American Journal of Plant Sciences
Vol.3 No.12(2012), Article ID:25783,9 pages DOI:10.4236/ajps.2012.312208
New Alleles of Rice ebisu dwarf (d2) Mutant Show Both Brassinosteroid-Deficient and -Insensitive Phenotypes
![]()
1Faculty of Bioresources and Environmental Sciences, Ishikawa Prefectural University, Nonoichi, Japan; 2Bioscience and Biotechnology Center, Nagoya University, Nagoya, Japan; 3RIKEN Advanced Science Institute, Wako, Japan.
Email: *sakamoto@ishikawa-pu.ac.jp
Received September 25th, 2012; revised October 19th, 2012; accepted November 18th, 2012
Keywords: Brassinosteroids; Brassinosteroid Biosynthetic Enzymes; Brassinosteroid Sensitivity; Cytochrome P450 Monooxygenase; Mutant Allele; Rice
ABSTRACT
ebisu dwarf (d2) is a mutant caused by mutation in a rice brassinosteroid biosynthetic enzyme gene, CYP90D2/D2, thereby conferring a brassinosteroid-deficient dwarf phenotype. Three newly isolated d2 alleles derived from a Nipponbare mutant library (d2-3, d2-4, and d2-6) produced more severe dwarf phenotypes than the previously characterized null allele from a Taichung 65 mutant library, d2-1. Linkage analysis and a complementation test clearly indicated that the mutant phenotypes in d2-6 were caused by defects in CYP90D2/D2, and exogenous treatment with brassinolide, a bioactive brassinosteroid, rescued the dwarf phenotype of three Nipponbare-derived d2 mutants. However, the content of endogenous bioactive brassinosteroid, castasterone, and the expression of brassinosteroid-response genes indicated that partial suppression of the brassinosteroid response in addition to a brassinosteroid deficiency has occurred in the Nipponbare-derived d2 mutants. Based on these results, we discuss the possibility that wild-type Nipponbare has some defects in an unknown factor or factors related to the brassinosteroid response in rice.
1. Introduction
Brassinosteroids are endogenous phytohormones that are involved in the regulation of various growth and developmental processes in higher plants, including cell and stem elongation, dark-adapted morphogenesis (skotomorphogenesis), environmental stress responses, and the differentiation of tracheary elements [1-3]. Brassinosteroids have been shown to enhance various aspects of crop production in rice (Oryza sativa L.), including brassinosteroid-induced disease resistance [4] and abiotic stress tolerance [5]. Interestingly, overproduction and deficiency of brassinosteroids both increased grain and biomass yield, although they did this by different mechanisms [6-8]. These findings suggest that genetic improvement of crop production is feasible by modulating brassinosteroid metabolism and the content of bioactive brassinosteroids.
Several brassinosteroid-related mutants have been isolated in rice. The brassinosteroid-insensitive mutant d61 is caused by defects in the brassinosteroid receptor gene, OsBRI1 [9,10]. In addition, brassinosteroid-deficient dwarf1 (brd1) is caused by mutations in the brassinosteroid biosynthetic enzyme gene that encodes C-6 oxidaseCYP85A1 [11,12]. ebisu dwarf (d2) is a loss-of-function mutant of the brassinosteroid biosynthetic enzyme gene that encodes C-23 hydroxylase, CYP90D2/D2 [13,14]. osdwarf4 and d11 are mutants of brassinosteroid biosynthetic enzyme genes that encode two C-22 hydroxylases, CYP90B2 and CYP724B1, respectively [6,15]. brassinosteroid-deficient dwarf2 (brd2) is caused by mutations in the sterol biosynthetic enzyme gene DIMINUTO/ DWARF1, which functions in the conversion of 24-methylenecholesterol to campesterol [16]. Characterization of these brassinosteroid-deficient mutants has helped us to understand the regulation of brassinosteroid biosynthesis in rice and the functions of brassinosteroids in rice growth and development. However, there are several enzymes involved in rice brassinosteroid biosynthesis for which mutants have not yet been isolated.
Although the major pathway for brassinosteroid biosynthesis has been established [17-19], recent studies demonstrated the complexity of the pathway by revealing new branches and shortcut routes [20,21]. To clarify the function of the other biosynthetic enzymes involved in brassinosteroid biosynthesis for which mutants have not been identified, we screened rice mutant libraries and isolated three new candidate lines. Here, we describe the characterization of these three mutants, which have unique phenotypes that exhibit both brassinosteroid deficiency and insensitivity. Based on the results of our analysis, we discuss the possibility that the well-known wild-type rice, Nipponbare, has some previously unreported defect in its brassinosteroid response.
2. Materials and Methods
2.1. Characterization of Mutants
To obtain brassinosteroid-related mutants, we performed a large-scale screening of rice mutant collections that were produced using a retrotransposon (Tos17), chemical mutagens, and γ-ray irradiation. Three candidate lines (d2-3, d2-4, and d2-6) were obtained from a Nipponbare library with tissue culture–induced mutations [22]. Seeds of wild-type rice (Oryza sativa L. “Nipponbare”) and the mutants were sterilized in 1% NaClO for 30 min and sown on Murashige and Skoog agar medium. For the initial phytohormone treatment, seedlings were grown in a growth chamber at 28˚C under continuous light for 1 week, then transplanted into media containing 10 nM brassinolide. For the subsequent field experiments, seedlings were grown for 1 month in a greenhouse, and then transplanted into a paddy field. For the gene expression analysis, we selected 2-week-old seedlings that exhibited uniform growth and adapted them to hydroponic culture for 2 days before treatment. Brassinolide treatment (100 nM) was carried out by adding the pure chemical to the culture medium. Whole seedlings were harvested 3 h after the treatment.
