Advances in Microbiology
Vol.4 No.1(2014), Article ID:42073,6 pages DOI:10.4236/aim.2014.41007
Understanding Laboratory Methods and Their Impact on Antimicrobial Resistance Surveillance, at Muhimbili National Hospital, Dar es Salaam, Tanzania
1Department of Public Health, School of Health Sciences, Monash University, Ruimsig, South Africa
2School of Public Health, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
3Department of Microbiology, Central Pathology Laboratory, Muhimbili National Hospital, Dar es Salaam, Tanzania
4Muhimbili Orthopaedic Institute, Muhimbili National Hospital, Dar es Salaam, Tanzania
Email: *peter.nyasulu@monash.edu
Copyright © 2014 Peter Nyasulu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Peter Nyasulu et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received November 28, 2013; revised December 28, 2013; accepted January 5, 2014
KEYWORDS
Antimicrobial Resistance; Microbiology; Surveillance; Laboratory; Tanzania
ABSTRACT
The study sought to describe laboratory methods and blood culture procedures and their impact on antimicrobial resistance surveillance among nosocomial bacteria. We conducted a systematic audit of blood culture procedures and practices in the Department of Microbiology, Central Pathology Laboratory at Muhimbili National Hospital, between 19th and 23rd March 2012. A total of 25 - 30 blood culture specimens were received each day as an indication of low volumes of blood culturing at this site. More blood culture requests came from the neonatal unit of the hospital, and were performed manually with high culture negative specimens. The laboratory performed antibiotic susceptibility testing as per the CLSI guidelines. No vancomycin resistance was ever reported at this site. All blood culture results were entered into the JEEVA laboratory information system, where results could be accessed by clinicians in the wards and data could be retrieved to assess patterns of antimicrobial resistance. Blood culture data entry system lacked quality control checks hence numerous errors and missing data were observed. Our results support the relevance of having improved laboratory procedures and good quality blood culture since surveillance of antimicrobial resistance primarily depends on good laboratory procedures, good quality and reliable blood culture data. This would essentially minimise imprecise estimates of rates of antimicrobial resistance at this hospital.
1. Introduction
Muhimbili National Hospitals’ (MNH) existence dates back to 1910 when it was known as Sewahaji. It is a 900- bed specialised National Referral and University Teaching Hospital [1] that provides tertiary health services to inhabitants of Dar es Salaam region, Tanzania, which harbours an estimated population of 2.5 million people [2]. The hospital admits 1000 to 1200 in-patients per day. Blood cultures at this hospital are not routinely conducted because clinical diagnosis of bacteraemia and empirical antibiotic therapy is the main approach to clinical management. Blood culture is only requested in special circumstances, mostly due to treatment non-response or in neonatals due to non-specificity of clinical symptoms in this age group. The aim of this study was to describe in details laboratory methods and procedures relating to blood cultures and their potential impact on antimicrobial resistance surveillance among nosocomial bacteria.
1.1. The Central Pathology Laboratory
The Medical Laboratory Services in Tanzania (called Tanganyika during the colonial era) were established in the late 19th Century during the German administration. The first Government Health Laboratory was established in 1897, at Ocean Road in Dar es Salaam. Historically, this laboratory was the first site of a medical laboratory in Tanzania. The laboratory was often visited by Dr Robert Koch who worked in the laboratory on several occasions as he was investigating tropical diseases such as malaria, sleeping sickness etc, which were then a major health problem in the country. Laboratory services have grown and been expanded countrywide. The Ocean Road Laboratory became the Central Pathology Laboratory (CPL) in the early 1960s and is still operational under the Ministry of Health and Social Welfare [3]. The CPL located at the MNH is a key player in the provision of high quality of laboratory services to all patients referred to and admitted at MNH or attended to as out-patients.
1.2. Departments and Laboratory Information System
The CPL is the leading provider of diagnostic laboratory services in Tanzania. In addition, the CPL offers referral laboratory services for tests requests from other public and private hospitals within Dar es Salaam and surrounding regions. The services provided by the CPL are: microbiology, histopathology, parasitology, haematology and blood transfusion, clinical chemistry etc. The CPL uses a laboratory information system (LIS) fully interfaced with all automated diagnostic machines and hospital information management system (HIMS), the Jeeva system 2000 [3]. The system was established as an attempt to improve laboratory services such as turnaround time for laboratory results.
