Advances in Microbiology
Vol.06 No.14(2016), Article ID:72918,9 pages
10.4236/aim.2016.614098
The Evidence of Clade 7.1 Avian Influenza Virus (H5N1) in Qinghai Lake
Wen Wang1*, Kirill Sharshov2*, Zhuo Li1*, Sisi Zheng1,3, Hao Sun1, Fang Yang1, Xuelian Wang1, Laixing Li1#
1Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xi’ning, China
2Research Institute of Experimental and Clinical Medicine, Novosibirsk, Russia
3University of Chinese Academy of Sciences, Beijing, China

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: December 1, 2016; Accepted: December 19, 2016; Published: December 22, 2016
ABSTRACT
The highly pathogenic influenza A virus subtype H5N1 spread throughout Asia since 2003, reached to Europe in 2005, and the Middle East, as well as Africa and caused a global concern for a potential pandemic threat last decade. A Clade 2.3.2 H5N1 virus became dominate in the Qinghai Lake region in 2009 with sporadic mammal cases of infection and transferred to Russia and Europe through wild migratory birds. Currently, HPAI H5N1 of clades 2.3.4, 2.3.2, and 7 are the dominant co-circulating H5N1 viruses in poultry in Asia. 2.3.2 Clade is dominant in wild birds through the world whereas there is no evident data about Clade 7 circulation in wild birds. We detected HPAI H5N1 virus of Clade 7.1 in Qinghai Lake, that closely related to Shanxi-like and Vietnam viruses co-circulating in poultry. This is the first report of Clade 7.1 H5N1 in wild birds. Based on phylogenetic analyses, the virus can be originated from Clade 7.1 virus gene pool that spread in Vietnam and Chinese poultry and could spread with migratory birds to Qinghai Lake. The Qinghai Lake continues to be significant hotspot for H5N1 surveillance since the regular outbreaks occurred there in wild birds and mammals. Based on these facts and findings, the related researchers should pay more attention to the Qinghai Lake basin as significant hotspot for H5N1 avian influenza surveillance since the regular H5N1 outbreaks occurred there in wild birds with sporadic mammal cases of infection.
Keywords:
Highly Pathogenic Avian Influenza, H5N1, Clade 7.1, Qinghai Lake, Wild Birds

1. Introduction
Avian influenza is a viral disease adapted to birds, drawing more attention all over the world [1] . Recent outbreaks of an avian-origin H5N8, H7N9 influenza virus raise concern of the emergence of novel reassortant viruses in Eurasia and the potential threat to the human population [2] [3] . The highly pathogenic influenza A virus subtype H5N1 (HPAI H5N1) spread throughout Asia since 2003, reached to Europe in 2005, and the Middle East, as well as Africa and caused a global concern for a potential pandemic threat last decade [4] [5] [6] [7] . In 2005, a great amount of migratory birds were infected by HPAI H5N1 in Qinghai Lake region (northwest of China) which resulted the death of over 6000 birds [4] [6] [8] . Based on their phylogenetic analysis, these viruses were classified as Clade 2.2 [9] . The Clade 2.2 has been found in Qinghai Lake region for over 4 years and circulated worldwide through birds’ migration [10] . However, in 2009, a novel H5N1 virus lineage Clade 2.3.2 became dominate in the Qinghai Lake region with sporadic mammal cases of infection [11] [12] , and transferred to Russia and Europe through wild migratory birds [13] . Currently HPAI H5N1 of clades 2.3.4, 2.3.2, and 7 are the dominant co-circulating H5N1 viruses in poultry in China [14] . At the same time, 2.3.2 clade is dominant in wild birds through the world but there is no evident data about clade 7 isolation from wild birds reported.
For better understanding of the virus spreading in wild migratory birds of Qinghai Lake region, we provide annual HPAI surveillance in wild birds [15] including sentinel Bar-headed goose (Anser indicus), which shared the same habitat with that of the wild ones.
2. Materials and Methods
2.1. Samples
Before study, all Bar-headed geese have been tested for influenza virus infection by both serological and PCR tests; and the ones with negative results were placed into Qinghai Lake region in special reserve park. To avoid the possible viral transmission from human to wild birds, the place for feeding sentinel Bar-headed geese was close to wild birds habitats but isolated from human interference. Experimental procedures such as collection, transportation, storage and tests were performed by following the WHO and OIE manuals [16] .
