American Journal of Plant Sciences
Vol.3 No.2(2012), Article ID:17552,7 pages DOI:10.4236/ajps.2012.32027
The Pathogen and Wound Induces Expression of Genes Related to Proanthocyanidins (PAs) Synthesis in Cotton Leaves

1The College of Life Science, Chongqing Normal University, Chongqing, China; 2Key Laboratory of Eco-Environments in Three Gorges Reservoir Region (Ministry of Education), School of Life Science, South-West University, Chongqing, China.
Email: *yangxy94@swu.edu.cn
Received October 11th, 2011; revised November 15th, 2011; accepted November 25th, 2011
Keywords: Cotton; Flavanone 3-Hydroxylase; Proteomics; Verticillium dahliae
ABSTRACT
Responses to biotic and abiotic stress have been extensively studied in plants. In the current proteomic study, the cotton (Gossypium hirsutum L.) seedlings were infected with Verticillium dahliae by root-dip inoculation using suspension of fungal conidia. The different proteins were analyzed by two-dimensional gel elactrophoresis (2-DE), and flavanone 3-hydroxylase (F3H) showed a significantly up-regulation in cotton leaf after V. dahliae infection. Further research revealed F3H and the downstream genes of F3H in proanthocyanidins (PAs) biosynthesis were also significantly induced and showed coordinate expression patterns during wounding. The results indicate that PAs in cotton act an important role in response to infection V. dahliae and wounding.
1. Introduction
Proanthocyanidins (PAs), also called condensed tannins, are a class of flavanols that result from the condensation of flavan-3-ol units and are oligomeric or polymeric flavonoid end products of the flavonoid pathway [1].
Research has shown that PAs perform many physiological functions. In plants, the major function of PAs is to provide protection against microbial pathogens, insect pests and larger herbivores [2]. These can be mostly attributed to free radical scavenging and antioxidant activities of PAs [3]. In our research, flavanone 3-hydroxylase (F3H) significantly increased after cotton (Gossypium hirsutum L.) was infected by Verticillium dahliae and by wounding, and further research revealed the relationship of up-regulated expression level of F3H and the biosynthesis of PAs during wounding.
2. Materials and Methods
2.1. Conidial Inoculum, Plant Growth, and Inoculation System
V. dahliae was cultured in potato dextrose broth (PDB) (200 g potato, 20 g glucose, distilled water 1 L) at 25˚C for 20 days. Germinated upland cotton seeds were grown in sterile vermiculite under artificial light (12 h photoperiod at 500 μmole·m–2·s–1) at 25˚C and watered with auto claving Murashige and Skoog basal liquid medium (MS) [4] every 3 days. At 4th true leaf stage, the seedlings were infected with V. dahliae using root-dip inoculation into suspension of fungal conidia [5]. Cotton roots were slightly injured with a knife to ensure infection efficiency before root-dip inoculation. The 100 mL of a suspension of 106 conidia/mL was poured in the sterile vermiculite. The seedlings were incubated at 25˚C (12 h photoperiod), harvested at 0 h, 24 h and 72 h after inoculation and rinsed in deionized water. For wounding experiments, a strip of cotton stems were slightly scraped away using knife to simulate wounding.
2.2. Protein Extraction, 2-D Gel Electrophoresis and Gel Imaging
The samples were placed in a prechilled mortar and ground to a fine powder in liquid nitrogen. The finely ground powder (~0.5 g per tube) was immediately transferred into a 10 mL centrifuge tube and weighed, then stored in a –80˚C freezer until protein extraction. Protein extraction was carried out based on our previously established effective protein extraction method [6] with some modification. Briefly, the grounded sample was resuspended in 3 mL of extraction buffer (0.1 M Tris-HCl, pH 8.0, containing 30% w/v sucrose, 2% w/v SDS, 1 mM PMSF, 2% v/v β-mercaptoethanol), and then an equal volume of 1 M Tris-saturated phenol (pH 8.0) was added. The mixture was then homogenized on ice for 5 min. The upper phenol phase was collected after centrifugation at 12,000 g for 10 min (4˚C), and this extraction step was repeated once. The total phenol phase was extracted with an equal volume of the above extraction buffer. The upper phenol phase was then precipitated with five volumes of 0.1 M ammonium acetate in methanol overnight at –20˚C. After centrifugation at 12,000 g for 5 min (4˚C), the collected protein pellets were washed once with 3 mL of methanol and then washed once with 3 mL of cold 80% acetone (v/v) in water. The pellets were dried in a freeze vacuum dryer, and then resuspended in lysis buffer. The protein concentration was determined using the Bradford method [7] with bovine serum albumin as a standard. First dimensional electrophoresis was performed using immobilized pH gradient (IPG) on an IPGphor IEF system (Amersham Pharmacia, San Francisco, CA). The second dimensional electrophoresis (SDS-PAGE) was performed on Amersham Hoefer SE600 system (Amersham Pharmacia) with 10 mA for 1 h and 20 mA for 6 h at 15˚C. The 2-DE gels were stained using blue silver [8]. The gels were scanned using an image scanner (Amersham Pharmacia) following the user’s manual. To control for subtle differences in sample loading and gel staining, three replicates gels per time point were performed, and the volume of each spot was normalized to generate a relative volume. Automated spot matching was performed using Image Master 2D Platinum Version 5.0 Analysis Software (Amersham Pharmacia), and significantly (p < 0.05) altered spots were identified with the Student’s t-test feature of the software.