All clinical departments are computerised and interlinked to the LIS and the results are entered and posted on the Jeeva LIS for clinicians to access directly in the wards and other clinical departments through logging into the system with their username and password. The clinicians view the results online in the wards, and this expedites the clinical decision regarding treatment modalities for bacteraemia cases. Hard copies of the laboratory results are sent to the wards afterwards for filing in the patient’s files and cross referencing in case of a future episode of an illness. The microbiology unit at the CPL handles high volumes of laboratory results ranging from samples of blood, cerebral spinal fluids, pus swabs, urine specimens, stool etc. The microbiology unit does the following tests among others: bacterial identification, antibiotics susceptibility testing and serological tests. The LIS helps to ensure that results are captured in time and transmitted or released to the patients within acceptable time limits [4].
2. Methodology
2.1. Design and Study Setting
A systematic audit of blood culture procedures and practices was carried out in the department of microbiology of the Central Pathology Laboratory of Muhimbili National Hospital. The audit lasted 3 days and focussed on the procedures and practices carried out in the process of dealing with blood cultures i.e. tracing a pathway from receipt of blood culture specimen in the microbiology laboratory to processing the blood culture to communicating results to the clinicians in the wards and entering results on LIS. Our study focussed on blood culture from bacteraemia caused by Staphylococcus aureus (SA) and Klebsiella pneumoniae (KP).
2.2. Data Collection Procedures
We used a standard guide as we went through different sections of the microbiology department focussing on how blood cultures are done and how data are gathered in the laboratory and utilised for surveillance. The audit involved 1) a comprehensive orientation on the activities of the bacteriology section to familiarise with standard routines and laboratory practice, 2) observation of how blood culture procedures are done in the laboratory and, 3) individual discussions with staff involved in technical procedures of blood culturing and data entry of blood culture results.
3. Results
3.1. Blood Culture Specimen Flow
We schematically describe in the chart below specimen flow of blood cultures and related procedures pertaining to blood cultures at MNH.
The narration of Figure 1 is provided by the following steps:
Step 1: Two blood culture bottles are collected and sent to the laboratory. For children, only a single specimen is collected into a special blood culture bottle. Duplicate specimens in this laboratory are rare as blood cultures are collected on special request only not as a routine test.
Step 2: From the wards, all specimens are delivered to the laboratory reception area where they are sorted out based on the type of the specimen.
Step 3: At the reception area, blood culture specimens are isolated from the pool of other specimens by the laboratory clerk responsible for all microbiology specimens. A serial number is allocated and pasted onto each of the blood culture specimen bottle.
Step 4: The laboratory clerk then enters patient demographic details from a specimen order form into a register
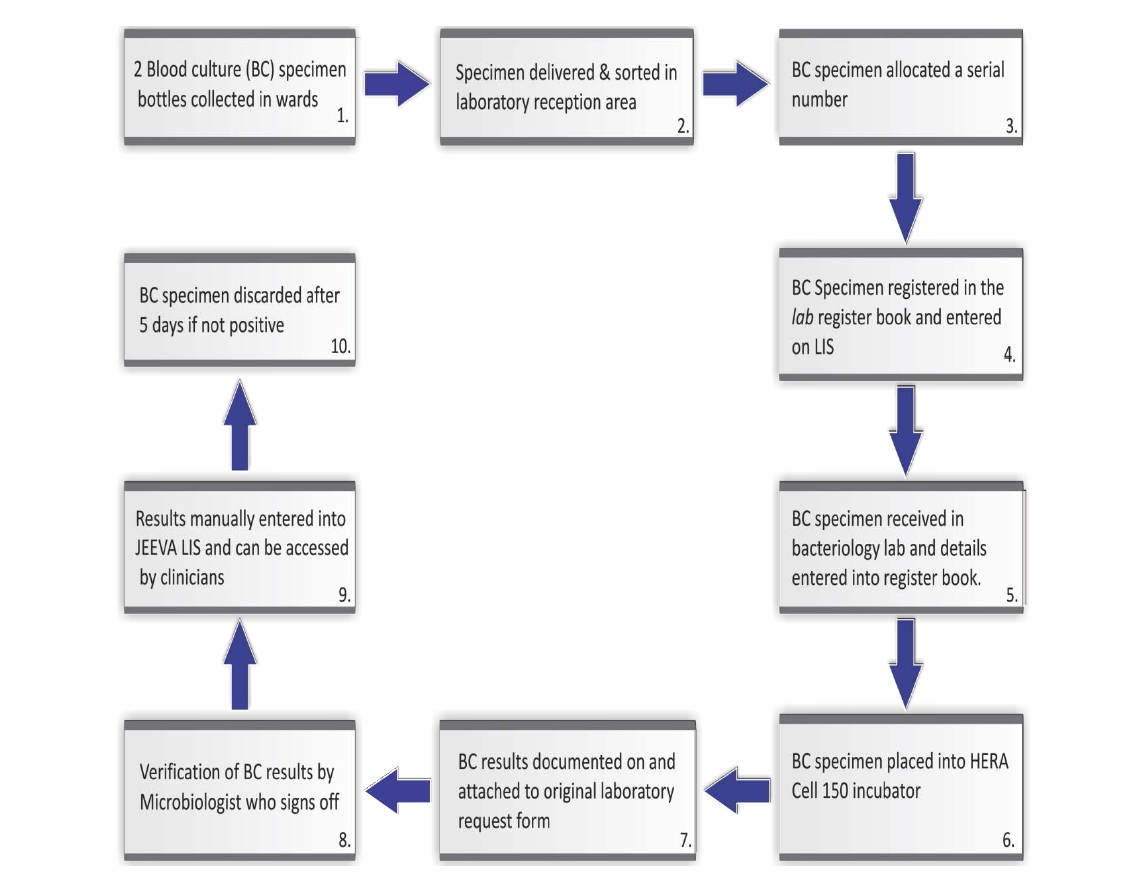