2.2. Virus Isolation and RT-PCR
After collection, all the samples were sent to China Animal Health and Epidemiology Center (Qingdao) which can support BSL 3 laboratory facilities. The swab samples were tested for influenza viruses by inoculating in allantoic cavity of 9-days old Specific Pathogen Free (SPF) chicken embryos and RT-PCR [16] . Each sample was incubated in SPF embryos with at least 3 passages; the virus isolation was checked by RT-PCR targeting M gene [16] . Viral RNA was extracted from positive allantoic fluid by trizol- chloroform method by following the manual provided by Ambion Company. (http://tools.lifetechnologies.com/content/sfs/manuals/trizol_ls_reagent.pdf), and reverse transcription (RT) was carried out under standard conditions using the Uni12 (5-AGC GAA AGC AGG-3) primer.
2.3. Intravenous Pathogenicity Test to Chickens
Intravenous pathogenicity of isolates for chickens was determined according to guidelines established by the World Health Organization [17] .
2.4. Sequencing and Genetic Analysis
PCR was performed using specific primers as described by Hoffmann et al., 2001 [18] . All the amplicons were purified by TaKaRa Agarose Gel DNA Purification Kit (Ver 2.0) and sequenced by Sangon Biotech (Sangon, Shanghai, China). Samples were analyzed using the Vector NTI 10.0 (Invitrogen, Carlsbad, CA) software package; Full-genome sequence was deposited to the GISAID database (accession numbers EPI463598 - EPI463605). For analysis all the sequences were aligned with MEGA5 (ClustalW alignment), and CLUSTAL X (with MUSCLE option) software [19] [20] complemented by manual tenements, following the similarity criterion [21] . The phylogeny among all eight segments was reconstructed using MEGA5.0 with the maximum likelihood algorithm. Bootstrap (BS) values for both data sets were estimated by using 1,000 replicates [19] .
3. Results
In April of 2011, we took cloacal, oropharyngeal swab samples from 234 sentinel Bar-head geese from which we found one H5 positive by RT-PCR. We successfully isolated A/Bar-headed goose/Qinghai/0604/2011 (H5N1) strain in chicken embryos. The virus caused death in chicken embryos within 2 days and we evaluated its virulence in chickens. 6-week-old (SPF) chickens were inoculated intravenously with 0.1 ml of a 1:10 dilution of allantoic fluid containing the virus and were observed clinically for 10 days. IVPI was determined by the OIE standard procedure and showed the virus to be highly pathogenic for chicken (IVPI = 2, 1).
Analysis based on HA and NA genes showed that the A/Bar-headed goose/Qinghai/ 0604/2011 is closely related to the Vietnam virus A/Chick/Vietnam/NCVD-016/2008 with values of 100% and 99% identity for HA and NA genes, respectively. We found that the viral HA was not clustered with Clade 2.2 and Clade 2.3.2 which only have been isolated from Qinghai Lake Region (Figure 1). Phylogeny based on last 2014-year H5N1 nomenclature showed that our strain belongs to Clade 7.1 together with viruses isolated from poultry in Vietnam and Shanxi and Ningxia provinces (2006-2008) [9] .
The HA alignment of the Clade 7.1 showed the same cleavage sites PQREGRRRKRG at position 337 to 347, however most of the viruses in other Clades possessed the cleavage sites as PQRERRRKKRG. The Glycine at position 341 could be considered as an obvious feature of Clade 7 group [5] [14] . Moreover, the Clade 7 has the same “QSG” avian-like motif in receptor-binding site as most of the Avian H5N1 viruses.
Figure 1. Phylogenetic tree based on nucleotide sequences of HA genes (1-1700) of H5N1 influenza A viruses. The tree was generated by the distance-based Maximum likelihood method using MEGA 5.1 program. The reliability of the tree was assessed by bootstrap analysis with 1000 replicates. A branch length scale bar is shown beneath the tree. The Qinghai 0604 H5N1 virus in the study is shown triangle-marked.
