. Wang and X. Y. Chen, “A Cotton cDNA (GaPR10) Encoding a PathogenesisRelated 10 Protein with in Vitro Ribonuclease Activity,” Plant Science, Vol. 162, No. 4, 2002, pp. 629-636. doi:10.1016/S0168-9452(02)00002-X
- M. E. Mace, A. A. Bell and C. H. Beckman, “Histochemistry and Identification of Disease-Induced Terpenoid Aldehydes in Verticillium-Wilt-Resistant and -Susceptible Cottons,” Canadian Journal of Botany, Vol. 54, No. 18, 1976, pp. 2095-2099. doi:10.1139/b76-225
- Y. H. Cheong, H. Chang, R. Gupta, X. Wang, T. Zhu and S. Luan, “Transcriptional Profiling Reveals Novel Interactions between Wounding, Pathogen, Abiotic Stress and Hormonal Responses in Arabidopsis,” Plant Physiology, Vol. 129, No. 2, 2002, pp. 661-677. doi:10.1104/pp.002857
- P. Constabel and C. A. Ryan, “A Survey of Woundand Methyl Jasmonate-Induced Leaf Polyphenol Oxidase in Crop Plants,” Phytochemistry, Vol. 47, No. 4, 1998, pp. 507-511. doi:10.1016/S0031-9422(97)00539-6
- I. A. Dubery, L. G. Teodorczuk and A. E. Louw, “Early Responses in Methyl-Jasmonate Preconditioned Cells toward Pathogen-Derived Elicitors,” Molecular Cell Biology Research Communications, Vol. 3, No. 2, 2000, pp. 105-110. doi:10.1006/mcbr.2000.0198
NOTES
*Corresponding author.