2.3. Protein Identification and Database Search
Spots of interest were cut out of the gels, and then the gel particles were transferred to a microcentrifuge tube. The gel particles were washed twice with deionized water, and 50 μl of decolorized solution [50 mM NH4HCO3/ CH3CN (1:1)] was added and the mixture was votexed for 20 min at 37˚C, repeated decolorization step until blue color disappeared. Shrink the gel particles with 50 μl acetonitrile until they become white. Dry the gels particles in a vacuum centrifuge for 10 min, and then mix the gel particles with 20 μl solution of 10 mM DTT in 25 mM NH4HCO3 for 1 h at 56˚C to reduce the proteins. At ambient temperature, the gels were dried in a vacuum centrifuge for 30 min prior to incubating with 20 μl alkylation solution (50 mM iodoacetamide in 25 mM NH4HCO3) for 45 min in the dark. The several wash step were performed as follows: 25 mM NH4HCO3 (2 × 10 min), 25 mM NH4HCO3 in 50% CH3CN (2 × 10 min), CH3CN (10 min). The gel particles were completely dried in a vacuum centrifuge for 10 min, and were enzymatic digested at 37˚C overnight with 5 ng/mL trypsin. Digested peptides were extracted with 0.1% TFA in 50% CH3CN and analyzed by Ultraflex TOF/TOF (Bruker, Germany). The results of mass spectra were submitted online to the Mascot search engine for searching against various databases [9].
2.4. RNA Isolation and RT-PCR
The total RNAs were extracted using a modified cetyltriethylammnonium bromide (CTAB) method [10]. First strand cDNA was synthesized from 1 μg of total RNA using reverse transcription system (Promega, USA). The gene-specific primers are showed in the Table 1. The quantitative RT-PCR was performed using iQ™ SYBR Green Supermix (Bio-RAD, USA) in the iQ™5 RealTime PCR Detection System (Bio-RAD, USA). To ensure reproducibility, the experiment was carried out three times. The semi-quantitative RT-PCR was performed in a 25 μl reaction mixture containing 1 μl cDNA solution and 10 μM primers. The number for PCR amplification was conducted for 25 cycles.
2.5. Extraction and Relative Quantification of PAs
Extraction and Quantification of soluble PAs was carried out according to Pang’s method [11] with some modification. Briefly, the leaf tissues were ground in liquid nitrogen and 0.1 g samples were extracted with 5 mL of extraction solution (70% acetone: 0.5% acetic acid) by vortexing followed sonication for 1 h. After centrifugation, residues were re-extracted once as above. Pooled supernatants were extracted three times with 30 mL of chloroform and hexane respectively, the volume of supernatants was determined using a graduated cylinder. The soluble PAs content was determined using dimethylaminocinnamaldehyde (DMACA) reagent. Briefly,
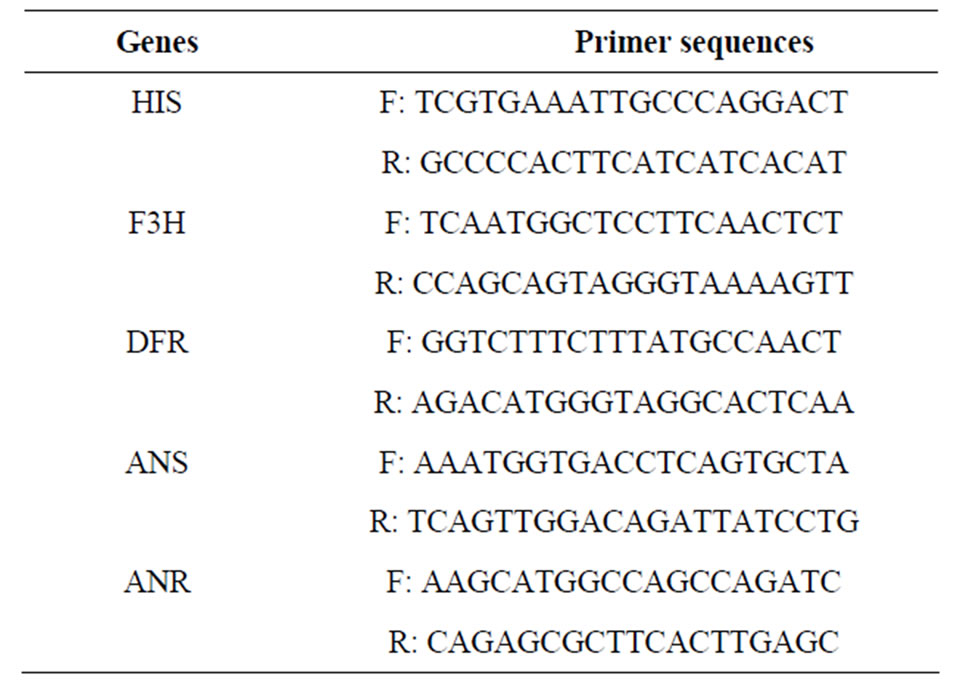
Table 1. Primer sequences used in this study.
20 μl samples were mixed with 20 μl of DMACA reagent [0.2% (w/v) DMACA in methanol-3 M HCl (1:1)], for blanks, the samples were replaced with 20 μl ddH2O. Absorption of the mixture was measured within 15 min at 640 nm using a spectrophotometer.