2.2. Mapping and Sequence Analyses
For the mapping of each mutant, we performed linkage analysis using an F2 population of about 2000 plants derived from the cross between the Nipponbare-derived mutant (japonica) and Kasalath (indica) varieties. The mutation sites of all three candidate lines were mapped onto the short arm of chromosome 1 and were tightly linked to the d2 locus. The nucleotide sequence of the wild-type CYP90D2/D2 gene from these three lines was determined using a dideoxynucleotide chain-termination method using an automated sequencing system (ABI377; Applied Biosystems, Foster City, CA, USA), and was analyzed using the Lasergene software (DNAStar, Inc., Madison, WI, USA). We also determined the nucleotide sequence of a 9.3-kb genomic segment containing the CYP90D3 gene (from positions –4191 to +5156, taking the translation initiation site as +1) from Nipponbare and Taichung 65.
2.3. Complementation of the d2 Mutant
For complementation of the d2-6 mutation, a 15.2 kb restriction fragment (from –6063 to +9182 bp; taking the translation initiation site as +1) between SmaI and EcoRI sites was obtained from PCR products which amplified the genomic sequence of the D2 gene from wild-type rice Nipponbare. The fragment was cloned into pBluescript II SK (Stratagene, La Jolla, CA, USA) to confirm the sequence, and then inserted between SmaI and EcoRI sites of the hygromycin-resistance binary vector pCAMBIA1300 (Cambia, Canberra, Australia). The resulting construct was introduced into Agrobacterium tumefaciens strain EHA105, and Agrobacterium-mediated transformation of the d2-6 mutant was performed as described [23]. Transgenic plants were selected on media containing 50 mg·L–1 hygromycin.
2.4. Analysis of Endogenous Brassinosteroid Levels
Shoots from wild-type and mutant plants were harvested 4 weeks after germination. To analyze the endogenous brassinosteroids, lyophilized shoots (equivalent to 20 g fresh weight) were extracted twice with 250 mL of MeOH, and deuterium-labeled internal standards (1 ng/g fresh weight) were added to the extract. After evaporation of the solvent in vacuo, the extract was partitioned between CHCl3 and water three times. The CHCl3-soluble fraction was purified with a silica gel cartridge column (Sep-Pak Vac 10 g; Waters, Milford, MA, USA). The column was subsequently eluted with 100 mL of CHCl3, 2% (v/v) MeOH in CHCl3, and 7% (v/v) MeOH in CHCl3. Each of 2% MeOH and 7% MeOH fractions was purified by Sephadex LH-20 column chromatography (column volume of 200 mL; column eluted with MeOH-CHCl3 [4:1]). The effluents of the elution volume to total column volume 0.6 to 0.8 were collected as BR fractions. The eluates were subjected to ODS-HPLC (Senshu Pak Pegasil ODS, 10 × 30 mm + Senshu Pak Pegasil ODS, 20 × 250 mm; Senshu Scientific, Tokyo) at a flow rate of 8 mL/min. Ninety percent acetonitrile was used as a solvent for the eluate derived from the 2% MeOH fraction, and 70% acetonitrile was used as a solvent for the eluate derived from the 7% MeOH fraction. HPLC purification from the 7% MeOH fraction yielded a brassinolide fraction (retention time (Rt) from 8 to 10 min), a castasterone fraction (Rt from 10 to 14 min), a teasterone fraction (Rt from 17 to 20 min), a typhasterol fraction (Rt from 26 to 32 min), and a 6-deoxocastasterone fraction (Rt from 38 to 44 min). HPLC purification from the 2% (v/v) MeOH fraction yielded 3-dehydroteasterone fraction (Rt from 12 to 16 min), a cathasterone fraction (Rt from 16 to 20 min), a 6-deoxoteasterone fraction (Rt from 28 to 32 min), a 3-dehydro- 6-deoxoteasterone fraction (Rt from 32 to 36 min), and a 6-deoxotyphasterol fraction (Rt from 44 to 48 min). To analyze 6-deoxocathasterone, a portion (1 g fresh weight equivalent) of CHCl3 fraction from a silica gel cartridge column was purified with an octadecyl silane (ODS) cartridge column (Sep-Pak Plus C18, Waters), which was eluted with MeOH, and subjected to ODS-HPLC (Senshu Pak ODS 1151-D, 4.6 ´ 150 mm, Senshu Scientific, Tokyo) at a flow rate of 1 mL/min with 100% MeOH. The HPLC purification yielded a 6-deoxocathasterone fraction (Rt from 3 to 4 min). Each BR fraction was analyzed by GC-MS after derivatization. GC-MS analysis was carried out on a mass spectrometer (JMS-AM SUN200, JEOL, Tokyo) connected to a gas chromatograph (6890A, Agilent Technologies, Wilmington, DE) with a capillary column DB-5 (0.25 mm ´ 15 m, 0.25 μm film thickness; J & W Scientific, Folsom, CA, USA). The analytical conditions were the same as previously described [13].
2.5. Gene Expression Analysis
Total RNA was extracted from whole seedlings using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). Single-strand cDNAs were synthesized by using the Advantage RT-for-PCR Kit (Clontech, Palo Alto, CA, USA). Quantitative RT-PCR was performed with an iCycler iQ real-time PCR system (Bio-Rad Laboratories, Hercules, CA, USA). The primer sequences were 5’- GTAGCCAGCTTGATCTCATCTC-3’ and 5’-GGGA CGACTCTACTGCATCA-3’ for BU1, 5’-TGATCCATTCCTGTACCCTG-3’ and 5’-TACCTTCTTCCTC CCATCTG-3’ for CYP85A1, 5’-CCACTAGCCACCAC TACTAC-3’ and 5’-GGTGTGTGCAACTTGGCTTG-3’ for CYP90D2/D2, 5’-CGAGGAATGCCCATCAAGG T-3’ and 5’-ACCTTGATGGGCATTCCTCG-3’ for CYP90D3, and 5’-CGCCAGTTTGGTCGCTCTCGATT TCG-3’ and 5’-TCAGGAGCTCCGTGCTCTTCTGGTA C-3’ for Histone H3. These primers specifically amplified the target gene sequences. Expression levels were normalized against the value obtained for Histone H3, which was used as the internal reference gene in each analysis.