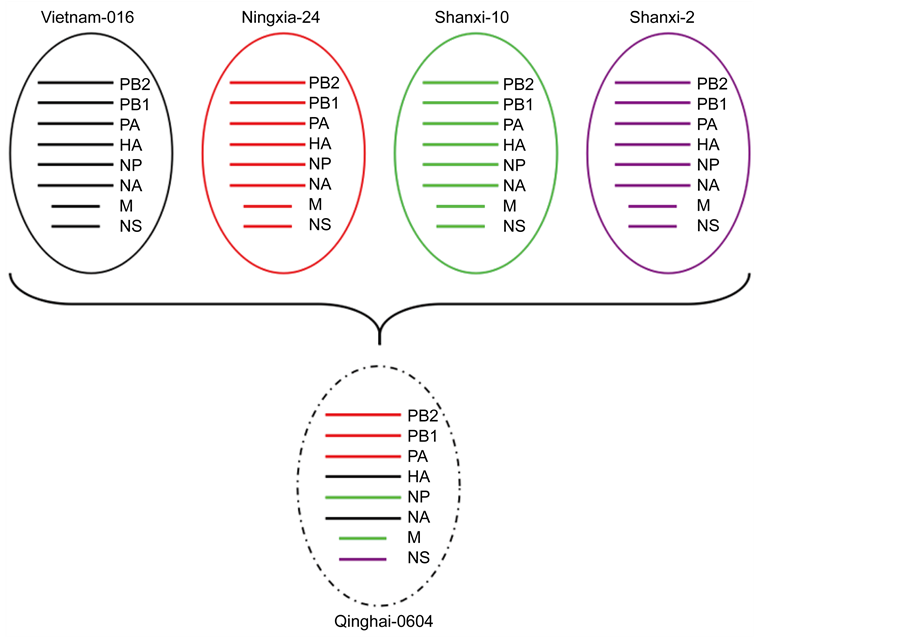
Phylogenetic analysis of other 6 internal genes confirmed that the virus belongs to Clade 7.1 and closely related to Vietnam and Chinese viruses isolated from poultry earlier (data not shown). According to phylogenetic analysis of all the segments the Qinghai0604 H5N1 virus belongs to Eurasian Avian-like clades. The nucleotide sequences of PA, PB1 and PB2 showed 99.5%, 99.8% and 99.5% similarities to virus A/Chicken/ Ningxia/24/2006 respectively (data not shown). For M and NP genes our virus had closest similarity to a north Chinese virus A/Chicken/Shanxi/10/2006 (98.7% and 98.3% respectively); NS gene had closest relation with the other Shanxi like virus A/Chicken/ Shanxi/2/2006 with 98.5% similarity. All these results suggested that the Qinghai 0604 virus contains gene pool that spread among Vietnam and Chinese Clade 7.1 viruses in poultry. Putative genomic compositions of the Clade 7.1 avian influenza (H5N1) virus from wild birds are showed in Figure 2.
4. Discussion
Based on the current study, our H5N1 Clade 7.1 virus has segments most likely originated from viruses of Chinese and Vietnam poultry. This is the first report of H5N1 clade 7.1 virus in wild birds at Qinghai Lake. Clade 7.1 is mostly distributed in Vietnam and neighbor Chinese provinces that are located on the East Asian bird migratory flyway. We suggest that the Clade 7 gene pool from poultry could spread to migratory
Figure 2. Putative genetic constitution of Clade 7.1 H5N1 Qinghai 0604 Virus, with four viruses i.e. Ningxia, Shanxi and Vietnam showed in different colors. There are eight genetic segments for each virus from top to bottom, they are polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), polymerase acidic protein (PA), hemagglutinin (HA), nucleocapsid Protein (NP), neuraminidase (NA), matrix protein (M), and nonstructural protein (NS).