3. Results and Discussion
3.1. The Up-Regulated F3H after the Infection of V. dahliae
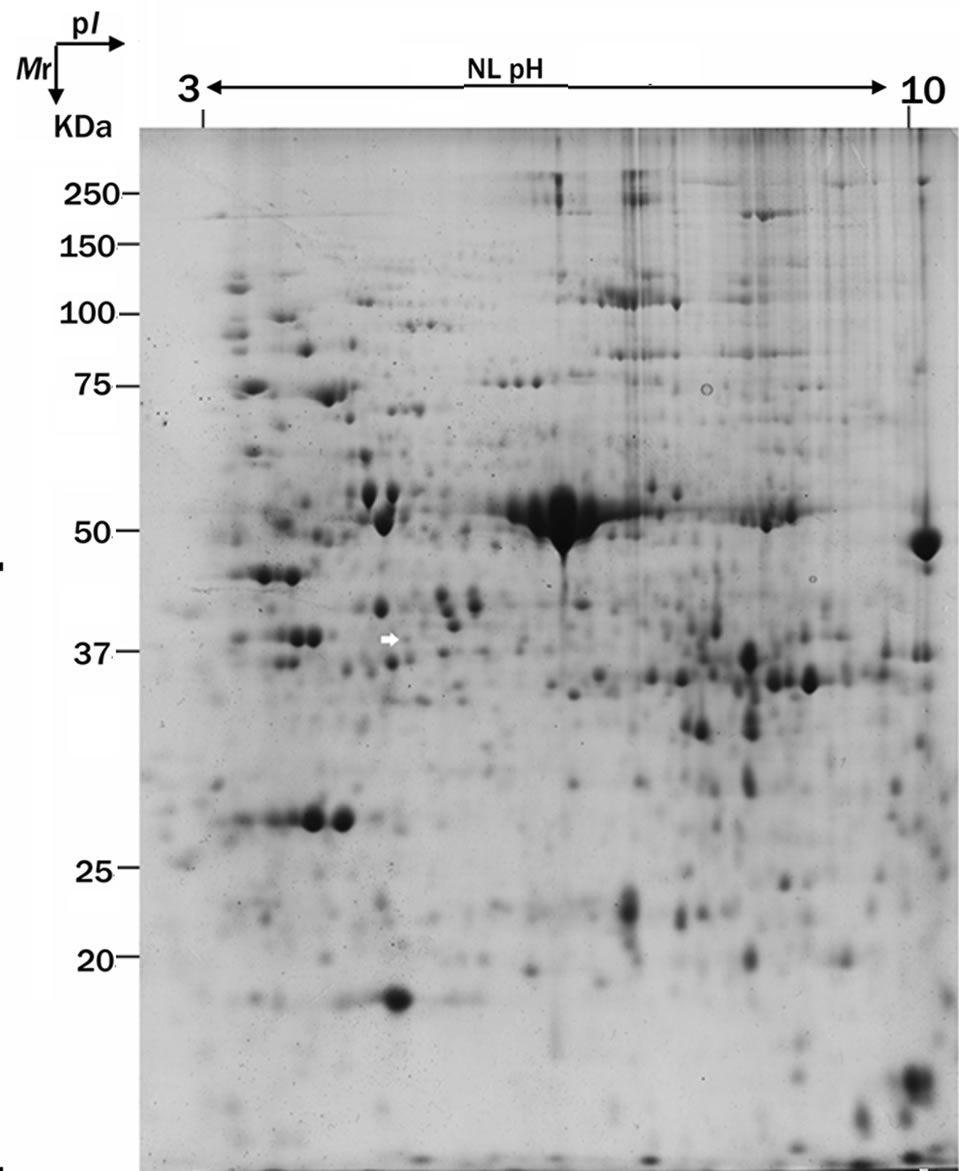
In order to study the interactions between cotton seedling and V. dahliae, we establish a proteomic analysis method compatible with cotton [6], and a well-resolved 2-DE gel pattern in the pI range of non-linear 3 - 10 was established (Figure 1(a)). Differential expression of protein profiles was analyzed at several time points (0 h, 24 h and 72 h) after cotton tissues were infected.
One of the different proteins showed significantly up-regulation pattern (Figure 1(b)), and at 24 h and 72 hits expression levels increased 1.8 and 2.5 times, respectively, compared with 0 h after inoculation (Figure 1(c)). The protein was identified as flavanone 3-hydroxylase (F3H) by peptide mass fingerprinting and searched on the Mascot. F3H is one of the key enzymes acting in the biosynthesis of flavonoids, and converts (2S)-flavanones to (2R, 3R)-dihydroflavonols that are the direct intermediates in the biosynthesis of flavonols [12]. Several studies have suggested that F3H gene expression is associated with resistance in plant [13], and activity of F3H was higher in resistant cultivars than susceptible ones [14].