2.6. Statistics
For gene expression experiments, we performed 3 biological repeats for wild-type and mutant plants. Statistical analysis of the data (the ratio of the target value to the internal reference) was performed using Version 5.1.2 of the JMP statistical package (SAS Institute Japan Ltd., Tokyo, Japan). Differences in the relative expression levels were analyzed by ANOVA followed by Holm’s method for multiple comparisons.
3. Results and Discussion
3.1. Isolation of Three Mutants with Brassinosteroid-Related Phenotypes
As the result of a large-scale screening of rice mutant collections, we identified three lines that showed the morphological characteristics of brassinosteroid-related mutants, namely dwarf and erect-leaf phenotypes. These mutants were obtained from a Nipponbare library with tissue culture-induced mutations [22]. Exogenously applying a bioactive brassinosteroid, brassinolide, rescued the dwarfism of these mutants and restored plant heights similar to that of the wild-type Nipponbare (Figure 1), suggesting that these mutants are brassinosteroid-deficient. Previously isolated brassinosteroid-related rice mutants have been categorized into three groups on the basis of leaf morphology and gross morphology. Plants exhibiting a group-1 phenotype formed only abnormal leaves with stiff, tortuous blades, and their leaf sheaths were scarcely developed. These plants did not flower, exhibit internode elongation, or bear seeds. These phenotypes were observed in the brassinosteroid-insensitive mutants, d61-3, d61-4, d61-5, and d61-6 [10]. Plants exhibiting a group-2 phenotype had severe dwarfing (about 10 cm in height) and malformed leaves with twisted leaf blades. The leaves of these plants were erect, and the ratio of leaf sheath length to leaf blade length was reduced. The floral organs did not form and internodes did not elongate in the group-2 phenotype plants, and they did not bear any seeds. These phenotypes were observed in the brassinosteroid-deficient mutants, brd1-1 and brd1-2 [11,12]. Plants exhibiting a group-3 phenotype showed a range of semi-dwarf phenotypes (about 70% ~ 95% height of wild-type). The leaves of these plants were erect, but the other abnormal phenotypes such as twisted leaf blades were not observed. Reproductive development of group-3 phenotype plants seems normal. These phenotypes were observed in the brassinosteroid-

Figure 1. Phenotypic rescue of the abnormal mutant phenotype by treatment with exogenous brassinolide. We transplanted 1-week-old seedlings of three Nipponbare-derived mutants, d2-3, d2-4, and d2-6, into MS medium containing 10 nM brassinolide (right three plants) or solvent (ethanol, left three plants), and grew them for 3 weeks. Although the root growth was inhibited by high concentration of brassinolide, above-ground morphology was rescued by the brassinolide treatment. Bar, 5 cm.
deficient mutants, brd2-1, d2-1, d2-2, d11-1, d11-2, and osdwarf4-1 [6,13,15,16], and in the brassinosteroid-insensitive mutants, d61-1 and d61-2 [9]. Because the gross morphology of newly isolated three mutants were ranged between group-2 and group-3 (discuss below), we hypothesized that these mutants had defects in one or more novel brassinosteroid biosynthetic enzymes or had novel defects in one or more previously identified brassinosteroid biosynthetic enzymes.
3.2. The Three Brassinosteroid-Deficient Mutants Were New d2 Alleles
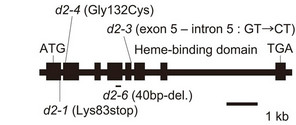
For the mapping of each mutant, we performed linkage analysis using an F2 population of about 2000 plants derived from the cross between the Nipponbare-derived mutant (japonica) and Kasalath (indica) varieties. The mutation sites of all three lines were mapped onto the short arm of chromosome 1 and were tightly linked to CYP90D2/D2. CYP90D2/D2 encodes a cytochrome P450 monooxygenase that functions in brassinosteroid biosynthesis, and its loss-of-function mutation causes the ebisu dwarf (d2) mutant [13]. Sequence analysis of CYP90D2/D2 from the three lines revealed that d2-3 had a single nucleotide substitution at the junction of exon 5 and intron 5 (G to C), d2-4 had a single nucleotide substitution (G to T) in exon 2 that induced an amino acid residue change (from Gly to Cys), whereas d2-6 had a 40-bp deletion in exon 4 (Figure 2(a)). Because this deletion caused a frame shift after Lys-313, the mutant protein lacks most of the enzyme, including the catalytic domain. The mutant phenotype of d2-6 was complemented (Figure 2(b)) by the introduction of a 15.2-kb genomic segment containing the CYP90D2/D2 gene (from positions –6063 to +9182 bp, taking the translation initiation site as +1). Based on these results, we concluded that d2-6 is a null allele of d2, and considered that all three lines are novel alleles of the d2 mutant.
3.3. Phenotypic Variation in the d2 Mutants
Previously, two d2 alleles had been isolated from the Taichung 65 mutant library, and one of them (d2-1) was considered to be a null allele because it had a single nucleotide substitution to produce a stop codon at Lys-83 (Figure 2(a)) [13]. d2-1 showed a semi-dwarf phenotype, in which plants reached 70% of the height of the wild-type, Taichung 65 (Figure 3) [13]. Interestingly, the plant heights of the d2-3, d2-4, and d2-6 mutants were about 30 cm, whereas that of Nipponbare, the wild-type that gave rise to d2-3, d2-4, and d2-6, was about 90 cm (Figure 3). These results indicate that the three newly obtained alleles, which included two knock-down alleles, showed a more severe dwarf phenotype than that shown by plants with the previously identified null allele, d2-1.
 (a)
(a) (b)
(b)
Figure 2. Characterization of the d2 mutants. (a) Genomic structure of the CYP90D2/D2 gene and positions of the mutations in the three d2 alleles discovered in the present study (d2-3, d2-4, and d2-6). Black boxes represent exons. (b) Complementation of the abnormal morphology of the d2-6 mutant by introduction of the CYP90D2/D2 gene. The d2-6 mutant containing the empty vector (left) and containing the DNA fragment encompassing the entire CYP90D2/D2 gene (right). Bar, 20 cm.