birds and might be connected with the East Asia and Australia flyway last 3 years. The lack of poultry at Qinghai Lake suggests the most possible way of Clade 7.1 appearance at Qinghai Lake due to bird migrations. We can hypothesized that sentinel Bar-headed geese most likely could be infected by wild birds. It is in accordance with the fact that sampling period was April, when wild birds migrate back to Qinghai Lake from Southeast Asia for breeding [5] [22] . Vietnam virus which is closely related to HA and NA segment of Qinghai 0604 was isolated from seized chicken at China-Vietnam border control station of Lang Son Province that confirms circulation of Clade 7.1 gene pool in poultry at that region [23] . Important that recent re-emergence of clade 7.1/7.2 at present, has urged Vietnam to the need for dynamically applied antigenicity-matching vaccines [24] . The Shanxi and Ningxia Provinces are situated within the East Asian flyway migratory bird flyway and the poultry transportation between these provinces and Vietnam was rare, suggesting that the migratory birds might also play an important role for the virus transmission to Qinghai Lake. Our results confirm that the M, NP and NS2 of Qinghai 0604 segments are closely related with Shanxi-like viruses. Similar results were previously suggested in which A/chicken/Shanxi/2/2006 like HA gene is the closest ancestor of clade 7 virus group [23] . It should be noticed that the Shanxi-like viruses, represented by A/chicken/Shanxi/2/06, are antigenically drifted H5N1 viruses that contain the reassortant HA, NA and PB2 genes [10] . Clade 7 H5N1 viruses have been recorded in a little number of avian outbreaks in China. However, some studies showed the Clade 7 virus circulation in healthy swine in Eastern China since 2008, which rises a concern for human threat [25] .
We believe that the gene pool of clade 7.1 can be spread by migrating birds in different provinces as we found in this study. The lack of poultry in Qinghai province could be a reason for eliminating of this virus―there is no data of further spreading. Most likely that migratory birds stop in Qinghai lake region, contact and infect sentinel Bar- headed geese. From the Generalized Flyway map [26] , we can conclude that the Qinghai Lake is located in the intersection place between Central Asian and East Asian birds’ flyways, which means that the migratory birds from both flyways could transfer virus into Qinghai lake region [22] [26] [27] . However, the Vietnam, Shanxi and Ningxia are located into the East Asian birds flyway, therefore, the transmission have more possibility to be occurred from that region by birds within East Asian and Australian flyway.
5. Conclusion
In conclusion, during annual surveillance, we detected HPAI H5N1 virus of Clade 7.1 in wild birds, that closely related to Shanxi-like and Vietnam viruses co-circulating in poultry. This is the first report of Clade 7.1 H5N1 in Qinghai Lake. Based on phylogenetic analyses, we believe that the virus can be originated from gene pool that spread in Vietnam and Chinese Clade 7.1 viruses in poultry. We suggest that the Clade 7.1 gene pool from poultry could spread to migratory birds and might be transferred to Qinghai Lake. The lack of poultry at Qinghai Lake suggests the most possible way of Clade 7.1 appearance with birds’ migrations and its further elimination. Based on these facts and findings, the related researchers should pay more attention to the Qinghai Lake basin as significant hotspot for H5N1 avian influenza surveillance since the regular H5N1 outbreaks occurred there in wild birds with sporadic mammal cases of infection.
Acknowledgements
This work was supported by the State Forestry Administration Program (No. Y31I351B01), the National Key Basic Research Program of China (973 Program, No. 2010CB530301), the China Biodiversity Observation Networks (Sino BON) and the State Administration of Foreign Experts Affairs of China (No. 20166300005). We are very thankful to Sheng Cao, Ping Li, Hui Chu, Lang Feng, Cunlian Kui and Jian Cao for their help in the routinely surveillance.
Cite this paper
Wang, W., Sharshov, K., Li, Z., Zheng, S.S., Sun, H., Yang, F., Wang, X.L. and Li, L.X. (2016) The Evidence of Clade 7.1 Avian Influenza Virus (H5N1) in Qinghai Lake. Advances in Microbiology, 6, 1053-1061. http://dx.doi.org/10.4236/aim.2016.614098
References
- 1. Webster, R.G. and Govorkova, E.A. (2014) Continuing Challenges in Influenza. Annals of the New York Academy of Sciences, 1323, 115-139.
https://doi.org/10.1111/nyas.12462 - 2. Ge, F.F., Ju, H.B., Yang, D.Q., Liu, J., Wang, J., Lu, J., Li, X., Zhang, W.Y., Liu, P.H. and Zhou, J.P. (2014) Epidemiological Situation and Genetic Analysis of H7N9 Influenza Viruses in Shanghai in 2013. Archives of Virology, 159, 3029-3041.