3.2. Transcript Accumulation of F3H in Leaf in Response to Wound
In order to further verify that the F3H expression pattern is a response to the infection of V. dahliae, quantitative RT-PCR assays was used. As expected, the expression patterns of genes were similar to the corresponding protein, and F3H mRNA expression was highest at 24 h followed by gradual down-regulation. Obviously, the F3H show temporal expression patterns during infection of V. dahliae. An unexpected result, however, was observed in control experiments using the sterile distilled water instead of V. dahliae conidia, and a significant up-regulated F3H was also observed in the control plant at 24 h (Figure 2(a)). One explanation for the unexpected result is that F3H gene expression is induced by wounding. The hypothesis was based on the followed reason: in order to ensure the V. dahliae infection efficiency by the rootdrenching method, cotton roots are slightly injured with a knife in our inoculation experiment. To validate our hypothesis, a strip of cotton stems were slightly scraped away using a knife to simulate the wounding experiment, and a leaf was quickly stored in –80˚C freezer as control. After 24 h of treatment, the other leaf was harvested. The
 (a)
(a) (b)
(b)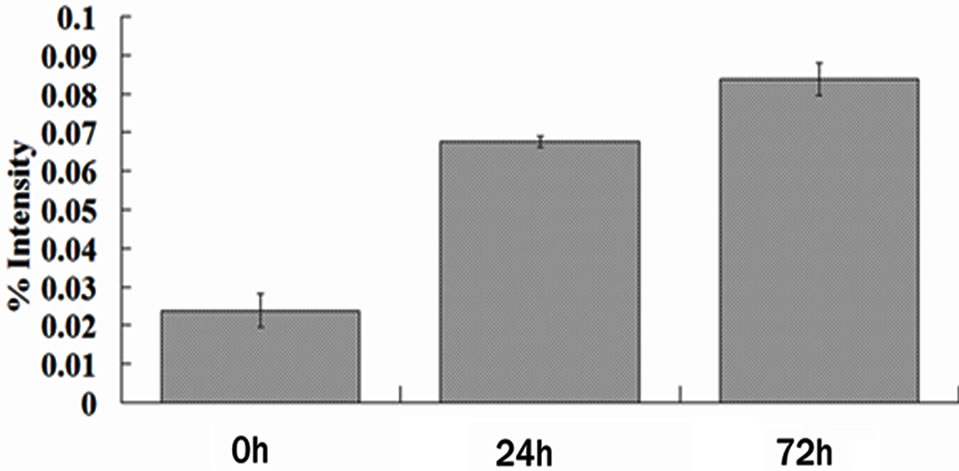 (c)
(c)
Figure 1. F3H protein expression is up-regulated in the cotton leaf after the infection by V. dahliae. (a) Represents 2-DE patterns of proteins from cotton (4th leaves of seedlings) inoculated with V. dahliae (72 h). White arrow indicates the position of F3H protein; (b) Magnified views of the up-regulated F3H in the cotton after the infection by V. dahliae; (c) Average expression intensity of normalized protein was calculated, and results are mean ± SE from three independent experiments.
 (a)
(a)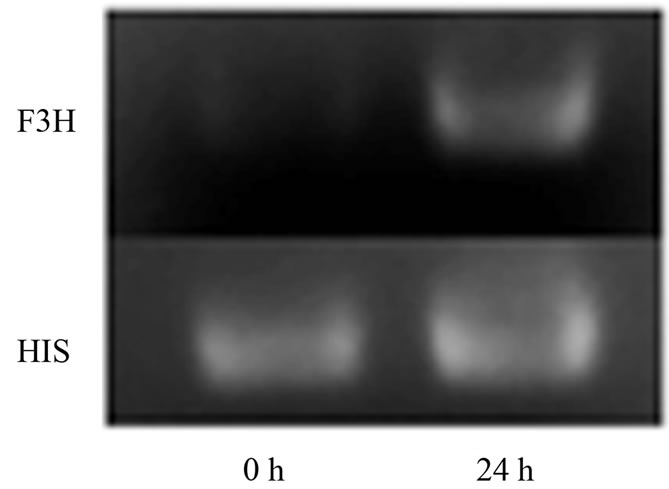 (b)
(b)
Figure 2. Differential expression of F3H using RT-qPCR and semi-quantitative RT-PCR. (a) Results of real-time PCR exprssion analysis of F3H gene, and F3H gene showed temporally up-regulated expression pattern after the infection by V. dahliae, and the control plants were inoculated with sterile water and harvested at 72 h; (b) Expression analysis of F3H in mock wounding leaf using semi-quantitative RTPCR, and F3H showed significant up-regulation in the leaf of wounded plant.
relative expression level of F3H gene was detected by RT-PCR. The result is consistent with the hypothesis that the F3H gene can be induced by wounding, and a significant up-regulation was observed in the wounded plant (Figure 2(b)). The result clearly indicated that the F3H is a wounding inducible gene. Several previous studies have reported the F3H gene was induced by sugars [15] and ultraviolet-B irradiation [16] and pathogens [13,14].