Figure 3. Phenotypic variation in the d2 mutants. Comparison of gross morphology between the wild-type (Nipponbare and Taichung 65) and d2 mutants. Nipponbare is the wild-type for d2-3, d2-4, and d2-6; Taichung 65 is the wild-type for d2-1. Bar, 20 cm.
Because the three newly obtained alleles and the previously identified d2-1 were derived from two different japonica varieties, Nipponbare and Taichung 65, respectively, our results suggest that the genetic background of the mutant plants affects the magnitude of the change in the phenotype.
3.4. Two CYP90D Genes in Rice
There are two Arabidopsis CYP90D1 orthologs, CYP90- D2/D2 and CYP90D3, in the rice genome, and D2 encodes CYP90D2/D2 [13]. In wild-type Nipponbare, our quantitative reverse-transcription PCR analysis revealed that both CYP90D genes were expressed at different levels in different organs (Figure 4). CYP90D2/D2 was expressed in all the organs of wild-type rice, but at a lower level in the roots and higher levels in the elongating internode and inflorescence, whereas CYP90D3 was expressed at the highest level in the roots and at lower levels in the other organs. These results suggest that both CYP90D2 and CYP90D3 are functional in wild-type rice.
3.5. CYP90D3 in the d2 Mutants
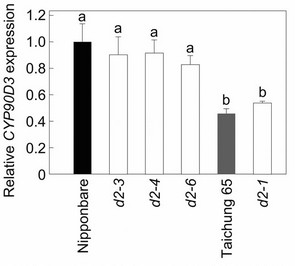
Because CYP90D2/D2 and CYP90D3 have similar sequences (75% nucleotide identity) and both CYP90D2 and CYP90D3 appear to be functional in wild-type rice (Figure 4), we hypothesized that the homeostatic system controlling the levels of bioactive brassinosteroids would up-regulate the expression of CYP90D3 in the d2-1 plants to compensate for the defects in brassinosteroid production caused by the loss-of-function mutation of CYP90D2/D2. CYP90D3 expression increased slightly (but not significantly) in d2-1 compared to its wild-type, Taichung 65 (about 1.2 times that in Taichung 65; Figure 5(a)), however, the steady-state level of CYP90D3 mRNA was significantly lower in Taichung 65 than in Nipponbare (about 46% that in Nipponbare; Figure 5(a)). In contrast, CYP90D3 mRNA did not show an increase in the d2-3, d2-4, and d2-6 mutants derived from wildtype Nipponbare (90%, 91%, and 83% of that in Nip-
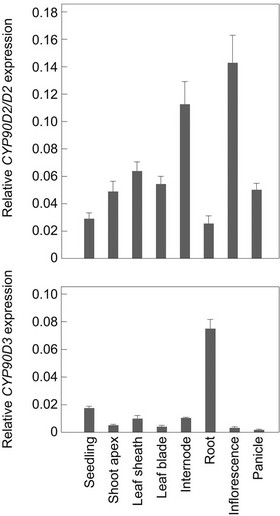
Figure 4. Expression of CYP90D2/D2 and CYP90D3 genes in various organs of wild-type Nipponbare. Expression levels were normalized against the values obtained for Histone H3. Each bar represents the mean ± s.d. of three biological repeats.
ponbare, respectively; Figure 5(a)). Although the steadystate level of CYP90D3 mRNA was lower in Taichung 65 than in Nipponbare, increase or decrease in the levels of CYP90D3 expression in mutants were consistent with the magnitude of the mutant phenotype; that is, d2-1 showed a semi-dwarf phenotype, whereas d2-3, d2-4, and d2-6 plants showed more severe dwarfing than d2-1 plants, and this difference in CYP90D3 expression pattern may explain why d2-1 showed a less extreme phenotype. These results led us to hypothesize that Nipponbare has a mutant CYP90D3 gene, and that the expression of CYP90D3 is misregulated in Nipponbare and in the Nipponbare-derived d2 mutants.
To test this hypothesis, we compared the nucleotide sequence of a 9.3-kb genomic segment containing the CYP90D3 gene (from positions –4191 to +5156, taking the translation initiation site as +1) from Nipponbare and Taichung 65. We found the insertion of a Tourist-like miniature inverted-repeat transposable element (MITE) of 430 bp, called miniature Ping (mPing) [24-26], in the first intron of CYP90D3 from the Taichung 65 genome (Figure 5(b)). This insertion was flanked on both ends by a three-nucleotide sequence, TAA, which is the typi-
 (a)
(a)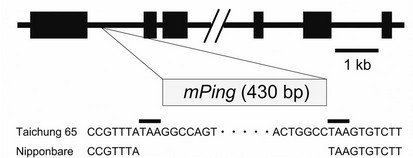 (b)
(b)
Figure 5. Expression of CYP90D3 gene in the d2 mutants. (a) Expression of CYP90D3 in wild-type rice and the d2 mutants. Expression levels were normalized against the values obtained for Histone H3. The values obtained from the wild-type Nipponbare were then normalized to a value of 1.0 to permit a comparison with the associated mutants. Nipponbare is the wild-type for d2-3, d2-4, and d2-6; Taichung 65 is the wild-type for d2-1. Each bar represents the mean ± s.d. of three biological repeats. Bars labeled with different letters differ significantly (The Holm-adjusted Student’s t test, P < 0.05); (b) Genomic structure of CYP90D3 and the position of the mPing insertion in Taichung 65. Black boxes represent exons.
cal target sequence for duplication upon mPing transposition. These structural features suggest that mPing was transposed into the first intron of CYP90D3 in Taichung 65. Ohmori et al. [27] reported that the insertion of Ping, an autonomous transposon that can activate mPing transposition, in the fourth intron of the DROOPING LEAF (DL) gene decreased the transcript level of DL. In our case, however, mPing was inserted into the Taichung 65 CYP90D3 gene, and the steady-state level of CYP90D3 mRNA was lower in Taichung 65 than in Nipponbare. Although the expression of CYP90D3 in Taichung 65 seemed to be regulated normally by the homeostatic system that controls the levels of bioactive brassinosteroids, these results indicate that the magnitude of the mutant phenotype caused by the mutations in CYP90D2/D2 could not be explained simply by the insertion of mPing in the Taichung 65 CYP90D3 gene or by the genetic redundancy of the CYP90D2/D2 and CYP90D3 genes in rice.