https://doi.org/10.1007/s00705-014-2177-1 - 3. Lee, Y.J., Kang, H.M., Lee, E.K., Song, B.M., Jeong, J, Kwon, Y.K., Kim, H.R., Lee, K.J., Hong, M.S., Jang, I., Cho,i K.S., Kim, J.Y., Lee, H.J., Kang, M.S., Jeong, O.M., Baek, J.H., Joo, Y.S., Park, Y.H. and Lee, H.S. (2014) Novel Reassortant Influenza A (H5N8) Viruses, South Korea, 2014. Emerging Infectious Diseases, 20, 1087-1089.
https://doi.org/10.3201/eid2006.140233 - 4. Chen, H., Smith, G.J.D., Zhang, S.Y., Qin, K., Wang, J. and Li, K.S. (2005) H5N1 Virus Outbreak in Migratory Waterfowl. Nature, 436, 191.
https://doi.org/10.1038/nature03974 - 5. Li, Y.B., Shi, J.Z., Zhong, G.X., Deng, G.H., Tian, G.B., Ge, J.Y., Zeng, X.Y., Song, J.H., Zhao, D.M., Liu, L.L., Jiang, Y.G., Guan, Y.T., Bu, Z.G. and Chen, H.L. (2010) Continued Evolution of H5N1 Influenza Viruses in Wild Birds, Domestic Poultry, and Humans in China from 2004 to 2009. Journal of Virology, 84, 8389-8397.
https://doi.org/10.1128/JVI.00413-10 - 6. Liu, J., Xiao, H., Lei, F., Zhu, Q., Qin, K., Zhang, X.W., Zhang, X.L., Zhao, D., Wang, G., Feng, Y., Ma, J., Liu, W., Wang, J. and Gao, G.F. (2005) Highly Pathogenic H5N1 Influenza Virus Infection in Migratory Birds. Science, 309, 1206.
https://doi.org/10.1126/science.1115273 - 7. Sharshov, K., Romanovskay, A., Uzhachenko, R., Durymanov, A., Zaykovskaya, A., Kurskaya, O., Ilinykh, P., Silko, N., Kulak, M., Alekseev, A., Zolotykh, S., Shestopalov, A. and Drozdov, I. (2010) Genetic and Biological Characterization of Avian Influenza H5N1 Viruses Isolated from Wild Birds and Poultry in Western Siberia. Archives of Virology, 155, 1145-1150.
https://doi.org/10.1007/s00705-010-0676-2 - 8. Zheng, J. and He, Y.B. (2006) Considerations on Prevention and Control of Avian Influenza Epidemic Situation of Wild Birds in Qinghai Lake Region. Chinese Journal of Wildlife, 27, 19-21.
- 9. World Health Organization/World Organization for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group (2014) Revised and Updated Nomenclature for Highly Pathogenic Avian Influenza A (H5N1) Viruses. Influenza and Other Respiratory Viruses, 8, 384-388.
- 10. Wang, G., Zhan, D., Li, L., Lei, F., Liu, B. and Liu, D. (2008) H5N1 Avian Influenza Re-Emergence of Lake Qinghai: Phylogenetic and Antigenic Analyses of the Newly Isolated Viruses and Roles of Migratory Birds in Virus Circulation. Journal of General Virology, 89, 697-702.
https://doi.org/10.1099/vir.0.83419-0 - 11. Hu, X.D., Liu, D., Wang, M.Y., Yang, L., Wang, M., Zhu, Q., Li, L. and Gao, G.F. (2010) Clade 2.3.2 Avian Influenza Virus (H5N1), Qinghai Lake Region, China. Emerging Infectious Diseases, 17, 560-561.
https://doi.org/10.3201/eid1703.100948 - 12. Zhou, J., Sun, W., Wang, J., Guo, J., Yin, W., Wu, N., Li, L., Yan, Y., Liao, M., Huang, Y., Luo, K., Jiang, X. and Chen, H. (2009) Characterization of the H5N1 Highly Pathogenic Avian Influenza Virus Derived from Wild Pikas in China. Journal of Virology, 83, 8957-8964.