3.3. The Up-Regulated Expression of Genes Involved in Pas Biosynthesis in Response to Wounding
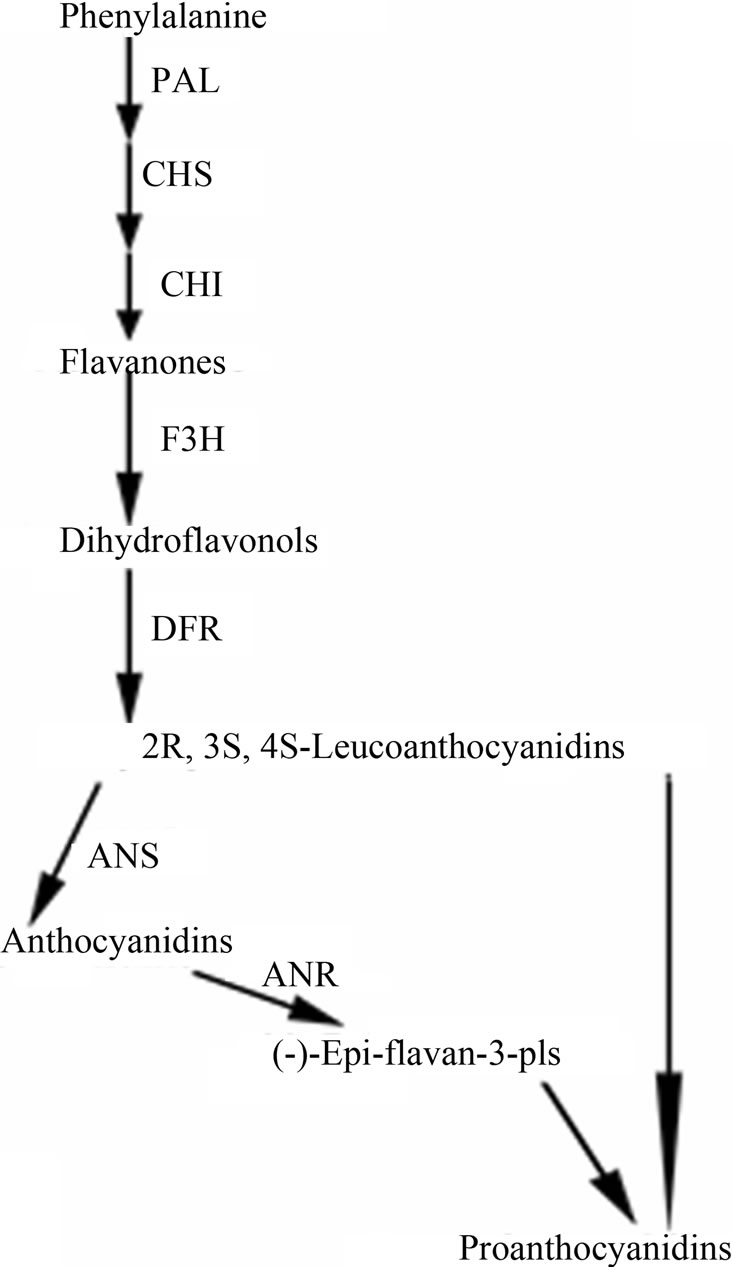
Plants anchor to the ground through the root system for acquisition of nutrients and water, and thus are inevitably injured by any possible mechanism wound such as wind, sand, hail, and rain [17]. Plants respond to mechanical wounding have been extensively studied, and the first identified wound-inducible defense proteins in plants include proteinase inhibitors I and II from potato and tomato [18,19]. In recent advances in plants wound signaling, the central role of jasmonic acid in plant responses to wounding is well established [17,20,21]. Furthermore, other compounds, including the oligopeptide systemin, oligosaccharides, and other phytohormonesalso play a important role in wound response [20]. F3H showed a wound-induced gene in this study, however F3H is merely a “core” enzyme of the flavonoid pathway, and its downstream metabolic products include anthocyanins and PAs [22]. Anthocyanins are responsible for the red, blue and purple coloration of many organs of angiosperms [23], and PAs play a protective function against microbial pathogens, insect pests and larger herbivores in plants [2]. The genes, involved in F3H downstream pathway in cotton, have been cloned [24] (Figure 3(a)), and their expression levels were detected at 24 h after wounding by RT-PCR analysis (primers are shown in Table 1). With the exception of dihydroflavonol-4- reductase (DFR), the other two genes, anthocyanidin synthase (ANS) and anthocyanidin reductase (ANR), both showed significantly up-regulated expression patterns (Figure 3(b)). DFR is involved in multi-step reactions, and research have reported that AtDFR was not activated by PAs biosynthesis related transcription factor [25]. The ANS catalyzes the last step in the biosynthesis of the anthocyanin from the colorless leucoanthocyanidins to the colored anthocyanidins [26]. The ANR converts anthocyanidins to (-)-Epi-flavan-3-ols, and ectopic expression of ANR in tobacco flower petals and Arabidopsis leaves results in loss of anthocyanins and accumulation of PAs [27]. The result of PAs content also showed that PAs transiently accumulate after wounding (the content is related to growth status of the leaf, with a younger leaf having higher content of PAs) (Figure 3(c)). We have also measured the content of anthocyanin in wounded leaves; however no significant regularity was found (data not shown). Taken together, our results imply that the up-regulated genes were involved in PAs biosynthesis. Robin et al. [28] have showed that a transcription factor, MYB134 activats PAs biosynthetic related genes expression leading to PAs accumulation in poplar following mechanical wounding, pathogen infection, and ultraviolet B irritation. In the present study, our results suggest that PAs biosynthesis related genes were significantly induced and showed coordinate expression patterns during wounding.