3.6. Bioactive Brassinosteroids Accumulate in the Nipponbare-Derived d2 Mutants
To clarify what happens in the Nipponbare-derived d2 mutants, we compared the endogenous levels of brassinosteroids in d2-3 and d2-6 to those in wild-type Nipponbare. Because the most biologically active brassinosteroid, brassinolide, has been detected in Arabidopsis and some dicot plants but not in rice, Kim et al. [28] proposed that the precursor of brassinolide, castasterone, acts as the primary bioactive brassinosteroid in rice. D2 gene encodes a cytochrome P450 monooxygenase, CYP- 90D2, that catalyze C-23 hydroxylation of various 22- hydroxylated brassinosteroids [14]. The levels of 6-deoxoteasterone and teasterone, which are synthesized only by the C-23 hydroxylation of 6-deoxocathasterone and cathasterone, respectively, were decreased in d2 mutants (Table 1). These results are consistent with the fact that CYP90D2 act as brassinosteroid C-23 hydroxylases in rice. Surprisingly, castasterone did not decrease in d2-3 and d2-6; on the contrary, it accumulated slightly (Table 1). This result is opposite to the previous report that castasterone content decreased in d2-1 [13], and is inconsistent with the dwarf phenotype of the Nipponbare-derived d2 mutants if these mutants are simply caused by a brassinosteroid deficiency. Because accumulation of castasterone was observed in the brassinosteroid-insensitive dwarf mutants d61-2 and d61-4, which are caused by knock-down mutations of the rice brassinosteroid receptor gene OsBRI1 [9,10], the accumulation of castasterone in d2-3 and d2-6 plants suggests that these dwarf mutants are brassinosteroid-insensitive. However, as described above, exogenous brassinolide treatment rescued the dwarf phenotype of the Nipponbarederived d2 mutants, indicating that these mutants retain an ability to respond to bioactive brassinosteroids, at least when they are applied constitutively in excess amounts. Based on these results, we hypothesized that the biosynthesis of brassinosteroids and the response to these compounds are partially suppressed by the mutations in CYP90D2/D2 and by an unknown factor or unknown factors, respectively, in the Nipponbare-derived d2 mutants.
3.7. Brassinosteroid Response Is Partially Suppressed in the Nipponbare-Derived d2 Mutants
To confirm whether the response to brassinosteroids is partially suppressed in the Nipponbare-derived d2 mutants, we monitored the effect of brassinolide treatment on the expression of two brassinosteroid-responsive genes, CYP85A1 and BU1. CYP85A1 encodes brassinosteroid C-6 oxidase, and its expression is regulated by a homeostatic system that controls bioactive brassinosteroid levels; i.e. CYP85A1 expression was increased in the brassinosteroid-related mutants, and was decreased by the brassinolide treatment [11]. BU1 encodes a helixloop-helix protein that participates in the regulation of rice lamina inclination [29]. BU1 is thought to be a primary brassinosteroid-response gene, because brassinosteroids increase BU1 expression even in the presence of the protein synthesis inhibitor cycloheximide.
The level of CYP85A1 mRNA in brassinolide-treated Taichung 65 seedlings significantly decreased to about 35% of the untreated control level (Figure 6). Similarly, CYP85A1 expression decreased in brassinolide-treated d2-1 plants (to 43% of that in untreated d2-1), although the steady-state level of CYP85A1 mRNA in the absence of brassinolide treatment was significantly higher in d2-1 than in wild-type Taichung 65 (about 1.2 times that in untreated Taichung 65; Figure 6). This expression pattern of CYP85A1 in d2-1 plants can be explained by the homeostatic system as described above. In d61-1, a Taichung 65-derived brassinosteroid-insensitive mutant [9], CYP85A1 expression was also significantly higher than wild-type in the absence of brassinolide treatment (1.1 times that in untreated Taichung 65) and unaffected by brassinolide treatment (Figure 6). Expression of CYP- 85A1 in brassinolide-treated Nipponbare seedlings significantly decreased to about 44% of the untreated Nipponbare (Figure 6). Interestingly, the level of CYP85A1 mRNA in untreated d2-3, d2-4, and d2-6 was lower than that of Nipponbare (89%, 82%, and 84% of that in untreated Nipponbare, respectively), and was further decreased by brassinolide treatment (77%, 81%, and 81% of that in untreated mutants, respectively; Figure 6). This expression pattern of CYP85A1 in Nipponbare-derived d2 mutants cannot be explained simply by brassinosteroid-deficiency, as was the case of Taichung 65-derived
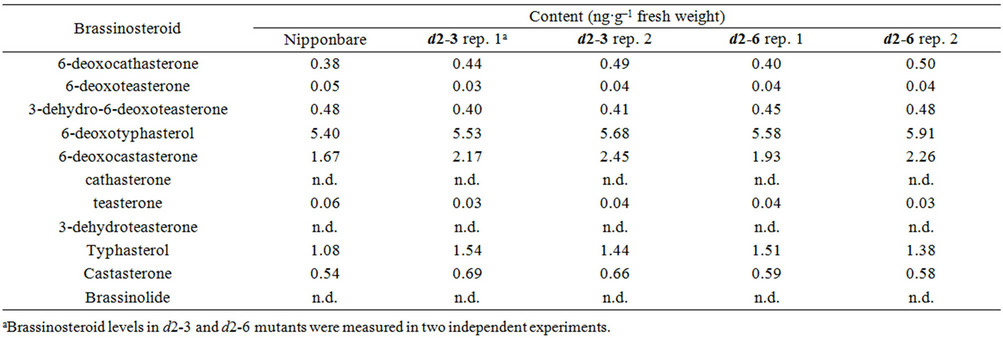
Table 1. Endogenous content of brassinosteroids in the wild-type nipponbare and d2 mutants.