Figure 1. Blood culture data flow and interlinkage with the LIS at MNH microbiology laboratory.
book and LIS database. Once this is done the specimen is then delivered to the bacteriology laboratory for processing.
Step 5: In the bacteriology laboratory, the technician receiving the specimen then enters the patients’ details into yet another register book so as to track samples and minimise loss.
Step 6: Blood culture specimen is then placed into the HERA CELL 150 incubator and physically monitored each morning to detect bacterial growth. If visible signs of positive culture are noted, the specimen is taken out for gram staining and susceptibility testing.
Step 7: The results of blood cultures (both positive and negative specimens), are documented on the blood culture results form, which is then attached to the original laboratory request form.
Step 8: Verification of blood culture results is done by the Microbiologist who heads the department or his immediate representative. Once results are signed off, the results are ready to be released to the wards.
Step 9: The blood culture results are handed back to the laboratory clerk who manually enters them into the JEEVA LIS. The electronic record is linked to the ward such that the clinicians in the ward can access the results directly online through the LIS computer network installed in the wards. The hard copies of the results are also sent to the requesting clinician.
Step 10: The blood culture specimen is discarded after 5 days once no indication of positivity is observed. All necessary protocols for blood cultures are followed so as to minimise error.
3.2. Sample Volumes
Muhimbili National hospital is a large and busy hospital, however the number of blood culture specimens received each day by the microbiology department is in the region of 25 - 30, this gives a clear indication that blood culturing is not a routine practice. In the wards, blood culture is only requested in specific clinical circumstances such as failed empirical antibiotic treatment. As per information from the laboratory register, it was clear that more blood culture requests originate from the paediatrics departments’ neonatal unit.
3.3. Blood Culture Processing
Susceptibility testing of isolates is dependent on the availability of disk panels and therefore not all isolates are tested for resistance to all antibiotics. Sometimes testing is only done on second line drugs which are not the standard of care in the hospital. The results of these tests are therefore of little help to clinicians who manage patients with blood born infections. It was observed that all Staphylococcus aureus isolates were tested for vancomycin resistance as a way of monitoring emerging vancomycin resistant Staphylococcus aureus (VRSA) [5,6].
3.4. Common Antibiotics Tested
The most common antibiotics subjected to susceptibility testing of blood culture isolates at this hospital were: 1) Staphylococcus species: amikacin, penicillin, ampicillin, cloxacillin, tetracycline, erythromycin, gentamicin, cephalothin, chloramphenicol, vancomycin; 2) Klebsiella species: ampicillin, chloramphenicol, tetracycline, amikacin, amoxyclav, cefuroxime and imipenem; 3) Pseudomonas species: ampicillin, amikacin, ciprofloxacin, chloramphenicol, gentamicin and cotrimoxazole; 4) Escherichia coli: chloramphenicol, gentamicin, amikacin, ampicillin and cefuroxime [7].