https://doi.org/10.1128/JVI.00793-09 - 13. Sharshov, K., Silko, N., Sousloparov, I., Zaykovskaya, A., Shestopalov, A. and Drozdov, I. (2010) Avian Influenza (H5N1) Outbreak among Wild Birds, Russia, 2009. Emerging Infectious Diseases, 16, 49-51.
https://doi.org/10.3201/eid1602.090974 - 14. Sun, H., Sun, Y., Pu, J., Zhang, Y., Zhu, Q., Li, J., Gu, J., Chang, K.C. and Liu, J. (2014) Comparative Virus Replication and Host Innate Responses in Human Cells Infected with Three Prevalent Clades (2.3.4, 2.3.2, and 7) of Highly Pathogenic Avian Influenza H5N1 Viruses. Journal of Virology, 88, 725-729.
https://doi.org/10.1128/JVI.02510-13 - 15. Sharshov, K., Marchenko, V., Yang, F., Alekseev, A., Cao, J., Li, Z., Shestopalov, A. and Li, L. (2014) Avian Influenza H5N1 Surveillance in Geese of Qinghai Province, China (2012). Advances in Infectious Diseases, 4, 18-21.
https://doi.org/10.4236/aid.2014.41003 - 16. OIE (2004) Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 5th Edition, OIE, Paris. www.oie.int/doc/ged/D6448.PDF
- 17. WHO (2002) Manual on Animal Influenza Diagnosis and Surveillance. World Health Organization (WHO), Harbin.
- 18. Hoffmann, E.J., Stech, Y., Guan, R.G. and Webster, D.R. (2001) Universal Primer Set for the Full-Length Amplification of All Influenza A Viruses. Archives of Virology, 146, 2275-2289.
https://doi.org/10.1007/s007050170002 - 19. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731-2739.
https://doi.org/10.1093/molbev/msr121 - 20. Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Research, 25, 4876-4882.
https://doi.org/10.1093/nar/25.24.4876 - 21. Simmons, M.P. (2004) Independence of Alignment and Tree Search. Molecular Phylogenetic and Evolution, 31, 874-879.
https://doi.org/10.1016/j.ympev.2003.10.008 - 22. Li, L.X. (1999) A System List and Status of Waterbirds in Qinghai-Xizang (Tibet) Plateau. Acta Biologica Plateau Sinica, 14, 116-128.
- 23. Nguyen, T., Davis, C.T., Stembridge, W., Shu, B., Balish, A., Inui, K., Do, H.T., Ngo, H.T., Wan, X.F., McCarron, M., Lindstrom, S.E., Cox, N.J., Nguyen, C.V., Klimov, A.I. and Donis, R.O. (2009) Characterization of a Highly Pathogenic Avian Influenza H5N1 Virus Sub-Lineage in Poultry Seized at Ports of Entry into Vietnam. Journal of Virology, 387, 250-256.
https://doi.org/10.1016/j.virol.2009.03.006 - 24. Le, T.H. and Nguyen, N.T. (2014) Evolutionary Dynamics of Highly Pathogenic Avian Influenza A/H5N1 HA Clades and Vaccine Implementation in Vietnam. Clinical and Experimental Vaccine Research, 3, 117-127.
https://doi.org/10.7774/cevr.2014.3.2.117 - 25. He, L., Zhao, G., Zhong, L., Liu, Q., Duan, Z., Gu, M., Wang, X., Liu, X. and Liu, X. (2013) Isolation and Characterization of Two H5N1 Influenza Viruses from Swine in Jiangsu Province of China. Archives of Virology, 158, 2531-2541.
https://doi.org/10.1007/s00705-013-1771-y - 26. Veen, J., Yurlov, A.K., Delany, S.N., Mihantiev, A.I., Selivanova, M.A. and Boere, G.C. (2005) An Atlas of Movements of Southwest Siberian Waterbirds. Wetlands International, Wageningen.
- 27. Li, L.X. (1997) Brief History of Vertebrate Study and Species List in Qinghai Lake Basin. Acta Biologica Plateau Sinica, 13, 133-146.
NOTES

*These authors contributed equally to this work.