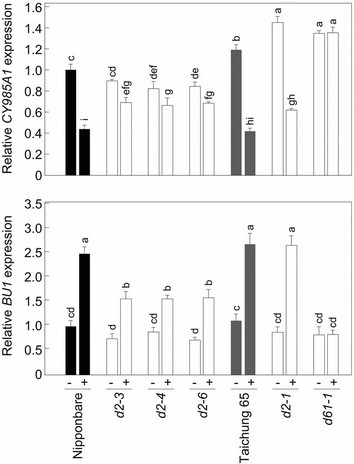
Figure 6. Expression of CYP85A1 and BU1 genes in the d2 mutants. We treated 2-week-old wild-type and mutant seedlings with 100 nM brassinolide (+) or a control solvent (ethanol, –) for 3 h. Expression levels were normalized against the values obtained for Histone H3. The values obtained from the wild-type Nipponbare seedlings treated with solvent were then normalized to a value of 1.0. Nipponbare is the wild-type for d2-3, d2-4, and d2-6; Taichung 65 is the wild-type for d2-1. Each column represents the mean ± s.d. of three biological repeats. Bars labeled with different letters differ significantly (The Holm-adjusted Student’s t test, P < 0.05).
d2-1. Because the bioactive brassinosteroid castasterone accumulated in Nipponbare-derived d2 mutants (Table 1), the expression of CYP85A1 may also be regulated directly by its catalytic product, castasterone.
The level of BU1 mRNA in Taichung 65 seedlings was significantly increased by brassinolide treatment (about 2.4 times that in untreated Taichung 65; Figure 6). Similarly, BU1 expression was increased by brassinolide treatment in the Taichung 65-derived d2-1 mutant (about 3.1 times that in untreated d2-1), reaching the same level as in the brassinolide-treated wild-type Taichung 65, although the steady-state level of BU1 mRNA in the absence of brassinolide treatment was slightly (but not significantly) lower in d2-1 than in Taichung 65 (about 78% that in Taichung 65; Figure 6). This expression pattern of BU1 in d2-1 plants can be explained by brassinosteroid deficiency. In d61-1, BU1 expression slightly (but not significantly) decreased both with and without brassinolide treatment (about 74% that in Taichung 65; Figure 6). Expression of BU1 in the Nipponbare seedlings was also significantly increased by brassinolide treatment both in the wild-type (about 2.5 times that in untreated plants) and in the d2-3, d2-4, and d2-6 mutants (about 2.1, 1.8, and 2.2 times that in untreated plants, respectively; Figure 6). Although, the steady-state levels of BU1 mRNA in all three Nipponbare-derived d2 mutants were lower than that of wild-type Nipponbare (74%, 88%, and 72% of that in untreated Nipponbare, respectively), and the increase in BU1 expression caused by the brassinolide treatment was to lower level in these mutants than in wild-type Nipponbare. These results support our hypothesis that the brassinosteroid response is partially suppressed in the Nipponbare-derived d2 mutants.
The difference in the severity of the mutant phenotype between the Taichung 65-derived d2-1 mutant and the Nipponbare-derived d2 mutants (d2-3, d2-4, and d2-6) can be explained by the finding that d2-1 showed a typical brassinosteroid-deficient phenotype, which was confirmed by the endogenous brassinosteroid content and expression of brassinosteroid-response genes, whereas d2-3, d2-4, and d2-6 represent partial suppression of the brassinosteroid response in addition to a brassinosteroid deficiency. Because our linkage analysis and complementation test clearly indicated that the mutant phenoltypes in d2-3, d2-4, and d2-6 plants were caused by defects in the CYP90D2/D2 gene, we speculated that the parent of the d2-3, d2-4, and d2-6 plants, Nipponbare, has one or more defects in its brassinosteroid response. Although double mutants of brassinosteroid-biosynthesis and -response genes have been generated in Arabidopsis [30,31], details of what happened in these double mutants (i.e., the levels of endogenous brassinosteroids, levels and patterns of expression of brassinosteroid-response genes) have not been described. At present, we cannot determine what factor or factors affect the brassinosteroid response in a Nipponbare background, but our results suggest that the brassinosteroid-stimulated growth responses in rice will differ among genetic backgrounds and that possible changes in cellular sensitivity to brassinosteroids must be considered when the effects of brassinosteroids on the regulation of various growth and developmental processes are examined in different rice cultivars. We predict that isolation and characterization of the unknown factor or factors will help us to understand how brassinosteroids regulate rice growth and development and will let us utilize the genes related to biosynthesis of and responses to these phytohormones to improve rice crop production.
4. Conclusion
In this study, we characterized three new alleles of the rice d2 mutation. In contrast to the previously isolated null allele derived from Taichung 65, d2-1, which shows a semi-dwarf phenotype combined with a typical brassinosteroid-deficient phenotype, Nipponbare-derived d2-3, d2-4, and d2-6 plants showed severe dwarfing and partial suppression of the brassinosteroid response, in addition to a brassinosteroid deficiency. Because the mutant phenotypes in d2-3, d2-4, and d2-6 are caused by defects in the CYP90D2/D2 gene, we hypothesize that their wildtype parent, Nipponbare, has one or more defects in unknown factors related to the response of rice to brassinosteroids.
5. Acknowledgements
We thank Prof. Suguru Takatsuto (Joetsu University of Education) for supplying the deuterium-labeled internal standards, and Asako Tokida-Segawa for technical assistance. T.S. was supported by Grants-in-Aid for Young Scientists (nos. 19688001 and 24780005) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. S.F. was supported by Grantsin-Aid for Scientific Research (B) (nos. 19380069 and 23380066) from MEXT.