In current study, we first identified a significantly different expression protein (F3H) in cotton leaf after infection of V. dahliae, and the mRNA expression level of F3H was consistent with protein level. A significant up-regulation, however, was observed in control plant at 24 h, and then we demonstrate that F3H is a wound-induced gene. The downstream genes of F3H, involved in PAs biosynthesis, also are significantly up-regulated in cotton after wounding. Some investigators have demonstrated that several steps of the F3H downstream pathway were coordinately regulated by conserved transcription factors such as bHLH and MYB [25,29], which are
 (a)
(a)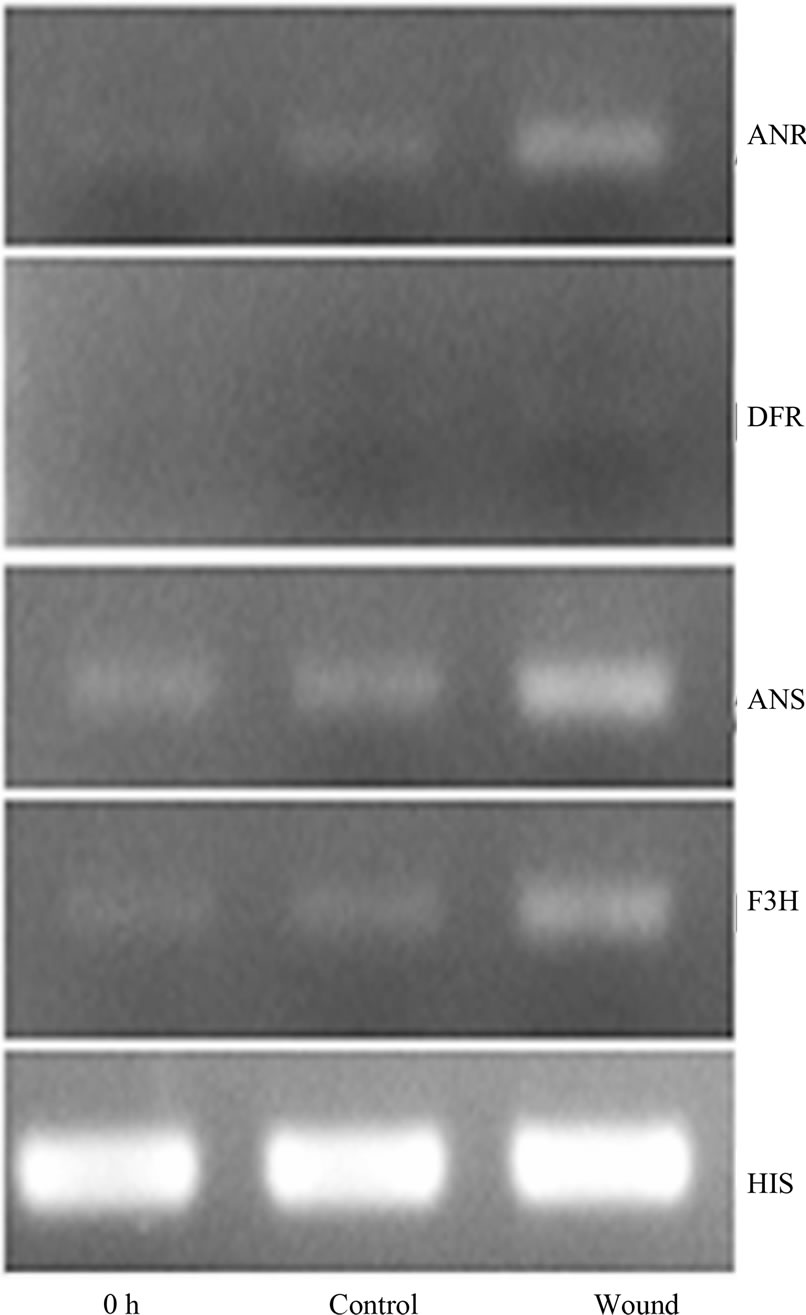 (b)
(b)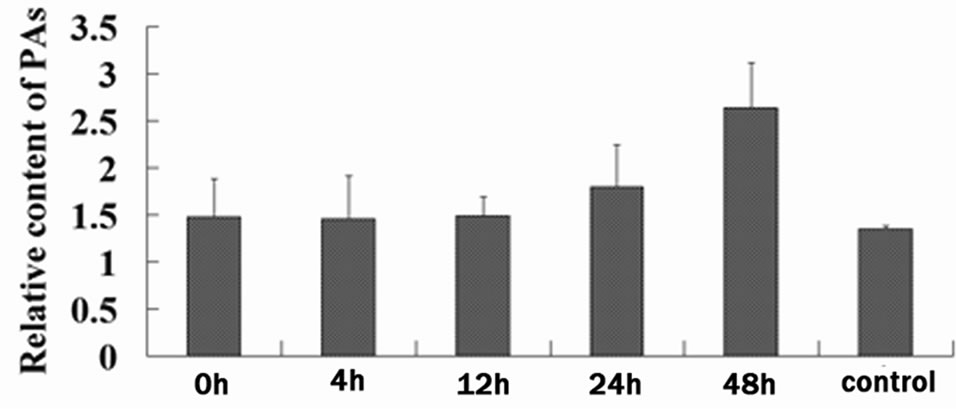 (c)
(c)
Figure 3. Wound-induced activation of the PAs biosynthetic pathway. (a) Schematic of the major branch pathways of PA biosynthesis. Enzyme names are abbreviated as follows: phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol-4-reductase (DFR), anthocyanidin synthase (ANS) and anthocyanidin reductase (ANR); (b) Expression of F3H, ANS and ANR represented significant up-regulation after the wounding treatment of cotton. RTPCR was conducted with the cotton histone (HIS) gene as a control. PCR was conducted for 25 cycles; (c) PAs in cotton leaf was temporally up-regulated after mock wounding treatment, average content was calculated, and results are mean ± SE from four independent experiments.
consistent with our experiments. PAs have an extensively physiologic effect on plants, animals and human, however, the specific mechanism has been not verified. The oxidative burst generated in response to wounding can be detected at wound sites and in distal leaf veins within 1 h after wounding, and the reactive oxygen species (ROS) may been capable to induce the metabolic cascade for active defense as signaling molecule [30,31].
However, as a substrate for oxidative, they are also capable of injuring cells. Thus, the steady-state level of ROS in cells needs to be tightly regulated [32]. Others have suggested that the PAs can help to prevent oxidative stress and may reduce ROS [33]. Taken together, these results suggest that PAs play an important role to maintain homeostasis of plants.
4. Acknowledgements
We are grateful to Prof. Xianming Deng (Department of Plant Protection, Southwest University, China) for the donation of the V9-strain of Verticillium dahliae and to Dr. Wangfeng Zhang (Shihezi University, China) for kindly providing the acid-delinted upland cotton seeds. This work was supported by the Natural Science Foundation Project of CQ CSTC (2009BB1123), and the Foundation for Excellent Doctor of Southwest University (kb2009018).