3.5. Antibiotic Susceptibility Testing
The antibiotic susceptibility testing procedures at this site are done in accordance with the Clinical Laboratory Standards Institute (CLSI), 2010 guidelines [7]. The minimum inhibitory concentration (MIC) for each antibiotic to determine cutoff for antibiotic resistance as outlined in these guidelines are followed. External quality control is done on a regular basis using specimens from the Centres for Disease Control (CDC) to ascertain validity and reliability of antibiotic susceptibility results produced by this laboratory.
3.6. Challenges in Blood Culturing
3.6.1. Automated Laboratory Equipment
Lack of a functioning automated microbial detection system was observed to be important obstacles to effective blood culturing at this laboratory. The automated blood culturing equipment is often not in good working order and servicing takes long time to be done. The laboratory often relies on manual blood culturing technique which has its own limitations such as subjective determination of a positive culture through visual assessment. Accuracy is dependent on individual technicians’ visual acuity, hence subject to over or under estimations of true positives. However, it should be noted that manual blood culturing are still the most common mode of blood culturing in most resource constrained countries.
3.6.2. Blood Cultures Results
As per records entered into the blood culture register book, a high rate of negative cultures was observed. This might be due to prior antibiotics use before a blood culture specimen was taken or it might be a true representation of negative blood cultures. In addition, we also noted that there was a high rate of coagulase negative S.aureus which might be due to contamination of the blood culture samples at the point of collection.
3.7. Common Challenges and Errors in Blood Culture Data Recording
The outline here give some of the common challenges and errors in data recording found at MHN, microbiology laboratory.
• There was lack of a standardised way of entering data. For example, age was entered as date of birth, age in months, days, years, etc. It was also often just documented as adult or child. This created confusion in terms of knowing the exact ages of the patients who had blood culture done.
• Missing data was a big issue as information on gender, age, hospital ward; type of organism and clinical data was often not available.
• Lack of standardised reporting of the blood culture results i.e. results would be reported differently yet it meant the same: “no bacterial growth; Negative, NBG” etc.
• Different data types were entered into the register and also only a few of the positive blood cultures had sensitivity results entered.
• Lack of specific dates that specimens were taken e.g. the record would just show month i.e. July but no specific date mentioned.
3.8. Standard Operating Procedures
The microbiology department operates on principles laid down in the standard operating procedures (SOPs) manual. These procedures are overseen by a Quality Control Officer, who is a member of the team in the microbiology laboratory. The controlling officer is responsible for effecting and approving any changes to the SOPs. Implementation of the SOPs is overseen by the Microbiologist heading the department. The manuals are kept in the microbiology laboratory for ease of reference by all team members.
3.9. Challenges with Data Quality
3.9.1. Laboratory Information System (LIS) Data Entry Format
Jeeva (“Life”) Informatics Solutions LLC is a Bioinformatics solutions provider. The software company was founded by Dr Harsha K Rajasimha and is situated in Montgomery County, Maryland, United States of America and is specialised in providing “On-Demand Virtual Bioinformatics Core Facility”, genomics bigdata management, analysis, and interpretation [8]. Entry of blood culture results into the JEEVA LIS database was done by a single individual. There was no verification of data entered by a second individual to check for accuracy of data entered and to allow for timely correction of errors. The system does not have check codes to control data that is being entered. For example, a characteristic such as “age” the system could take in data in any numerical format such as absolute age, year/date/month, year, months and days. This was certainly problematic and a huge source of error.
3.9.2. Clinical Data
There was often no documentation of patients’ prior antibiotics use before a blood culture sample was taken. No provisional diagnosis was captured on the laboratory request form. Should the laboratory request form have some clinical history documented, such information would not be captured onto the system as the database structure of the LIS was not programmed to capture such information.
3.9.3. Determination of Nosocomial Bactereamia
There was no documentation on laboratory request forms on duration of in hospital stay prior to blood culture specimen being taken. Lack of this information makes it difficult to separate nosocomial from community acquired bacteraemia. In so doing the burden of antimicrobial resistance due to nosocomial infection becomes difficult to effectively ascertain.