REFERENCES
- S. D. Clouse and J. M. Sasse, “Brassinosteroids: Essential Regulators of Plant Growth and Development,” Annual Review of Plant Physiology and Plant Molecular Biology, Vol. 49, No. 1, 1998, pp. 427-451. doi:10.1146/annurev.arplant.49.1.427
- J. M. Sasse, “Physiological Action of Brassinosteroids: An Update,” Journal of Plant Growth Regulation, Vol. 22, No. 4, 2003, pp. 276-288. doi:10.1007/s00344-003-0062-3
- P. Krishna, “Brassinosteroid-Mediated Stress Responses,” Journal of Plant Growth Regulation, Vol. 22, No. 4, 2003, pp. 289-297. doi:10.1007/s00344-003-0058-z
- H. Nakashita, M. Yasuda, T. Nitta, T. Asami, S. Fujioka, Y. Arai, K. Sekimata, S. Takatsuto, I. Yamaguchi and S. Yoshida, “Brassinosteroid Functions in a Broad Range of Disease Resistance in Tobacco and Rice,” Plant Journal, Vol. 33, No. 5, 2003, pp. 887-898. doi:10.1046/j.1365-313X.2003.01675.x
- S. Koh, S. C. Lee, M. K. Kim, J. H. Koh, S. Lee, G. An, S. Choe and S. R. Kim, “T-DNA Tagged Knockout Mutation of Rice OsGSK1, an Orthologue of Arabidopsis BIN2, with Enhanced Tolerance to Various Abiotic Stresses,” Plant Molecular Biology, Vol. 65, No. 4, 2007, pp. 453- 466. doi:10.1007/s11103-007-9213-4
- T. Sakamoto, Y. Morinaka, T. Ohnishi, H. Sunohara, S. Fujioka, M. Ueguchi-Tanaka, M. Mizutani, K. Sakata, S. Takatsuto, S. Yoshida, H. Tanaka, H. Kitano and M. Matsuoka, “Erect Leaves Caused by Brassinosteroid Deficiency Increase Biomass Production and Grain Yield in Rice,” Nature Biotechnology, Vol. 24, No. 1, 2006, pp. 105-109. doi:10.1038/nbt1173
- Y. Morinaka, T. Sakamoto, Y. Inukai, M. Agetsuma, H. Kitano, M. Ashikari and M. Matsuoka, “Morphological Alteration Caused by Brassinosteroid Insensitivity Increases the Biomass and Grain Production of Rice,” Plant Physiology, Vol. 141, No. 3, 2006, pp. 924-931. doi:10.1104/pp.106.077081
- C. Y. Wu, A. Trieu, P. Radhakrishnan, S. F. Kwok, S. Harris, K. Zhang, J. Wang, J. Wan, H. Zhai, S. Takatsuto, S. Matsumoto, S. Fujioka, K. A. Feldmann and R. I. Pennell, “Brassinosteroids Regulate Grain Filling in Rice,” Plant Cell, Vol. 20, No. 8, 2008, pp. 2130-2145. doi:10.1105/tpc.107.055087
- C. Yamamuro, Y. Ihara, X. Wu, T. Noguchi, S. Fujioka, S. Takatsuto, M. Ashikari, H. Kitano and M. Matsuoka, “Loss of Function of a Rice brassinosteroid insensitive1 Homolog Prevents Internode Elongation and Bending of the Lamina Joint,” Plant Cell, Vol. 12, No. 9, 2000, pp. 1591-1606.
- A. Nakamura, S. Fujioka, H. Sunohara, N. Kamiya, Z. Hong, Y. Inukai, K. Miura, S. Takatsuto, S. Yoshida, M. Ueguchi-Tanaka, Y. Hasegawa, H. Kitano and M. Matsuoka, “The Role of OsBRI1 and Its Homologous Genes, OsBRL1 and OsBRL3, in Rice,” Plant Physiology, Vol. 140, No. 2, 2006, pp. 580-590. doi:10.1104/pp.105.072330
- Z. Hong, M. Ueguchi-Tanaka, S. Shimizu-Sato, Y. Inukai, S. Fujioka, Y. Shimada, S. Takatsuto, M. Agetsuma, S. Yoshida, Y. Watanabe, S. Uozu, H. Kitano, M. Ashikari and M. Matsuoka, “Loss-of-Function of a Rice Brassinosteroid Biosynthetic Enzyme, C-6 Oxidase, Prevents the Organized Arrangement and Polar Elongation of Cells in the Leaves and Stem,” Plant Journal, Vol. 32, No. 4, 2002, pp. 495-508. doi:10.1046/j.1365-313X.2002.01438.x
- M. Mori, T. Nomura, H. Ooka, M. Ishizaka, T. Yokota, K. Sugimoto, K. Okabe, H. Kajiwara, K. Satoh, K. Yamamoto, H. Hirochika and S. Kikuchi, “Isolation and Characterization of a Rice Dwarf Mutant with a Defect in Brassinosteroid Biosynthesis,” Plant Physiology, Vol. 130, No. 3, 2002, pp. 1152-1161. doi:10.1104/pp.007179
- Z. Hong, M. Ueguchi-Tanaka, K. Umemura, S. Uozu, S. Fujioka, S. Takatsuto, S. Yoshida, M. Ashikari, Kitano and M. Matsuoka, “A Rice Brassinosteroid-Deficient Mutant, ebisu dwarf (d2), Is Caused by a Loss of Function of a New Member of Cytochrome P450,” Plant Cell, Vol. 15, No. 12, 2003, pp. 2900-2910. doi:10.1105/tpc.014712
- T. Sakamoto, T. Ohnishi, S. Fujioka, B. Watanabe and M. Mizutani, “Rice CYP90D2 and CYP90D3 Catalyze C-23 Hydroxylation of Brassinosteroids in Vitro,” Plant Physiology and Biochemistry, Vol. 58, 2012, pp. 220-226. doi:10.1016/j.plaphy.2012.07.011
- S. Tanabe, M. Ashikari, S. Fujioka, S. Takatsuto, S. Yoshida, M. Yano, A. Yoshimura, H. Kitano, M. Matsuoka, Y. Fujisawa, H. Kato and Y. Iwasaki, “A Novel Cytochrome P450 Is Implicated in Brassinosteroid Biosynthesis via the Characterization of a Rice Dwarf Mutant, dwarf11, with Reduced Seed Length,” Plant Cell, Vol. 17, No. 3, 2005, pp. 776-790. doi:10.1105/tpc.104.024950