REFERENCES
- F. He, Q. H. Pan, Y. Shi and C. Q. Duan, “Biosynthesis and Genetic Regulation of Proanthocyanidins in plants,” Molecules, Vol. 13, No. 10, 2008, pp. 2674-2703. doi:10.3390/molecules13102674
- R. A. Dixon, D. Y. Xie and S. B. Sharma, “Proanthocyanidins—A Final Frontier in Flavonoid Research?” New Phytologist, Vol. 165, No. 1, 2005, pp. 9-28. doi:10.1111/j.1469-8137.2004.01217.x
- V. S. Neergheen, M. A. Soobrattee, T. Bahorun and O. I. Aruoma, “Characterization of the Phenolic Constituents in Mauritian Endemic Plants as Determinants of Their Antioxidant Activities in Vitro,” Journal of Plant Physiology, Vol. 163, No. 8, 2006, pp. 787-799. doi:10.1016/j.jplph.2005.09.009
- T. Murashige and F. Skoog, “A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures,” Physiologia Plantarum, Vol. 15, No. 43, 1962, pp. 473-497. doi:10.1111/j.1399-3054.1962.tb08052.x
- S. Freeman and R. J. Rodriguez, “A Rapid Inoculation Technique for Assessing Pathogenicity of Fusarium oxysporum f.sp. niveum and F. o. melonis on Cucurbits,” Plant Disease, Vol. 77, 1993, pp. 1198-1201. doi:10.1094/PD-77-1198
- C. J. Xie, D. Wang and X. Y. Yang, “Protein Extraction Methods Compatible with Proteomic Analysis for the Cotton Seedling,” Crop Science, Vol. 49, No. 2, 2009, pp. 395-402. doi:10.2135/cropsci2008.06.0367
- M. M. Bradford, “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding,” Analytical Biochemistry, Vol. 72, No. 1-2, 1976, pp. 248-254. doi:10.1016/0003-2697(76)90527-3
- G. Candiano, M. Bruschi, L. Musante, L. Santucci, G. M. Ghiggeri, B. Carnemolla, P. Orecchia, L. Zardi and P. G. Righetti, “Blue Silver: A Very Sensitive Colloidal Coomassie G-250 Staining for Proteome Analysis,” Electrophoresis, Vol. 25, No. 9, 2004, pp. 1327-1333. doi:10.1002/elps.200305844
- D. N. Perkins, D. J. Pappin, D. M. Creasy and J. S. Cottrell, “Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data,” Electrophoresis, Vol. 20, No. 18, 1999, pp. 3551- 3567. doi:10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2
- M. Luo, Y. H. Xiao, L. Hou, X. Y. Luo, D. M. Li and Y. Pei, “Cloning and Expression Analysis of a LIM-Domain Protein Gene from Cotton (Gossypium hirsuturm L.),” Journal of Genetics and Genomics, Vol. 30, No. 2, 2003, pp. 175-182.
- Y. Z. Pang, G. J. Peel, E. Wright, Z. Y. Wang and R. A. Dixon, “Early Steps in Proanthocyanidin Biosynthesis in the Model Legume Medicago truncatula,” Plant Physiology, Vol. 145, No. 3, 2007, pp. 601-615. doi:10.1104/pp.107.107326
- T. A. Holton and E. C. Cornish, “Genetics and Biochemistry of Anthocyanin Biosynthesis,” Plant Cell, Vol. 7, No. 7, 1995, pp. 1071-1083.
- S. Cho, W. Chen and F. J. Muehlbauer, “Constitutive Expression of the Flavanone 3-Hydroxylase Gene Related to Pathotype-Specific Ascochyta Blight Resistance in Cicer arietinum L,” Physiological and Molecular Plant Pathology, Vol. 67, No. 2, 2005, pp. 100-107. doi:10.1016/j.pmpp.2005.09.011
- R. Ardi, I. Kobiler, B. Jacoby, N. T. Keen and D. Prusky, “Involvement of Epicatechin Biosynthesis in the Activation of the Mechanism of Resistance of Avocado Fruits to Colletotrichum gloeosporioides,” Physiological and Molecular Plant Pathology, Vol. 53, No. 5-6, 1998, pp. 269-285. doi:10.1006/pmpp.1998.0181
- Y. J. Zheng, L. Tian, H. T. Liu, Q. H. Pan, J. C. Zhan and W. D. Huang, “Sugars Induce Anthocyanin Accumulation and Flavanone 3-Hydroxylase Expression in Grape Berries,” Plant Growth Regul, Vol. 58, No. 3, 2009, pp. 251-260. doi:10.1007/s10725-009-9373-0
- B. G. Kim, J. H. Kim, J. Kim, C. Lee and J. H. Ahn, “Accumulation of Flavonols in Response to Ultraviolet-B Irradiation in Soybean is Related to Induction of Flavanone 3-Beta-Hydroxylase and Flavonol Synthase,” Molecules and Cells, Vol. 25, No. 2, 2008, pp. 247-252.
- P. Reymond, H. Weber, M. Damond and E. E. Farmer, “Differential Gene Expression in Response to Mechanical Wounding and Insect Feeding in Arabidopsis,” Plant Cell, Vol. 12, No. 5, 2000, pp. 707-720.
- E. Nelson, C. M. Walker-Simmons, D. Makus, G. Zuroske, J. Graham and A. C. Ryan, “Regulation of Synthesis and Accumulation of Proteinase Inhibitors in Leaves of Wounded Tomato Plants,” Plant Resistance to Insects, American Chemical Society, Washington, DC, 1983, pp. 103-122.