4. Discussion
Surveillance of antimicrobial resistance monitors changes in microbial populations, allows for the early detection of resistant microbial strains of public health relevance, and supports the prompt notification and active investigation of outbreaks of resistant bacteria [9]. Surveillance of antimicrobial resistance is primarily dependant on good laboratory procedures, good quality and reliable routine blood culture data. To improve the quality of blood culture data and minimise improper estimates of antimicrobial resistance, it is essential that important steps be taken to improve the system of specimen collection at the point of care, registration and blood culture procedures in the microbiology laboratory [10]. High rates of specimen contamination as evidenced by more coagulase negative culture results, this calls for the need to proactively improve blood culture specimen collection procedures as this would ultimately lead to a reduction in blood culture contamination, and provide proper estimates of bacteraemia episodes and rates of antimicrobial resistance.
There is a need to place special emphasis on appropriate completion of blood culture request forms by clinicians in the wards, specimen registration by laboratory clerk and accurate entry of blood culture results by laboratory technicians. Accuracy of blood culture results could also be improved if the automated blood culture machines were functioning properly. Improvements in quality of data could also be enhanced through improved data entry process into LIS either by introducing another software such as WHONET free access software developed since 1989 by the WHO Collaborating Centre for Surveillance of Antimicrobial Resistance specifically for antimicrobial susceptibility monitoring plus introducing a system of validating data entered into the LIS [11].
5. Conclusion
The LIS needs check codes so that the system is able to track errors on data entry. Simple improvements in the current system could update the system to be an effective surveillance tool to help monitor development and spread of antimicrobial resistance [12] among blood borne pathogens in Tanzania. Such information in the long run will help in policy formulation around antimicrobial usage to contain the growing crisis of antimicrobial resistance in the country.
Acknowledgements
We would like to thank the Chief Executive Officer of the Muhimbili National Hospital (MNH)-Dr Marina Njelekela and the Director of Clinical Support of MNHDr Praxeda Ogweyo. This work would not have been possible without their support. We would also like to acknowledge the Consortium for Advanced Research Training in Africa (CARTA) for financial support for this study.
REFERENCES
- Muhimbili National Hospital. http://www.mnh.or.tz (Accessed March 30 2013)
- United Republic of Tanzania, Prime Minister’s Office, “Regional Administration and Local Government. Strategic Plan for 2010/11-2012/2013.”
- MNH, “Directorate of Clinical Support Services Profile.” http://www.mnh.or.tz/index.php/directorates/clinical-services (Accessed March 30 2013)
- United Republic of Tanzania, Ministry of Health and Social Welfare, “National Health Laboratory Strategic Plan, 2009-2015. p. ix-52p.”
- T. Mazzulli, “Vancomycin Resistant Staphylococcus aureas (VRSA). Canadian Antimicrobial Resistance Alliance.” http://www.can-r.com/mediaResources/VRSA.pdf (Accessed November 10 2013)
- P. C. Applebaum, “The Emergence of Vancomycin-Intermediate nd Vancomycin-resIstant Staphylococcus aureus,” Clinical Microbiology and Infection, Vol. 12, No. S1, 2006, pp. 16-23. http://dx.doi.org/10.1111/j.1469-0691.2006.01344.x
- Clinical and Laboratory Standards Institute, “Performance Standards for Antimicrobial Susceptibility Testing. Twentieth Information Supplement,” Wayne, PA.
- “Jeeva (‘Life’) Informatics Solutions.” http://www.jeevadx.com/ (Accessed October 31 2013)
- WHO, “Surveillance of Antimicrobial Resistance.” http://www.who.int/drugresistance/surveillance/en/ (Accessed November 11 2013)
- WHO, “Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World,” 2003. http://www.who.int/csr/resources/publications/drugresist/en/IAMRmanual.pdf
- WHONET, “Microbiology Laboratory Database Software [Computer Programme],” World Health Organisation, and Boston (MA): WHO Collaborating Centre for Surveillance of Antimicrobial Resistance, Microbiology Laboratory, Brigham and Women’s Hospital, Geneva, 1999.
- WHO, “The WHO Global Strategy for Containment of Antimicrobial Resistance.” http://www.who.int/csr/resources/publications/drugresist/en/EGlobal_Strat.pdf (Accessed November 11 2013)
NOTES
*Corresponding author.