- Z. Hong, M. Ueguchi-Tanaka, S. Fujioka, S. Takatsuto, S. Yoshida, Y. Hasegawa, M. Ashikari, H. Kitano and M. Matsuoka, “The Rice brassinosteroid-deficient dwarf2 Mutant, Defective in the Rice Homolog of Arabidopsis DIMINUTO/DWARF1, Is Rescued by the Endogenously Accumulated Alternative Bioactive Brassinosteroid, Dolichosterone,” Plant Cell, Vol. 17, No. 8, 2005, pp. 2243- 2254. doi:10.1105/tpc.105.030973
- A. Sakurai, “Brassinosteroid Biosynthesis,” Plant Physiology and Biochemistry, Vol. 37, No. 5, 1999, pp. 351- 361. doi:10.1016/S0981-9428(99)80041-2
- S. Fujioka, T. Noguchi, T. Watanabe, S. Takatsuto and S. Yoshida, “Biosynthesis of Brassinosteroids in Cultured Cells of Catharanthus roseus,” Phytochemistry, Vol. 53, No. 5, 2000, pp. 549-553. doi:10.1016/S0031-9422(99)00582-8
- T. Noguchi, S. Fujioka, S. Choe, S. Takatsuto, F. E. Tax, S. Yoshida and K. A. Feldmann, “Biosynthetic Pathways of Brassinolide in Arabidopsis,” Plant Physiology, Vol. 124, No. 1, 2000, pp. 201-210. doi:10.1104/pp.124.1.201
- S. Fujioka, S. Takatsuto and S. Yoshida, “An Early C-22 Oxidation Branch in the Brassinosteroid Biosynthetic Pathway,” Plant Physiology, Vol. 130, No. 2, 2002, pp. 930-939. doi:10.1104/pp.008722
- S. Fujita, T. Ohnishi, B. Watanabe, T. Yokota, S. Takatsuto, S. Fujioka, S. Yoshida, K. Sakata and M. Mizutani, “Arabidopsis CYP90B1 Catalyzes the Early C-22 Hydroxylation of C27, C28, and C29 Sterols,” Plant Journal, Vol. 45, No. 5, 2006, pp. 765-774. doi:10.1111/j.1365-313X.2005.02639.x
- H. Hirochika, K. Sugimoto, Y. Otsuki, H. Tsugawa and M. Kanda, “Retrotransposons of Rice Involved in Mutations Induced by Tissue Culture,” Proceedings of the National Academy of Sciences of the United States of America, Vo. 93, No. 15, 1996, pp. 7783-7788. doi:10.1073/pnas.93.15.7783
- Y. Hiei, S. Ohta, T. Komari and T. Kumashiro, “Efficient Transformation of Rice (Oryza sativa L.) Mediated by Agrobacterium and Sequence Analysis of Boundaries of the T-DNA,” Plant Journal, Vol. 6, No. 2, 1994, pp. 271- 282. doi:10.1046/j.1365-313X.1994.6020271.x
- N. Jiang, Z. Bao, X. Zhang, H. Hirochika, S. R. Eddy, S. R. McCouch and S. R. Wessler, “An Active DNA Transposon Family in Rice,” Nature, Vol. 421, No. 6919, 2003, pp. 163-167. doi:10.1038/nature01214
- K. Kikuchi, K. Terauchi, M. Wada and H. Y. Hirano, “The Plant MITE mPing Is Mobilized in Anther Culture,” Nature, Vol. 421, No. 6919, 2003, pp. 167-170. doi:10.1038/nature01218
- T. Nakazaki, Y. Okumoto, A. Horibata, S. Yamahira, M. Teraishi, H. Nishida, H. Inoue and T. Tanisaka, “Mobilization of a Transposon in the Rice Genome,” Nature, Vol. 421, No. 6919, 2003, pp. 170-172. doi:10.1038/nature01219
- Y. Ohmori, M. Abiko, A. Horibata and H. Y. Hirano, “A Transposon, Ping, Is Integrated into Intron 4 of the DROOPING LEAF Gene of Rice, Weakly Reducing Its Expression and Causing a Mild Drooping Leaf Phenotype,” Plant and Cell Physiology, Vol. 49, No. 8, 2008, pp. 1176-1184. doi:10.1093/pcp/pcn093
- B. K. Kim, S. Fujioka, S. Takatsuto, M. Tsujimoto and S. Choe, “Castasterone Is a Likely End Product of Brassinosteroid Biosynthetic Pathway in Rice,” Biochemical and Biophysical Research Communications, Vol. 374, No. 4, 2008, pp. 614-619. doi:10.1016/j.bbrc.2008.07.073
- A. Tanaka, H. Nakagawa, C. Tomita, Z. Shimatani, M. Ohtake, T. Nomura, C. J. Jiang, J. G. Dubouzet, S. Kikuchi, H. Sekimoto, T. Yokota, T. Asami, T. Kamakura and M. Mori, “BRASSINOSTEROID UPREGULATED1, Encoding a Helix-Loop-Helix Protein, Is a Novel Gene Involved in Brassinosteroid Signaling and Controls Bending of the Lamina Joint in Rice,” Plant Physiology, Vol. 151, No. 2, 2009, pp. 669-680. doi:10.1104/pp.109.140806
- J. Li, K. A. Lease, F. E. Tax and J. C. Walker, “BRS1, a Serine Carboxypeptidase, Regulates BRI1 Signaling in Arabidopsis thaliana,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 98, No. 10, 2001, pp. 5916-5921. doi:10.1073/pnas.091065998
- J. Li, J. Wen, K. A. Lease, J. T. Doke, F. E. Tax and J. C. Walker, “BAK1, an Arabidopsis LRR Receptor-Like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling,” Cell, Vol. 110, No. 2, 2002, pp. 213-222. doi:10.1016/S0092-8674(02)00812-7
NOTES
*Corresponding author.