- C. A. Ryan, “Protease Inhibitors in Plants: Genes for Improving Defenses against Insects and Pathogens,” Annual Review of Phytopathology, Vol. 28, No. 1, 1990, pp. 425-449. doi:10.1146/annurev.py.28.090190.002233
- J. Leon, E. Rojo and J. J. Sanchez-Serrano, “Wound Signalling in Plants,” Journal of Experimental Botany, Vol. 52, No. 354, 2001, pp. 1-9. doi:10.1093/jexbot/52.354.1
- C. A. Ryan and D. S. Moura, “Systemic Wound Signaling in Plants: A New Perception,” Proceedings of the National Academy of Sciences USA, Vol. 99, No. 10, 2002, pp. 6519-6520. doi:10.1073/pnas.112196499
- B. Winkel-Shirley, “Flavonoid Biosynthesis: A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology,” Plant Physiol, Vol. 126, No. 2, 2001, pp. 485-493. doi:10.1104/pp.126.2.485
- M. Philpott, L. R. Ferguson, K. S. Gould and P. J. Harris, “Anthocyanidin-Containing Compounds Occur in the Periderm Cell Walls of the Storage Roots of Sweet Potato (Ipomoea batatas),” Journal of Plant Physiology, Vol. 166, No. 10, 2009, pp. 1112-1117. doi:10.1016/j.jplph.2008.12.007
- Y. H. Xiao, Z. S. Zhang, M. H. Yin, M. Luo, X. B. Li, L. Hou and Y. Pei, “Cotton Flavonoid Structural Genes Related to the Pigmentation in Brown Fibers,” Biochemical and Biophysical Research Communications, Vol. 358, No. 1, 2007, pp. 73-78. doi:10.1016/j.bbrc.2007.04.084
- R. Koes, W. Verweij and F. Quattrocchio, “Flavonoids: A Colorful Model for the Regulation and Evolution of Biochemical Pathways,” Trends in Plant Science, Vol. 10, No. 5, 2005, pp. 236-242. doi:10.1016/j.tplants.2005.03.002
- R. C. Wilmouth, J. J. Turnbull, R. W. D. Welford, I. J. Clifton, A. G. Prescott and C. J. Schofield, “Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis thaliana,” Structure, Vol. 10, No. 1, 2002, pp. 93-103. doi:10.1016/S0969-2126(01)00695-5
- D. Y. Xie, S. B. Sharma, N. L. Paiva, D. Ferreira and R. A. Dixon, “Role of Anthocyanidin Reductase, Encoded by BANYULS in Plant Flavonoid Biosynthesis,” Science, Vol. 299, No. 5605, 2003, pp. 396-399. doi:10.1126/science.1078540
- R. D. Mellway, L. T. Tran, M. B. Prouse, M. M. Campbell and C. P. Constabel, “The Wound-, Pathogen-, and Ultraviolet B-Responsive MYB134 Gene Encodes an R2R3 MYB Transcription Factor That Regulates Proanthocyanidin Synthesis in Poplar,” Plant Physiol, Vol. 150, No. 2, 2009, pp. 924-941. doi:10.1104/pp.109.139071
- A. Cultrone, P. S. Cotroneo and G. R. Recupero, “Cloning and Molecular Characterization of R2R3-MYB and bHLH-MYC Transcription Factors from Citrus sinensis,” Tree Genetics & Genomes, Vol. 6, No. 1, 2009, pp. 101- 112. doi:10.1007/s11295-009-0232-y
- M. Orozco-Cardenas and C. A. Ryan, “Hydrogen Peroxide Is Generated Systemically in Plant Leaves by Wounding and Systemin via the Octadecanoid Pathway,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 96, No. 11, 1999, pp. 6553-6557. doi:10.1073/pnas.96.11.6553
- E. Castro-Mercado, Y. Martinez-Diaz, N. Roman-Tehandon and E. Garcia-Pineda, “Biochemical Analysis of Reactive Oxygen Species Production and Antioxidative Responses in Unripe Avocado (Persea americana Mill var Hass) Fruits in Response to Wounding," Protoplasma, Vol. 235, No. 1-4, 2009, pp. 67-76. doi:10.1007/s00709-009-0034-y
- R. Mittler, S. Vanderauwera, M. Gollery and F. Van Breusegem, “Reactive Oxygen Gene Network of Plants,” Trends in Plant Science, Vol. 9, No. 10, 2004, pp. 490- 498. doi:10.1016/j.tplants.2004.08.009
- H. Fujii, T. Nakagawa, H. Nishioka, E. Sato, A. Hirose, Y. Ueno, B. X. Sun, T. Yokozawa and G. I. Nonaka, “Preparation, Characterization, and Antioxidative Effects of Oligomeric Proanthocyanidin-L-Cysteine Complexes,” Journal of Agricultural and Food Chemistry, Vol. 55, No. 4, 2007, pp. 1525-1531. doi:10.1021/jf062819n
Abbreviations:
F3H, flavanone 3-hydroxylase;
PAs, proanthocyanidins;
DFR, dihydroflavonol-4-reductase;
ANS, anthocyanidin synthase;
ANR, anthocyanidin reductase;
ROS, reactive oxygen species;
PAL, ammonia lyase;
CHS, chalcone synthase;
CHI, chalcone isomerase;
HIS, histone;
2-DE, two-dimensional gel electrophoresis;
2-ME, β-mercaptoethanol;
PMSF, Phenylmethanesulfonyl fluoride;
SDS, sodium dodecylsulphate.
NOTES
*Corresponding author.

